Abstract
Sperm are remarkably complex cells with a singularly important mission: to deliver paternal DNA and its associated factors to the oocyte to start a new life. The integrity of sperm DNA is a keystone of reproductive success, which includes fertilization and embryonic development. In addition, the significance in these processes of proteins that associate with sperm DNA is increasingly being appreciated. In this review, we highlight proteomic studies that have identified sperm chromatin proteins with fertility roles that have been validated by molecular studies in model organisms or correlations in the clinic. Up to 50% of male-factor infertility cases in the clinic have no known cause and therefore no direct treatment. In-depth study of the molecular basis of infertility has great potential to inform the development of sensitive diagnostic tools and effective therapies that will address this incongruity. Because sperm rely on testis-specific protein isoforms and post-translational modifications for their development and function, sperm-specific processes are ideal for proteomic explorations that can bridge the research lab and fertility clinic.
Sperm are intricate yet streamlined cells with the underlying purpose of producing a healthy baby. Sperm cells in every sexually reproducing animal are highly specialized delivery vehicles for the chromatin cargo within, which is composed of DNA and its associated proteins. It is clear that sperm chromatin is essential for sperm function and subsequent embryonic development because defects in sperm chromatin are linked to natural reproductive malfunctions like spontaneous abortion as well as assisted reproductive failure (1–3). These defects can include disrupted DNA integrity caused by genetic mutations, apoptotic DNA fragmentation, or exposure to environmental agents and free radicals (4). Furthermore, disruption of chromatin proteins contributes to decreased fertility (5). Despite this evidence, most clinical assays for sperm chromatin only detect gross defects in DNA integrity (4, 6). This, combined with the typical assessments of sperm production, including motility, morphology, and male hormone levels, still result in a striking 30–50% of male-factor infertility cases having no known cause (7). With 2.1 million couples in the United States facing infertility as of 2002, there is a pressing need for an expanded array of sensitive tests to improve diagnostic capabilities in the clinic (8).
The requirement for more research extends beyond diagnosis, however, because many of the basic cellular mechanisms that underlie male infertility remain unknown. Consequently, virtually no therapies exist that remedy the molecular causes of sperm dysfunction. Rather, prolonged or invasive assisted reproductive technology (ART)1 procedures and intracytoplasmic sperm injection (ICSI) are used to bypass male infertility, with a modest 42% success rate (9). In addition, the safety of widespread ICSI use has recently been called into question. It is speculated that sperm from infertile patients contain cytologically subtle chromatin abnormalities that can affect the resulting embryo (10, 11). Thus it is important to define the epigenetic features of paternal chromatin that can affect future generations. Further knowledge about sperm chromatin protein function can lead to the development of novel therapies that target only sperm, reducing possibilities for side effects or unintended consequences on resulting offspring.
Any comprehensive understanding of sperm biology must include proteomic analysis. Sperm are particularly well suited to proteomic approaches. Compared with most other cell types, they are easily isolated from other tissues and fluids. Importantly, sperm development and function rely heavily on sperm-specific protein isoforms of somatic counterparts, as well as the post-translational modification of key sperm proteins (12, 13). For example, during spermatogenesis, histone proteins in developing sperm are replaced by testis-specific histone variants important for fertility (14). Also, because de novo transcription in post-meiotic sperm is largely silenced, the cell depends on post-translational modifications to implement subsequent stages of sperm formation, maturation, and activation (15). The development of clinical applications arising from the proteomic discovery of sperm proteins and sperm-specific protein isoforms is a budding field, with abundant opportunities for innovation. Of particular promise is the identification of sperm-specific post-translational modifications that are functionally important during and after sperm development.
