Abstract
Clinical proteomics is an emerging field that deals with the use of proteomic technologies for medical applications. With a major objective of identifying proteins involved in pathological processes and as potential biomarkers, this field is already gaining momentum. Consequently, clinical proteomics data are being generated at a rapid pace, although mechanisms of sharing such data with the biomedical community lag far behind. Most of these data are either provided as supplementary information through journal web sites or directly made available by the authors through their own web resources. Integration of these data within a single resource that displays information in the context of individual proteins is likely to enhance the use of proteomic data in biomedical research. Human Proteinpedia is one such portal that unifies human proteomic data under a single banner. The goal of this resource is to ultimately capture and integrate all proteomic data obtained from individual studies on normal and diseased tissues. We anticipate that harnessing of these data will help prioritize experiments related to protein targets and also permit meta-analysis to uncover molecular signatures of disease. Finally, we encourage all biomedical investigators to maximize dissemination of their valuable proteomic data to rest of the community by active participation in existing repositories such as Human Proteinpedia.
Advancements in proteomics and its clinical applications have led researchers to exploit them to discover protein markers for cancer diagnosis, interrogate key components of signaling pathways, capture protein-protein interactions, dissect organellar proteomes, identify post-translational modifications and to catalog protein expression and subcellular localization profiles (1, 2). Clinical proteomics deals with the application of proteomic technologies to help decipher the changes that occur in cells, tissues, and organs under diseased conditions. With the increase in the use of recent high-throughput technologies such as mass spectrometry, data generation far outstrips the pace of data storage and dissemination. Data once generated can always be revisited and queried in new or different ways that could even lead to potential breakthroughs in terms of identifying diagnostic markers or therapeutic targets. Although proteomic data can be submitted to public repositories, this is neither popular nor mandated, even for published data. Given the high experimental and labor costs in addition to the precious nature of the data, it is imperative that there are concerted community efforts to capturing such data and making them available in formats that would be most useful to biomedical researchers.
Cancer Biomarkers and Disease Proteomics—
The potential of mass spectrometry to identify proteins in samples in a high throughput (3) manner with reduced sample requirements have made mass spectrometry an ideal tool to be deployed in clinical proteomics (4). Thus, use of proteomics for identification of cancer biomarkers for diagnostic, prognostic, or therapeutic applications is of substantial interest. In this regard, quantitative analysis of protein expression in normal and cancer tissues to identify proteins overexpressed in cancers has already been successfully reported by a number of groups (5–10). Because it has already been demonstrated that early diagnosis of breast, colorectal, and cervical cancers through screening approaches can lead to a reduction in mortality rates (11), there is sufficient justification for aggressive pursuit of novel biomarkers for early detection of all cancers.
In addition to the search for biomarkers, it is also of interest to identify proteomic changes that occur in diseases to gain insights into their pathogenesis. Such proteomic changes could include alterations in abundance of proteins or their post-translational modifications or subcellular localization, among others (12). In the future, it may even be possible to diagnose a particular disease condition from organ-specific proteomic signatures present in serum. For this, we must first systematically obtain proteomic data from individual organs. Such data can be archived, and meta-analysis can be carried out to decipher the signatures, as was recently reported for head and neck and colon cancers (13).
Is Proteomics Synonymous with Mass Spectrometry?—
The routine use of mass spectrometers to identify a multitude of proteins in a high-throughput fashion has led to a situation where the terms ‘“proteomics” and “mass spectrometry” are sometimes used interchangeably. A number of repositories have been developed that only accept data from mass spectrometry experiments. However, proteomics includes a broad array of techniques that are still in common use including Western blotting, immunohistochemistry, yeast two-hybrid, peptide and protein microarrays, x-ray crystallography, NMR spectroscopy, fluorescence microscopy, and flow cytometry. Among these techniques, antibody-based methods are especially used in the oncology field for diagnosis and classification of cancers (14). HUPO1 Antibody Initiative (15) was initiated to accelerate the production and use of validated antibodies against human proteins (16). With the availability of a large number of antibodies, assays such as immunohistochemistry and enzyme-linked immunosorbent assay can be used for biomarker validation. Therefore, it is important to remember the clinical platforms that are relevant to oncology research when proteomic platforms are being discussed.
Genomic Versus Proteomic Data—
In the case of genomic data, the International Nucleotide Sequence Consortium has already established a working principle according to which any sequence data that is submitted to any one of the 3 members, GenBank (17), European Molecular Biology Laboratory (EMBL) (18), or DNA Data Bank of Japan (DDBJ) (19), will automatically be reflected in the other data bases. Further, all sequences submitted to these data bases are freely available to the public without any restrictions. This method of data sharing has been in practice for over 20 years now. Further, if a manuscript contains novel sequences, submission of the nucleotide sequences to any one of the three major nucleotide sequence data bases prior to publication is mandatory. In fact, manuscripts are accepted subject to the condition that a unique data base accession number assigned by these data bases will be provided by the authors before publication.
Unlike genomic data, however, proteomic data is diverse with a multitude of experimental platforms and data types with the result that there are no general working principles for data submission that apply to all types of proteomic data. However, for specific data types such as mass spectrometry data, specific guidelines are beginning to emerge (20) although they are not universally adopted at the current time.