Functional analysis of sperm-specific factors following proteomic identification is crucial to our understanding of male reproduction. Direct experimental approaches can identify relevant variables and reduce experimental complexity. Such strategies are facilitated by the use of model organisms like sea urchins, fish, mice, worms, and flies. Model organisms often have simplified anatomical structures and streamlined developmental programs in comparison to humans. Their male germ cells can be obtained in large quantities in various developmental stages. Though some male reproductive proteins evolve rapidly (16), others involved in fundamental processes of sperm development like meiosis are conserved (17–19). As in humans, the chromatin of each of these animals is at its most compact in mature sperm. Ultimately, defining the molecular functions of conserved proteins can aid in identifying biologically relevant clinical markers of human infertility, as well as targets for infertility therapies.
One challenge currently facing proteomicists, basic biologists, and clinicians is how to make sense of the vast amount of data being generated through proteomic, genomic, functional, and clinical studies. Only through solving this challenge can the full potential of proteomics be translated into meaningful clinical options for male infertility. This review will highlight a selection of recent advances in our understanding of sperm chromatin biology that allow proteomic data to be interpreted within cellular or functional contexts. These studies include strategies for employing bioinformatics to generate testable hypotheses, increasing the specificity of proteomic analysis, analyzing function in model organisms, and correlating expression changes with detailed observations from the clinic. The proteomics of other sperm subcellular compartments, fluid components, or physiological reproductive structures are presented elsewhere (20, 21). By discussing sperm chromatin-based studies, we illustrate the contributions that chromatin studies can make toward understanding fertility, as well as the need to develop sophisticated molecular tools for the study of male-factor infertility. We also promote communication and integration between lab and clinic when designing and interpreting studies.
Sperm Are Ideal for Proteomic Analysis—
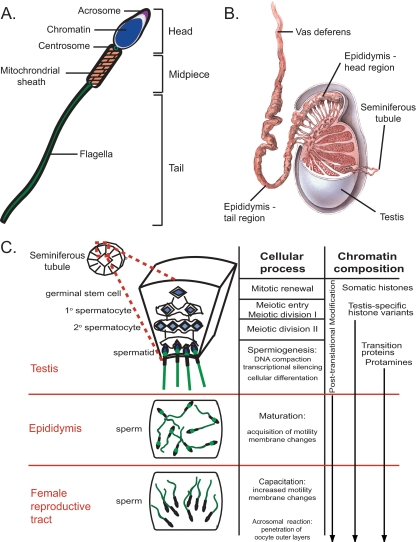
Sperm are arguably the most specialized cells in the human body (Fig. 1A). As such, a remarkable set of complex events must unfold to ensure the proper development and function of each cell. These include a) configuring chromatin for efficient delivery to the egg and preserving the epigenetic information needed for subsequent zygotic development (22); b) using a flagellated force-generating tail structure for long-distance travel to the egg (23, 24); and c) unleashing a series of membrane-associated cellular changes for penetrating the substantial corona and zona pellucida layers surrounding the egg (25–27). The sperm cell develops these capacities at distinct time points before fertilization (Fig. 1B). Chromatin changes occur in the testis during meiosis (in which copies of the genome are partitioned into haploid spermatid cells) and spermiogenesis (in which spermatids elongate to form sperm with fully compacted chromatin). Sperm motility is gained in the epididymis upon exit from the testis, and sperm capacitation for penetrating the zona pellucida occurs within the female reproductive tract. These events are largely controlled by post-translational events, for transcription and translation greatly subside as DNA becomes tightly compacted and cytoplasm is jettisoned during spermiogenesis (13, 15). Progression of subsequent developmental stages is mediated by existing signaling molecules (26, 27). For example, the phosphatase protein PP1γ is involved in multiple aspects of sperm development, including spermiogenesis and acquisition of sperm motility (28–30). Much is yet unknown regarding the molecular basis of sperm biology, particularly how these drastic changes in cell morphology and function occur in the absence of new protein synthesis.
Fig. 1.