Data in Centralized Repositories Versus Supplementary Information—
Given the current size of most proteomic data sets, the authors are often unable to accommodate them in the body of the article. Most of them end up publishing the majority of such data as supplementary information either at the web site of the journal or on their own web site (21). However, there are a number of disadvantages of submitting data as supplementary information instead of contributing them to centralized repositories as listed. 1) Most scientific articles are not freely available and preclude many scientists from accessing published articles. Even if the supplementary information is provided freely by the journals, it would be of no use without the original article that is only accessible by a fraction of the scientific community. 2) Data added as supplementary information might not be easily accessible, as most are in pdf or word document formats and cannot be searched readily. 3) The supplementary data provided by the authors generally does not follow a specific format. This makes it difficult to combine independent data sets for data mining or meta-analysis purposes. 4) Retrieving information on a specific gene from supplementary information is not a trivial task because the nomenclature system is often decided by the authors. 5) Supplementary information is most often limited to the web space provided by the journals and large raw mass spectrometry data (in the gigabyte range) are mostly left out.
On the contrary, data contributed to centralized repositories can be downloaded freely, is more searchable, and is often constrained so that common standard formats are used. Moreover, it is possible for information from diverse research articles to be integrated and presented to the user at the context of the protein or a biological pattern as is done in the case of Human Proteinpedia. With the recent advancements in semantic web (22) and data base interoperability (23), it will become even more fruitful for the scientific community to contribute their data to centralized repositories for optimal utilization of data.
Standardization and Vocabulary Issues in Proteomic Data—
Gene nomenclature is regulated by human genome organization, whereas naming of proteins is largely left to individual investigators. This is unfortunate because even literature searches are based on text and not sequences, which makes it almost impossible to retrieve the published literature on any given protein in a comprehensive fashion. Some features of proteins are beginning to be standardized using controlled vocabularies such as eVOC (24) for describing tissue expression, Gene Ontology (25) for cellular component, molecular function, and biological process, while RESID (26) and Proteomics Standard Initiative-Molecular Interaction (27) vocabularies are available for post-translational modifications and protein-protein interactions, respectively. Proteomics Standard Initiative-Mass Spectrometry (PSI-MS) vocabularies are used to standardize mass spectrometry-based experimental annotations. Nevertheless, even though these controlled vocabularies are available, they are by no means in common use as major data bases themselves do not always adhere to the available vocabularies (28).
A Need for Unified Information about Proteins—
Some of the most popular public repositories store information about specific aspects of proteins. For instance, Protein Data Bank (PDB) (29) is an archive of structural data of biological macromolecules. PRoteomics IDEntifications PRIDE data base (30) and PeptideAtlas (31) are some of the leading mass spectrometry-based data repositories. HPRD (32), IntAct (33), Mint (34), BioGrid (35), and data base of interacting proteins (36) are some of data bases capturing protein-protein interaction data. LifeDB (37) catalogs subcellular localization, whereas Human Protein Atlas (38) archives immunohistochemistry data. These data bases were designed to either collect or accommodate data only from specific experiment types; very few archive data from multiple platforms. Thus, it is currently impossible for a researcher to view all of these data stored in these specialized data bases in one location. Further, there is a lack of mechanisms to automatically exchange most proteomic data types between repositories without substantial manipulation and, in most instance, manual intervention or curation.
In developing a resource for housing proteomic data including that from clinical proteomics, two major issues should be considered. The first is that the data should be shared regardless of the size of the dataset (i.e. it is not just high-throughput data that are worth sharing; data from individual experiments is often even more valuable and should not be ignored). Second, there should be a central portal where the available data is compiled and displayed in the context of a gene/protein. The latter feature would permit users to construct complex queries such as “what are the post-translational modifications on my protein of interest, its interacting proteins, its subcellular localization, and if it is overexpressed in cancers”. Such queries cannot be made in any of the existing proteomic repositories although some provide links to other data bases for certain data types.
Human Proteinpedia as a Portal for Basic and Clinical Information about Proteins—
Human Proteinpedia (39) is a community portal for sharing human proteomic data that is developed with the active participation of more than 70 laboratories around the world. It allows researchers to share their human proteomic data in a manner that is somewhat similar to that of Wikipedia. However, experimental evidence is mandatory for inclusion of data in Human Proteinpediaand; the contributions are always linked to the investigator and the laboratory. Annotations pertaining to post-translational modifications, expression in cell lines or tissues, protein-protein interactions, enzyme substrate, and subcellular localization can be submitted. Human Proteinpedia includes data from diseases such as cancers thereby allowing the biomedical community to take a system's view of the disease proteome. Moreover, it can accommodate data from multiple experimental platforms such as yeast two-hybrid screens, peptide/protein arrays, immunohistochemistry, Western blots, mass spectrometry, co-immunoprecipitation, and fluorescence microscopy.