Human sperm and sperm development. A, schematic of a human sperm cell, which is composed of three regions. The head region contains the highly compacted sperm DNA and associated proteins (blue) surrounded by the cellular membrane (black) that in the head region have receptors for recognizing oocyte factors. The acrosome (purple) houses digestive enzymes used for penetrating the oocyte outer layers. The mid-piece region consists of the paternally contributed centrosome (green circle) and mitochondria (tan) used for energy generation. The flagellar tail (green) provides motility. B, diagram of testis, epididymis, and vas deferens. Sperm develop within the seminiferous tubules that are coiled within the testis. Sperm transit to the epididymis where they further mature and are then stored in the vas deferens. Contraction of the vas deferens propels sperm through the male reproductive tract during ejaculation. Image from LifeART (and/or) MediClip® (2008) Wolters Kluwer Health, Inc.- Lippincott Williams & Wilkins. All rights reserved. C, diagram of human sperm formation and associated processes. A cross-section of a seminiferous tubule found within the testis is illustrated. Germinal stem cells renew through repeated rounds of mitosis then shift to meiosis to form primary and secondary spermatocytes. After meiosis, haploid cells undergo spermiogenesis, where DNA is tightly compacted, and transcription is largely silenced. Cells also differentiate morphologically to form flagella. In the epididymis, sperm mature to become motile, and cellular membranes are prepared for fertilization. After transfer to the female reproductive tract, sperm cellular membranes undergo further changes, and sperm become highly motile, a process known as capacitation. Upon binding with the oocyte, acrosomes release enzymes to penetrate oocyte outer layers to allow sperm-egg membrane fusion for fertilization.
The wide array of changes in chromatin during sperm development provides an ideal platform for proteomic exploration of essential sperm-specific chromatin proteins. During meiosis, sperm chromosomes are segregated in a distinct fashion from that of oocyte chromosomes with unique timing and generated end products (31). After meiosis, sperm DNA experiences extreme chromosome compaction during spermiogenesis. This compaction is mediated by drastic changes at the most fundamental level of DNA packaging where a nucleosomal architecture shifts to a toroidal structure (32). Sperm nuclear basic proteins (SNBPs), which include variants of histone subunits, transition proteins, and protamine proteins, implement this change (33, 34). This transition occurs in a stepwise fashion, replacing somatic histones with testis-expressed histone variants, then transition proteins, and finally protamines (35). Deficits in SNBPs are known to cause male infertility (36–38). A growing body of work indicates that these chromatin proteins do not act exclusively to compact sperm DNA. Histone localization and post-translational modification of histones encode epigenetic information that may regulate transcription important for sperm development (22). They may also serve to mark the heterochromatic state of specific regions of the genome that may be important after fertilization, when somatic histones are incorporated back into paternal chromatin or during subsequent zygotic development (39). Thus achieving a deep understanding of the molecular basis of sperm chromatin composition and dynamics will impact multiple levels of fertility biology.
Lessons Learned from Whole Sperm Proteomics—
Any single component of a system must be understood within the context of the whole. To date, most proteomic studies on human sperm have concentrated on determining the protein content of the whole sperm cell. Estimates of sperm proteome size have been made based on two-dimensional poly-acrylamide gel electrophoresis (2D-PAGE), which creates a visual reference map of the sperm proteome by separating proteins according to isoelectric point and mass to ideally resolve protein isoforms (40). Using this technique, the number of sperm proteins has been estimated at 1000–2000 (41, 42). Subsequently, the resolution of 3872 separate protein spots was achieved via multiple 2D-PAGE, each corresponding to a restricted pH range for separation in the first dimension (43). Shotgun proteomic approaches have also contributed substantially to sperm proteome characterization (44, 45). Human sperm proteins were identified after proteolytic digestion and high-resolution liquid chromatography to separate peptides before tandem mass spectral identification (LC-MS2). To decrease sample complexity, sperm proteins were divided into Triton X-100 soluble or insoluble fractions, then further separated via one-dimensional SDS-PAGE before protease treatment and LC-MS2. Repeated rounds of LC-MS2 identified peptides corresponding to 1760 and 1056 proteins, respectively; though the identified protein list is only available for the latter (44, 45).
Such proteomic technologies are powerful and have yielded remarkably large lists of proteins. However, each still has its challenges, which include the detection of low abundance proteins (46). For 2D-PAGE, the excision and mass spectrometric characterization of spots to assign protein identity can be laborious. For example, Li et al. (43) verified the protein identities of 16 of 3872 spots, whereas Martinez-Heredia et al. (41) identified 131 of 1000 spots, providing just a glimpse of the proteins and their functions (47). While shotgun proteomic techniques can give a more thorough picture, the proteolytic digestion of whole proteins into individual peptides en masse may obfuscate detection of multiple isoforms or combinations of post-translational modifications of any one protein. The difference in the number of protein species detected via 2D-PAGE (3872) and the number of protein identities resolved by shotgun proteomics (1056) may attest to differences in technology. For example, the smaller number of proteins detected by shotgun proteomics may reflect that there are fewer proteins and protein isoforms in sperm because mature sperm have eliminated almost all cytoplasm. The larger number of protein spots detected by 2D-PAGE may suggest that there are more post-translationally modified versions of the proteins present. The development of new or combined proteomic approaches leading to more comprehensive identification of the sperm proteome places a full appreciation of the complexity of post-translational regulation within our grasp.
The elucidation of the sperm proteome provides a rich source of raw material for bioinformatic investigation. The use of Gene Ontology (GO) classifications to generate putative functional or subcellular localization groups provides a general snapshot of the proteome (48). In addition to GO analysis, more sophisticated classification methods can be employed to reveal meaningful patterns. For example, analysis of the sperm proteome of Drosophila melanogaster has generated new hypotheses about germline gene regulation and chromatin organization (49). First, shotgun mass spectrometric analysis leading to the identification of 381 proteins in Drosophila sperm was performed. By analyzing the chromosomal distribution of corresponding genes, Dorus et al. (49) determined that sperm proteins are underrepresented on the X chromosome. This result is consistent with the finding from DNA microarray analysis that male-expressed genes are also underrepresented on X (50). Sub-chromosomal clustering analysis also shows that individual chromosomes have regions with high densities of sperm-expressed genes, suggesting that the genome itself is organized into clusters of similarly expressed genes. This analysis led to the hypothesis that cessation of transcription during spermatogenesis occurs in a hierarchical fashion, with sperm-specific genes being silenced last (49). We can speculate further that SNBPs may also be located in sub-chromosomal clusters whose expression is regulated by chromatin-based mechanisms. In this way, existing large data sets can be mined for testable hypotheses using various bioinformatic strategies, contributing greatly to the potential of proteomics to address specific cellular processes.
Are studies of whole sperm the best application of proteomic technology to yield information about subcellular compartments like the nucleus? How are proteins categorized within GO groups actually connected in functional pathways? Although whole sperm analysis has increased our knowledge of the composition of a sperm cell, we still do not understand how most of these proteins contribute to specific aspects of fertility. Ultimately, to understand the significance of proteomic data, functional studies must be performed. The following section describes strategies that increase our knowledge of mechanisms important for fertility by combining directed proteomic approaches for identification of sperm-specific proteins with functional studies in model organisms.
Directing Proteomics Toward Functional Analysis—
Knowledge of male-factor infertility can be greatly expanded by in-depth analysis of each stage of sperm cell development in model organisms such as Caenorhabditis elegans, Drosophila, and mouse (18, 51). Because many essential elements of sperm development are evolutionarily conserved, the use of model organisms allows a reductionist approach to the syndrome of male infertility in the context of hypothesis-based experiments with controlled variables. In these organisms, sperm develop rapidly in tissues analogous to the human testis and are accessible to a variety of experimental approaches at each stage in the development (52–55). For example, cytological assays to view cells during all stages of sperm development are well established. Also, biochemical protocols for the stage-specific isolation of developing sperm are commonly employed, as are subcellular fractionation techniques to enrich for certain classes of proteins. Each of these organisms has extensively annotated genomes accessible through on-line databases like WormBase, FlyBase, and the Mouse Genome Informatics website (56–58). Loss-of-function analyses of specific proteins using genetic mutations or RNA interference (RNAi) are also straightforward to conduct.