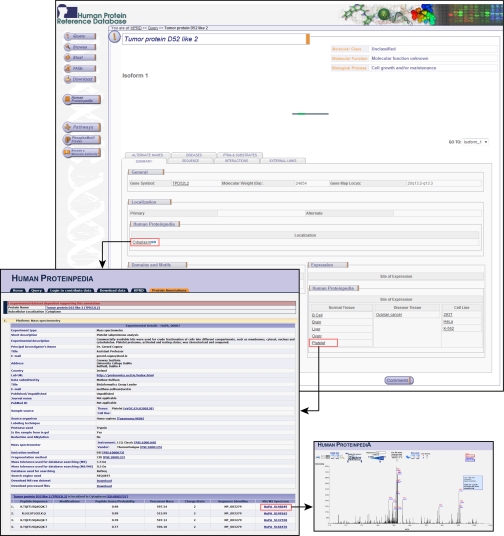
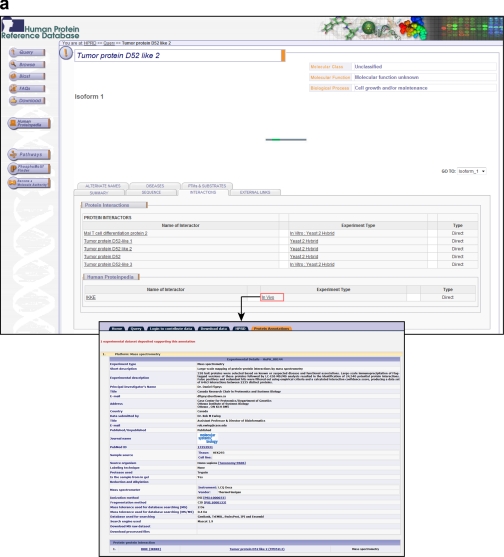
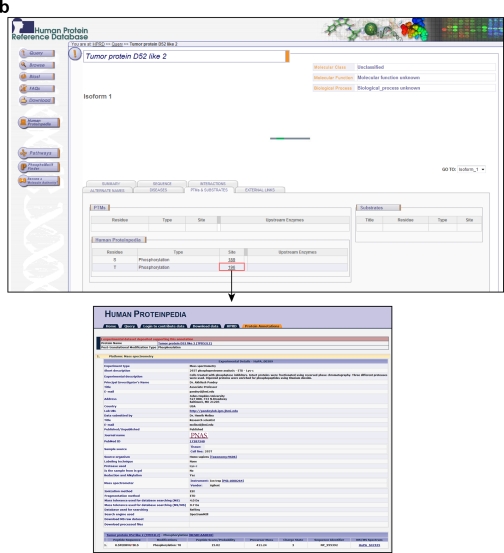
Thus, Human Proteinpedia represents an early attempt to unify human proteomic data under a single resource. An important feature of Human Proteinpedia is that it displays the data in the context of proteins that are annotated in HPRD, a literature curated data base for human proteins (32). An example of tumor protein D52-like 2, which is an uncharacterized protein, will illustrate how Human Proteinpedia can not only handle the complex query described above but provide meaningful answers that otherwise might be difficult to find or derive. Fig. 1 shows the expression of tumor protein D52-like 2 in normal tissues, diseases, and in cell lines along with its subcellular localization. These are all based on data submitted by the community, and the name of the contributing laboratory is clearly displayed when a user clicks on a link (the figure shows the link from the term “cytoplasm” and “platelet”). In addition, in this case, we would not know that this protein is expressed in ovarian cancer without the data contributed by the community. Similarly, Fig. 2a shows that tumor protein D52-like 2 interacts with I-Kappa-B Kinase-Epsilon, a kinase that phosphorylates IkappaB-α, based on a large-scale protein interaction mapping experiment. Finally, Fig. 2b shows that this protein is phosphorylated on serine and threonine residues with links to the primary data that can be explored by the users.
Fig. 1.
Display of expression and subcellular localization of tumor protein D52-like 2 in HPRD molecule page. Molecule page of tumor protein D52-like 2 in HPRD is displayed. Almost all information for this protein is derived from community annotations through Human Proteinpedia including subcellular localization and expression in tissues, cell lines, and diseases. The annotated data shows that this molecule is expressed in B cells, brain, liver, ovary, and platelets. It is also expressed in ovarian cancer and in several cell lines (293T, HeLa, and K-562). Clicking on any of these hyperlinked terms opens a pop-up window (e.g. cytoplasm or platelet, as shown), which provides additional experimental data and details about the contributing laboratory as well as any publications. For example, the window on the left shows peptide identification data, peptide scores, precursor mass, charge state, and sequence identifiers from this unpublished study. If available, the MS/MS spectra are hyperlinked to another window as shown in the right lower part that allows the users to manually inspect the data.
Fig. 2.
Display of post-translational modifications and protein interactors for tumor protein D52-like 2. a, the molecule page for tumor protein D52-like 2 is shown with several interacting proteins manually annotated in HPRD and one protein, I-Kappa-B Kinase-Epsilon, based on data contributed to Human Proteinpedia from a mass spectrometry experiment. The experimental details along with information about the contributing laboratory are also shown. b, no curated post-translational modifications exist for this protein in HPRD. However, the Human Proteinpedia tab shows that there are two phosphorylation sites that have been contributed based on a published study. The lower panel provides a description of the experiment, phosphopeptides identified, and the peptide score.