A traditional challenge encountered using model organisms is the discovery of candidate proteins to be analyzed for specific functions. Today, modern shotgun proteomic technology and sensitive methods of detection can create the opposite dilemma. Proteomic lists on the order of thousands of proteins present an overwhelming number of candidates for functional analysis. Several strategies can focus the scope of proteomic identification upon a process of interest. One is to employ subcellular fractionation techniques that focus on the organelles or fractions of interest (59). An additional method is the application of abundance criteria from mass spectrometry to prioritize likely resident proteins of subcellular organelles or regions (60). The utility of abundance criteria in prioritizing factors that make up various subcellular compartments of embryonic stem cells (cytoplasm, nucleoplasm, and chromatin/membrane) was validated by the finding that abundant proteins in each compartment corresponded to expected functional clusters based on GO (61). The refinement of the target protein list can be accomplished through subtractive or comparative analysis against a different cell type or developmental stage, thereby enriching for relevant proteins and eliminating general factors (62, 63).
One example of the use of these criteria is a study that prioritized the identification of sperm-enriched chromatin proteins with the goal of finding evolutionarily conserved fertility factors (17). Each of these three strategies was employed to generate a focused list that was subsequently used in functional studies. First, subcellular biochemical fractionation was used to purify sperm chromatin proteins, which reduced overall proteomic complexity. Large-scale protein identification was carried out using Multidimensional Protein Identification Technology (MudPIT), which employs a tandem set of liquid chromatographic steps (strong cation exchange and reverse phase) to separate proteolytic peptides before MS2 identification (64). The peptides identified corresponded to 1099 predicted C. elegans proteins (65). Second, reproducibility from multiple repetitions of MudPIT analysis was used to generate a prioritized list of 502 proteins, which represented 88% of the relative mass of all sperm chromatin proteins identified and were thus the most abundant proteins in the samples. Third, this group was subtracted against a list of all oocyte chromatin proteins generated in parallel, reducing the number of sperm chromatin-associated fertility candidates to 132. By taking this step, a large number of shared housekeeping and DNA proteins as well as canonical histones and shared meiotic proteins were eliminated. Thus, members of this list were likely to have sperm-specific chromatin localization and function.
The rationale for this experimental design was supported by functional analysis of these candidate proteins in worms (17). First, sperm chromatin localization was demonstrated for 11 candidates using immunolocalization. Subsequent functional analysis using RNAi and assays for fertility showed that 38% percent of these 132 proteins are important for germ cell formation or embryonic viability. The actual percentage is likely to be higher because functional redundancy between co-expressed protein isoforms may obscure detection of a fertility role for any single protein, and because genes required for spermatogenesis are often resistant to RNAi in C. elegans. Significantly, 59% percent of the 132 proteins have human homologs, whereas 37% of genes with mouse homologs that had been characterized showed male infertility in genetic knock-out strains. Through the application of directed proteomic approaches combined with functional and bioinformatic analysis, a focused and testable group of prime candidates was identified for future studies in C. elegans or other organisms.
Exploring Mechanism through Proteomics and Functional Studies—
Focused analyses of proteins that are expressed exclusively in sperm can further advance our knowledge of the molecular pathways required for chromatin function. For example, new SNBPs can be identified through proteomic analysis of biochemical fractions of sperm chromatin and characterized for roles in organizing chromatin. During spermatogenesis, a massive amount of epigenetic information is potentially being “wiped clean” by the histone replacement process that must be re-established in the zygote (22, 39). Although protamines ultimately displace the majority of somatic histones during spermiogenesis, 15% of histones in humans are retained, some at transcriptionally active sites (66, 67). This suggests that some epigenetic information is preserved in sperm. Therefore, elucidating not only the composition but also the spatio-temporal patterns of sperm nuclear basic proteins within chromatin has implications for understanding germline gene regulation and embryonic contributions of sperm chromatin.
To examine the stage-specific composition and organization of sperm chromatin undergoing condensation, a recent proteomic study utilized MS2 identification of SDS-PAGE-separated proteins from acid-extracted nuclei in later stages of spermatogenesis in mice (68). Acid-soluble fractions are expected to contain fundamental proteins mediating sperm chromosome condensation, which are highly basic. Five new histone variants with exclusive expression during late spermiogenesis were identified through proteomic analysis. Cytological analysis showed that in condensing spermatids, two of these histone variants, H2AL1 and H2AL2, are incorporated specifically into heterochromatin regions around centromeres, known as pericentromeres. The appearance of H2AL1 and H2AL2 coincides with the removal of post-translational modifications marking heterochromatin, such as histone H3 lysine 9 trimethylation and heterochromatin protein 1 (HP1) association to chromatin (69). H2AL1 and H2AL2 are also retained after protamine incorporation (68). Thus, sperm-specific histone variants may serve as potential epigenetic regulators for the re-establishment of heterchromatin at specific sites after fertilization. As such, the absence or reduced levels of these sperm-specific proteins may correlate with incompletely formed or defective sperm chromatin.
Although there are no direct human counterparts for these two histone variants, testis-specific human histone variants have been characterized and have potential clinical applications (14, 70). Because other types of epigenetic marks occur in many cell types, such as histone modifications, sperm-specific histone variants are ideally suited for further development as infertility biomarkers that reveal defects in chromatin packaging or sperm development that may have significant consequences on reproductive outcome or resulting offspring.
Functional Inquiry with an Eye on the Clinic—
Fertility research is at an important juncture, for technological innovations are allowing the convergence of large-scale proteomic studies and basic biological studies with clinical outcomes. Such connections are the major aim of biomedical research. The previous examples illustrate that proteomics can be harnessed to shed light on the composition of sperm chromatin through strategies that prioritize targets and reduce complexity, whereas biological function can be addressed by model organism research. However, the practical potential of these studies will not be realized if their results are not considered in the context of human infertility. This disconnect represents one of the fundamental challenges facing all fields involved. One story involving several groups, discussed below, is an example that makes the link between mechanistic characterization of a sperm chromatin component in mouse and predictors of reproductive outcome in humans.
In the mouse proteomic study described in the previous section, acid-soluble sperm chromatin fractions were shown to include previously unidentified stage-specific markers of pericentromeric heterochromatin (68). Using the same biochemical approach, another class of less basic proteins was identified to contain chaperone proteins, including HSPA2, a testis-specific member of the HSP70 chaperone family (71). Two separate roles in chromatin dynamics have been found for HSPA2 during sperm formation (71–73). During sperm meiosis, HSPA2 functions in desynapsis at the synaptonemal complex (74, 75). Mice lacking HSPA2 are male infertile, with meiotic arrest and apoptotic phenotypes (76, 77). A second role for HSPA2 was suggested by the observation that HSPA2 is present in both meiotic and post-meiotic sperm (71, 73, 77). HSPA2 becomes acid-soluble only after meiosis, during transition protein incorporation (71). Co-immunoprecipitation studies demonstrated that HSPA2 and transition proteins form acid-soluble complexes in elongating spermatids. Thus, HSPA2 is the first identified transition protein chaperone (71).