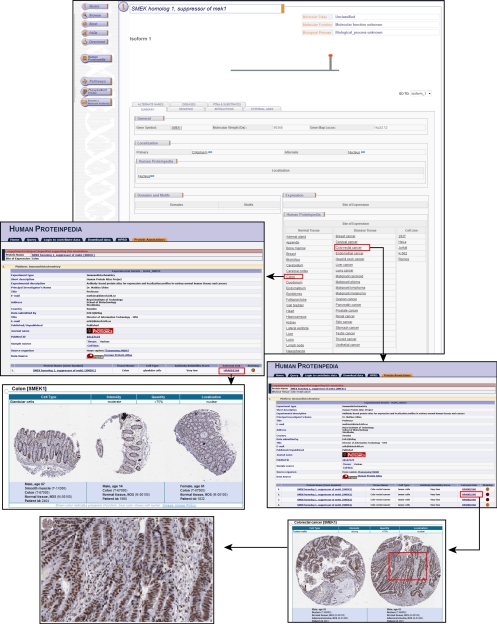
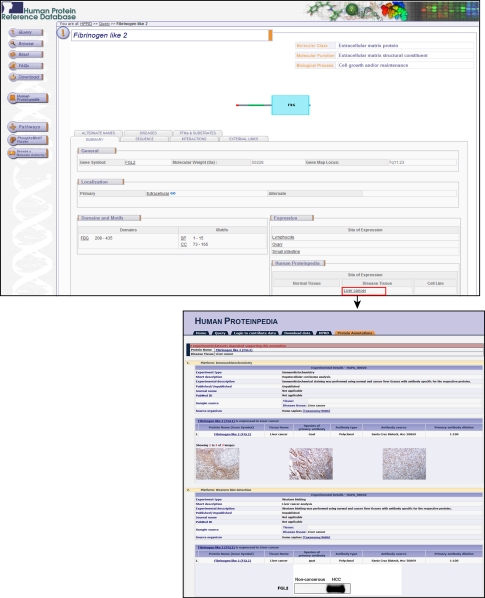
Likewise, Fig. 3 shows the molecule page of suppressor of mek1 (SMEK1) in HPRD. The molecule is unclassified and its site of expression in normal human tissues is also unknown in the literature. However, annotations contributed by the scientific community through Human Proteinpedia reveal the site of expression of SMEK1 in normal and disease tissue as well as cell lines (Fig. 3). These annotations reveal that SMEK1 is moderately expressed in glandular cells of normal colon tissue while being strongly expressed in tumor cells of colorectal cancer tissue. Fig. 4 shows the expression of an extracellular matrix protein, fibrinogen like 2 (FGL2), in hepatocellular carcinoma (HCC). This protein, similar to fibrinogen β and γ, was not previously reported to be involved in HCC. However, it is shown to be expressed in HCC by immunohistochemistry as well as by Western blotting (Fig. 4). Given the fact that early diagnosis will improve prognosis, it is important to pursue such overexpressed molecules, which could turn out to be potential biomarkers.
Fig. 3.
SMEK1 expression in colon and colorectal cancer. SMEK1 molecule page is shown with links to the Human Proteinpedia page indicating expression in colon and colorectal cancer (highlighted), among other sites and diseases. Links from colon displays the experiment description and the information of the contributing group. Human protein atlas links are provided from the Human Proteinpedia page, which indicate moderate expression of SMEK1 in the glandular cells of normal colon tissue. A hyperlink from colorectal cancer again leads to the same resource, which reveals strong expression of SMEK1 in the tumor cells in colorectal cancer tissue.
Fig. 4.
FGL2 expression in hepatocellular carcinoma. The molecule page of FGL2, a secreted protein, is shown. Based on unpublished data submitted to Human Proteinpedia, there are two entries based on two different experimental platforms showing that it is expressed in HCC2. Immunohistochemical staining shows that it is expressed in HCC; this is accompanied by information about the antibody used. The second entry shows that it is overexpressed based on Western blot analysis.2 R. Chaerkady and A. Pandey, unpublished data.
Human Proteinpedia have several advantages over other proteomic resources with respect to clinical proteomic data. Human Proteinpedia incorporates data from multiple experimental platforms, whereas most of the centralized repositories accumulate data from one or two experimental platforms. Given the advantages of each proteomic platform, integration of clinical data produced from all of them under a single banner was lacking. However, Human Proteinpedia displays such clinical information along with the literature-curated data in the context of a protein molecule. With gaining popularity, we expect that even more diverse clinical studies will be integrated and it will be possible to extract biologically meaningful patterns of molecules expressed in particular disease conditions. Further, such data could drive planning of new clinical studies.
Conclusions and Outlook—
To systematically take advantage of the explosion in proteomic data, it must be captured efficiently for the explicit purpose of sharing with the community. In this regard, the researchers should pursue depositing their data to any of the public repositories. In addition, the peer-reviewed journals should actively encourage the authors to submit their data to such proteomic repositories as proposed recently by Nature Biotechnology (40) and Nature Methods (41). Human Proteinpedia allows referees of submitted manuscripts to access the data anonymously if the authors have submitted the data prior to publication for this purpose.
To capture the proteomic data that has already been generated, our team at the Institute of Bioinformatics is scanning through the published issues to date in all of the major proteomic journals including Molecular & Cellular Proteomics, Proteomics, and Journal of Proteome Research for possible inclusion in Human Proteinpedia. The corresponding authors of the relevant articles are being contacted and requested to contribute the data. Those who volunteer work with the team so that the data submission is as simple and painless as possible for the contributor. In addition, the team will obtain data that is not present in Human Proteinpedia from other public proteomic repositories on a regular basis and integrate them with the existing information.
Cancer Genome Anatomy Project (42) aims to catalog the gene expression profiles of normal, precancer, and cancer tissue samples. The goal of this initiative is to improve detection, diagnosis, and treatment of patients through worldwide collaboration. While this project is mainly targeted toward genomic and transcriptomic analysis, future plans that include analyses of cancer proteomes are almost certain. Genomic analysis alone cannot predict the various proteomic alterations in cancers and a better understanding of these alterations will impact detection, diagnosis, and treatment. With additional initiatives being announced to dissect various aspects of the human proteome, including a recent one by HUPO, the need for a portal that allows effective sharing of data effectively among scientists is almost a prerequisite. We anticipate that Human Proteinpedia will be one such portal.
The day when biologists will have a single integrated portal to view data from genomics, transcriptomics, and proteomics data might not be too far off. An initial step to unify the human proteomic data has been taken with the development of Human Proteinpedia. However, this would not have been possible without the enthusiastic participation of the proteomics community. We hope that investigators will continue to share their data to maintain the momentum and anticipate that more and more laboratories will join. Future goals include the addition of protein structure information, and efforts are already on to allow users to view proteomic information submitted to Human Proteinpedia at the genomic level by mapping the peptides onto the genome. We anticipate that the availability of such data will spur the development of additional “omics” tools and newer bioinformatics approaches for harvesting the information provided by the datasets.
Footnotes
Published, MCP Papers in Press, June 23, 2008, DOI 10.1074/mcp.R800008-MCP200
The abbreviations used are: HUPO, human proteome organization; SMEK1, suppressor of mek1; FGL2, fibrinogen like 2; HCC, hepatocellular carcinoma; HPRD, human protein reference data base.
This work was supported, in whole or in part, by National Institutes of Health Grant U54 RR020839 (Roadmap Initiative for Technology Centers for Networks and Pathways). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Pandey, A., and Mann, M. ( 2000) Proteomics to study genes and genomes. Nature 405, 837–846 [DOI] [PubMed] [Google Scholar]
- 2.Nakamura, K., Aebersold, R., Bairoch, A., Dunn, M., Celis, J., Hanash, S., Hochstrasser, D., Humphrey-Smith, I., James, P., Klose, J., LaBaer, J., Langen, H., Mann, M., Parekh, R., Patterson, S., Pearce, C., Poepstorff, P., Simpson, R. J., Tomlinson, I., Tsugita, A., and Yates, J. ( 2004) From genome to proteome-aim of human proteomics. Seikagaku 76, 1271–1274 [PubMed] [Google Scholar]
- 3.Aebersold, R., and Mann, M. ( 2003) Mass spectrometry-based proteomics. Nature 422, 198–207 [DOI] [PubMed] [Google Scholar]
- 4.Cravatt, B. F., Simon, G. M., and Yates, J. R., 3rd ( 2007) The biological impact of mass spectrometry-based proteomics. Nature 450, 991–1000 [DOI] [PubMed] [Google Scholar]
- 5.Han, D. K., Eng, J., Zhou, H., and Aebersold, R. ( 2001) Quantitative profiling of differentiation-induced microsomal proteins using isotope-coded affinity tags and mass spectrometry. Nat. Biotechnol. 19, 946–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yao, X., Freas, A., Ramirez, J., Demirev, P. A., and Fenselau, C. ( 2001) Proteolytic 18O labeling for comparative proteomics: model studies with two serotypes of adenovirus. Anal. Chem. 73, 2836–2842 [DOI] [PubMed] [Google Scholar]