With such critical roles in spermatogenesis, the evolutionarily conserved chaperone HSPA2 is a prime candidate for fertility assessment in humans. In fact, two separate studies demonstrated that the ratio of HSPA2 levels to those of creatine kinase-B, another identified biochemical marker of sperm maturity, is predictive of in vitro fertilization outcome (72, 78, 79). Interestingly, levels of HSPA2, as well as 16 other proteins, were also found to be aberrant in a proteomic comparison of sperm from 10 normal donors and 20 patients that suffer from low motility of sperm (asthenozoospermia) (80). HSPA2 has been hypothesized to function in several post-meiotic sperm processes including cytoplasmic extrusion and plasma membrane remodeling, which may affect sperm morphology or cellular interactions (81). Additional data correlates HSPA2 expression with the formation of hyaluronic acid binding sites on the plasma membrane (82). In fact, ICSI performed with sperm selected for their affinity for hyaluronic acid resulted in 4- to 6-fold reduced incidence of chromosomal abnormalities (82–84). HSPA2 is therefore one example of a biologically relevant marker of reproductive potential that can be found integrating directed proteomics, functional characterization, and correlations with clinical observations. The body of work on HSPA2 demonstrates how clinical tools like biomarkers and effective protocols are validated from a full understanding of the biological significance of sperm or testis-specific proteins.
Dissecting the Complexity of Human Male Infertility—
Another exciting avenue that exploits the power of proteomics is the large-scale identification of biomarkers that correlate with clinical reproductive success or failure. For example, comparison of proteomes of human sperm samples from fertile and infertile individuals may identify many candidate proteins with altered expression (80, 85, 86). However, prioritizing these proteins for biomarker development is not a straightforward task. The sperm proteome of an individual may vary from sample to sample taken on different days (86). Likewise, fertility of individuals classed in either fertile or infertile categories can also span a large range (87). Thus it may be difficult to ascertain which proteins consistently differ in expression across studies that use different sample sizes and methodologies.
Similar to cell biological studies of the sperm proteome, clinical studies using proteomics can also benefit from reductionist approaches that focus on defined variables. Because infertility can be a complex syndrome with multiple causes, one such strategy is to first classify individuals based on known molecular markers of specific fertility defects then to identify other proteins that also show altered expression. These newly identified candidate proteins can then be further investigated for roles in the same processes mediated by the original molecular marker. Because changes in chromatin integrity and key sperm chromatin proteins such as protamines are already known to correlate with infertility, protamines represent an ideal starting point for such explorations (88, 89).
One such study by de Mateo et al. (47) applied proteomic analysis to find proteins showing altered expression in correlation with sperm DNA damage and protamine content. An altered ratio of the two protamine proteins, protamine 1 (P1) and protamine 2 (P2), correlates with male infertility (90–93). Sperm samples were obtained from 47 infertile patients and 10 sperm donors, all of those who presented with normal numbers of sperm (normozoospermia). Each sample was assessed for DNA fragmentation using terminal deoxynucleotidyl transferase-mediated nick end labeling (TUNEL) and extracted for protamines to determine the ratio of protamine proteins. First, the authors observed a range of differences in DNA fragmentation and P1/P2 ratios among all 57 samples. Each sample was then subjected to 2D-PAGE, resolving ∼1000 spots. MS2 analysis identified 33 new sperm proteins in addition to the 98 proteins the authors had found in previous studies (41). Of these, 101 were selected for comparison among the 57 sperm samples (47). Over half of the 101 proteins showed correlated differences with another protein among the 57 samples. These results could represent substantial individual variation between donors. Alternatively, they may represent functional associations. In support of the latter hypothesis, 8 proteins were found to vary in correlation with differences in TUNEL results and 7 with altered P1/P2 ratios.
This preliminary study indicates that proteomic analysis is able to detect a small number of protein expression differences within the sperm proteome and has identified two groups of proteins showing expression levels that co-vary with sperm chromatin assessments. Both TUNEL and P1/P2 ratios are known to correlate with fertility (4, 88); although it is not known whether the proteins found in this study do so. Further examination of larger numbers of sperm profiles will clarify which cohort differences are because of individual variation and which differences consistently correlate with these assays of sperm chromatin state. Such proteins can be further validated for their utility as biomarkers and functionally characterized. In summary, the strategic extension of correlative studies to these and other measures of infertility will yield relevant biomarkers that are useful in the clinic.