- 7.Everley, P. A., Krijgsveld, J., Zetter, B. R., and Gygi, S. P. ( 2004) Quantitative cancer proteomics: stable isotope labeling with amino acids in cell culture (SILAC) as a tool for prostate cancer research. Mol. Cell. Proteomics 3, 729–735 [DOI] [PubMed] [Google Scholar]
- 8.Gronborg, M., Bunkenborg, J., Kristiansen, T. Z., Jensen, O. N., Yeo, C. J., Hruban, R. H., Maitra, A., Goggins, M. G., and Pandey, A. ( 2004) Comprehensive proteomic analysis of human pancreatic juice. J. Proteome Res. 3, 1042–1055 [DOI] [PubMed] [Google Scholar]
- 9.Gagne, J. P., Ethier, C., Gagne, P., Mercier, G., Bonicalzi, M. E., Mes-Masson, A. M., Droit, A., Winstall, E., Isabelle, M., and Poirier, G. G. ( 2007) Comparative proteome analysis of human epithelial ovarian cancer. Proteome Sci. 5, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crnogorac-Jurcevic, T., Gangeswaran, R., Bhakta, V., Capurso, G., Lattimore, S., Akada, M., Sunamura, M., Prime, W., Campbell, F., Brentnall, T. A., Costello, E., Neoptolemos, J., and Lemoine, N. R. ( 2005) Proteomic analysis of chronic pancreatitis and pancreatic adenocarcinoma. Gastroenterology 129, 1454–1463 [DOI] [PubMed] [Google Scholar]
- 11.Cuzick, J. ( 1999) Screening for cancer: future potential. Eur. J. Cancer 35, 685–692 [DOI] [PubMed] [Google Scholar]
- 12.Kocher, T., and Superti-Furga, G. ( 2007) Mass spectrometry-based functional proteomics: from molecular machines to protein networks. Nat. Methods 4, 807–815 [DOI] [PubMed] [Google Scholar]
- 13.Muller, U., Ernst, G., Melle, C., Guthke, R., and von Eggeling, F. ( 2006) Convergence of the proteomic pattern in cancer. Bioinformatics 22, 1293–1296 [DOI] [PubMed] [Google Scholar]
- 14.Simon, R., and Sauter, G. ( 2003) Tissue microarray (TMA) applications: implications for molecular medicine. Expert Rev. Mol. Med. 5, 1–12 [DOI] [PubMed] [Google Scholar]
- 15.Haab, B. B., Paulovich, A. G., Anderson, N. L., Clark, A. M., Downing, G. J., Hermjakob, H., Labaer, J., and Uhlen, M. ( 2006) A reagent resource to identify proteins and peptides of interest for the cancer community: a workshop report. Mol. Cell. Proteomics 5, 1996–2007 [DOI] [PubMed] [Google Scholar]
- 16.Uhlen, M. ( 2007) Mapping the human proteome using antibodies. Mol. Cell. Proteomics 6, 1455–1456 [PubMed] [Google Scholar]
- 17.Benson, D. A., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J., and Wheeler, D. L. ( 2008) GenBank. Nucleic Acids Res. 36, D25–D30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cochrane, G., Akhtar, R., Aldebert, P., Althorpe, N., Baldwin, A., Bates, K., Bhattacharyya, S., Bonfield, J., Bower, L., Browne, P., Castro, M., Cox, T., Demiralp, F., Eberhardt, R., Faruque, N., Hoad, G., Jang, M., Kulikova, T., Labarga, A., Leinonen, R., Leonard, S., Lin, Q., Lopez, R., Lorenc, D., McWilliam, H., Mukherjee, G., Nardone, F., Plaister, S., Robinson, S., Sobhany, S., Vaughan, R., Wu, D., Zhu, W., Apweiler, R., Hubbard, T., and Birney, E. ( 2008) Priorities for nucleotide trace, sequence and annotation data capture at the Ensembl Trace Archive and the EMBL nucleotide sequence database. Nucleic Acids Res. 36, D5–D12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugawara, H., Ogasawara, O., Okubo, K., Gojobori, T., and Tateno, Y. ( 2008) DDBJ with new system and face. Nucleic Acids Res. 36, D22–D24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bradshaw, R. A., Burlingame, A. L., Carr, S., and Aebersold, R. ( 2006) Reporting protein identification data: the next generation of guidelines. Mol. Cell. Proteomics 5, 787–788 [DOI] [PubMed] [Google Scholar]
- 21.Santos, C., Blake, J., and States, D. J. ( 2005) Supplementary data need to be kept in public repositories. Nature 438, 738. [DOI] [PubMed] [Google Scholar]
- 22.Cannata, N., Schroder, M., Marangoni, R., and Romano, P. ( 2008) A Semantic Web for bioinformatics: goals, tools, systems, applications. BMC Bioinformatics 9, suppl. 4, S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marenco, L., Nadkarni, P., Martone, M., and Gupta, A. ( 2007) Interoperability across neuroscience databases. Methods Mol. Biol. 401, 23–36 [DOI] [PubMed] [Google Scholar]
- 24.Kelso, J., Visagie, J., Theiler, G., Christoffels, A., Bardien, S., Smedley, D., Otgaar, D., Greyling, G., Jongeneel, C. V., McCarthy, M. I., Hide, T., and Hide, W. ( 2003) eVOC: a controlled vocabulary for unifying gene expression data. Genome Res. 13, 1222–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., Davis, A. P., Dolinski, K., Dwight, S. S., Eppig, J. T., Harris, M. A., Hill, D. P., Issel-Tarver, L., Kasarskis, A., Lewis, S., Matese, J. C., Richardson, J. E., Ringwald, M., Rubin, G. M., and Sherlock, G. ( 2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garavelli, J. S. ( 2003) The RESID Database of Protein Modifications: 2003 developments. Nucleic Acids Res. 31, 499–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hermjakob, H., Montecchi-Palazzi, L., Bader, G., Wojcik, J., Salwinski, L., Ceol, A., Moore, S., Orchard, S., Sarkans, U., von Mering, C., Roechert, B., Poux, S., Jung, E., Mersch, H., Kersey, P., Lappe, M., Li, Y., Zeng, R., Rana, D., Nikolski, M., Husi, H., Brun, C., Shanker, K., Grant, S. G., Sander, C., Bork, P., Zhu, W., Pandey, A., Brazma, A., Jacq, B., Vidal, M., Sherman, D., Legrain, P., Cesareni, G., Xenarios, I., Eisenberg, D., Steipe, B., Hogue, C., and Apweiler, R. ( 2004) The HUPO PSI's molecular interaction format–a community standard for the representation of protein interaction data. Nat. Biotechnol. 22, 177–183 [DOI] [PubMed] [Google Scholar]
- 28.Mathivanan, S., Periaswamy, B., Gandhi, T. K., Kandasamy, K., Suresh, S., Mohmood, R., Ramachandra, Y. L., and Pandey, A. ( 2006) An evaluation of human protein-protein interaction data in the public domain. BMC Bioinformatics 7, suppl. 5, S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N., and Bourne, P. E. ( 2000) The protein data bank. Nucleic Acids Res. 28, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martens, L., Hermjakob, H., Jones, P., Adamski, M., Taylor, C., States, D., Gevaert, K., Vandekerckhove, J., and Apweiler, R. ( 2005) PRIDE: the proteomics identifications database. Proteomics 5, 3537–3545 [DOI] [PubMed] [Google Scholar]
- 31.Desiere, F., Deutsch, E. W., Nesvizhskii, A. I., Mallick, P., King, N. L., Eng, J. K., Aderem, A., Boyle, R., Brunner, E., Donohoe, S., Fausto, N., Hafen, E., Hood, L., Katze, M. G., Kennedy, K. A., Kregenow, F., Lee, H., Lin, B., Martin, D., Ranish, J. A., Rawlings, D. J., Samelson, L. E., Shiio, Y., Watts, J. D., Wollscheid, B., Wright, M. E., Yan, W., Yang, L., Yi, E. C., Zhang, H., and Aebersold, R. ( 2005) Integration with the human genome of peptide sequences obtained by high-throughput mass spectrometry. Genome Biol. 6, R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peri, S., Navarro, J. D., Amanchy, R., Kristiansen, T. Z., Jonnalagadda, C. K., Surendranath, V., Niranjan, V., Muthusamy, B., Gandhi, T. K., Gronborg, M., Ibarrola, N., Deshpande, N., Shanker, K., Shivashankar, H. N., Rashmi, B. P., Ramya, M. A., Zhao, Z., Chandrika, K. N., Padma, N., Harsha, H. C., Yatish, A. J., Kavitha, M. P., Menezes, M., Choudhury, D. R., Suresh, S., Ghosh, N., Saravana, R., Chandran, S., Krishna, S., Joy, M., Anand, S. K., Madavan, V., Joseph, A., Wong, G. W., Schiemann, W. P., Constantinescu, S. N., Huang, L., Khosravi-Far, R., Steen, H., Tewari, M., Ghaffari, S., Blobe, G. C., Dang, C. V., Garcia, J. G., Pevsner, J., Jensen, O. N., Roepstorff, P., Deshpande, K. S., Chinnaiyan, A. M., Hamosh, A., Chakravarti, A., and Pandey, A. ( 2003) Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 13, 2363–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kerrien, S., Alam-Faruque, Y., Aranda, B., Bancarz, I., Bridge, A., Derow, C., Dimmer, E., Feuermann, M., Friedrichsen, A., Huntley, R., Kohler, C., Khadake, J., Leroy, C., Liban, A., Lieftink, C., Montecchi-Palazzi, L., Orchard, S., Risse, J., Robbe, K., Roechert, B., Thorneycroft, D., Zhang, Y., Apweiler, R., and Hermjakob, H. ( 2007) IntAct–open source resource for molecular interaction data. Nucleic Acids Res. 35, D561–D565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chatr-aryamontri, A., Ceol, A., Palazzi, L. M., Nardelli, G., Schneider, M. V., Castagnoli, L., and Cesareni, G. ( 2007) MINT: the molecular INTeraction database. Nucleic Acids Res. 35, D572–D574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stark, C., Breitkreutz, B. J., Reguly, T., Boucher, L., Breitkreutz, A., and Tyers, M. ( 2006) BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 34, D535–D539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salwinski, L., Miller, C. S., Smith, A. J., Pettit, F. K., Bowie, J. U., and Eisenberg, D. ( 2004) The database of interacting proteins: 2004 update. Nucleic Acids Res. 32, D449–D451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mehrle, A., Rosenfelder, H., Schupp, I., del Val, C., Arlt, D., Hahne, F., Bechtel, S., Simpson, J., Hofmann, O., Hide, W., Glatting, K. H., Huber, W., Pepperkok, R., Poustka, A., and Wiemann, S. ( 2006) The LIFEdb database in 2006. Nucleic Acids Res. 34, D415–D418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uhlen, M., Bjorling, E., Agaton, C., Szigyarto, C. A., Amini, B., Andersen, E., Andersson, A. C., Angelidou, P., Asplund, A., Asplund, C., Berglund, L., Bergstrom, K., Brumer, H., Cerjan, D., Ekstrom, M., Elobeid, A., Eriksson, C., Fagerberg, L., Falk, R., Fall, J., Forsberg, M., Bjorklund, M. G., Gumbel, K., Halimi, A., Hallin, I., Hamsten, C., Hansson, M., Hedhammar, M., Hercules, G., Kampf, C., Larsson, K., Lindskog, M., Lodewyckx, W., Lund, J., Lundeberg, J., Magnusson, K., Malm, E., Nilsson, P., Odling, J., Oksvold, P., Olsson, I., Oster, E., Ottosson, J., Paavilainen, L., Persson, A., Rimini, R., Rockberg, J., Runeson, M., Sivertsson, A., Skollermo, A., Steen, J., Stenvall, M., Sterky, F., Stromberg, S., Sundberg, M., Tegel, H., Tourle, S., Wahlund, E., Walden, A., Wan, J., Wernerus, H., Westberg, J., Wester, K., Wrethagen, U., Xu, L. L., Hober, S., and Ponten, F. ( 2005) A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol. Cell. Proteomics 4, 1920–1932 [DOI] [PubMed] [Google Scholar]
- 39.Mathivanan, S., Ahmed, M., Ahn, N. G., Alexandre, H., Amanchy, R., Andrews, P. C., Bader, J. S., Balgley, B. M., Bantscheff, M., Bennett, K. L., Bjorling, E., Blagoev, B., Bose, R., Brahmachari, S. K., Burlingame, A. S., Bustelo, X. R., Cagney, G., Cantin, G. T., Cardasis, H. L., Celis, J. E., Chaerkady, R., Chu, F., Cole, P. A., Costello, C. E., Cotter, R. J., Crockett, D., DeLany, J. P., De Marzo, A. M., DeSouza, L. V., Deutsch, E. W., Dransfield, E., Drewes, G., Droit, A., Dunn, M. J., Elenitoba-Johnson, K., Ewing, R. M., Van Eyk, J., Faca, V., Falkner, J., Fang, X., Fenselau, C., Figeys, D., Gagne, P., Gelfi, C., Gevaert, K., Gimble, J. M., Gnad, F., Goel, R., Gromov, P., Hanash, S. M., Hancock, W. S., Harsha, H. C., Hart, G., Hays, F., He, F., Hebbar, P., Helsens, K., Hermeking, H., Hide, W., Hjerno, K., Hochstrasser, D. F., Hofmann, O., Horn, D. M., Hruban, R. H., Ibarrola, N., James, P., Jensen, O. N., Jensen, P. H., Jung, P., Kandasamy, K., Kheterpal, I., Kikuno, R. F., Korf, U., Korner, R., Kuster, B., Kwon, M. S., Lee, H. J., Lee, Y. J., Lefevre, M., Lehvaslaiho, M., Lescuyer, P., Levander, F., Lim, M. S., Lobke, C., Loo, J. A., Mann, M., Martens, L., Martinez-Heredia, J., McComb, M., McRedmond, J., Mehrle, A., Menon, R., Miller, C. A., Mischak, H., Mohan, S. S., Mohmood, R., Molina, H., Moran, M. F., Morgan, J. D., Moritz, R., Morzel, M., Muddiman, D. C., Nalli, A., Navarro, J. D., Neubert, T. A., Ohara, O., Oliva, R., Omenn, G. S., Oyama, M., Paik, Y. K., Pennington, K., Pepperkok, R., Periaswamy, B., Petricoin, E. F., Poirier, G. G., Prasad, T. S., Purvine, S. O., Rahiman, B. A., Ramachandran, P., Ramachandra, Y. L., Rice, R. H., Rick, J., Ronnholm, R. H., Salonen, J., Sanchez, J. C., Sayd, T., Seshi, B., Shankari, K., Sheng, S. J., Shetty, V., Shivakumar, K., Simpson, R. J., Sirdeshmukh, R., Siu, K. W., Smith, J. C., Smith, R. D., States, D. J., Sugano, S., Sullivan, M., Superti-Furga, G., Takatalo, M., Thongboonkerd, V., Trinidad, J. C., Uhlen, M., Vandekerckhove, J., Vasilescu, J., Veenstra, T. D., Vidal-Taboada, J. M., Vihinen, M., Wait, R., Wang, X., Wiemann, S., Wu, B., Xu, T., Yates, J. R., Zhong, J., Zhou, M., Zhu, Y., Zurbig, P., and Pandey, A. ( 2008) Human proteinpedia enables sharing of human protein data. Nat. Biotechnol. 26, 164–167 [DOI] [PubMed] [Google Scholar]
- 40.Editorial ( 2007) Democratizing proteomics data. Nat. Biotechnol. 25, 262. [DOI] [PubMed] [Google Scholar]
- 41.Editorial ( 2008) Thou shalt share your data. Nat. Methods 5, 209 [Google Scholar]
- 42.Strausberg, R. L., Buetow, K. H., Emmert-Buck, M. R., and Klausner, R. D. ( 2000) The cancer genome anatomy project: building an annotated gene index. Trends Genet. 16, 103–106 [DOI] [PubMed] [Google Scholar]