DISCUSSION
Though the application of proteomics to understanding fertility is still developing, our understanding of sperm development, function, and dysfunction has grown considerably through its use. To achieve continued progress and concrete results for couples battling infertility, the arenas of proteomics, basic biology, and the clinic must become integrated partners. Each discipline brings unique tools and perspectives to the table. Proteomics provides rapid and in-depth discovery of fertility factors and proteins with altered expression in infertile patients. Detailed molecular studies facilitated by model organisms can provide insight about the function or dysfunction of such proteins in sperm. Finally, clinical studies that correlate differences in protein expression with fertility measures can be used to translate molecular data into widespread patient benefit. The studies highlighted in this review focus on sperm chromatin to illustrate that the synergy of technically divergent fields increases knowledge of fundamental processes in sperm and facilitates development of new clinical tools.
The interdisciplinary sharing of information is integral for the incorporation of proteomics with basic biology and clinical research. Bioinformatic tools that are widely available to researchers and proteomicists need to be comprehensively developed and used in fertility studies. This includes the effective organization, management, and dissemination of standardized and detailed fertility data via accessible web-based portals, so that meaningful correlations can be ascertained and verified (94–96). In addition to making proteomic data available, proteomicists and basic biologists should describe their protocols and workflow thoroughly, promoting reproducibility of results. Likewise, it is necessary for clinical scientists to include the parameters and cut-off for fertility assessments with detailed characterizations of patients and clinical samples to aid in cross-study comparisons and meta-analyses. Because in today's world no one researcher can wear all hats, opportunities abound for productive and meaningful collaborations for scientific discovery as well as the translation of these discoveries into clinical application.
The full complexity of the sperm proteome will be revealed using rapidly advancing proteomic technologies applied to different subcellular or biochemical fractions of sperm cells. Even with the application of directed strategies to enrich for typical DNA-associated factors, the studies discussed yielded unexpected proteins that function in male fertility. For example, proteomic analysis of acid-solubilized sperm chromatin revealed not only sperm nuclear basic structural proteins, but also a set of chaperone proteins that included HSPA2 (71). Likewise, of the 132 proteins identified in sperm chromatin proteomics in C. elegans, the majority of these proteins were actually in the categories of RNA binding, signaling, and unknown proteins (17). Such findings underscore a significant benefit of large-scale proteomic identification as a nonbiased approach to find a broad range of functionally significant candidate proteins for further functional analysis.
The dissection of sperm function and dysfunction will benefit greatly from proteomic analysis because sperm rely heavily on post-translational modifications for their development and function. It will therefore be necessary to define specific protein isoforms as well as combinations of post-translational modifications, for each may have unique regulatory functions at different stages in sperm development. Unique sets of sperm-specific “signatures” of post-translational modifications may regulate chromatin structure and gene expression from spermatogenesis through zygotic development (70). Sperm are therefore an ideal testing ground for new strategies that combine rapid protein identification with ways to easily distinguish relevant combinations of post-translational modifications on individual proteins (97–102). Such studies will have a significant impact on our understanding of the epigenetic information stored within the paternal chromatin contribution that may have long lasting effects on future generations.
The complexity of protein species in sperm is only one of the challenges facing scientists interested in converting proteomic knowledge into clinical applications. Infertility is a multifactorial syndrome, and any one case of infertility likely is caused by multiple genetic and environmental insults. Therefore approaches that define how individual factors act in specific fertility processes allow the dissection of infertility from the bottom up. We have highlighted examples in C. elegans and mouse that have effectively used proteomics to molecularly define crucial aspects of sperm chromatin (17, 68, 71); though studies in other organisms, including agricultural models, have made significant contributions as well (103–105).
Although lack of knowledge about the molecular function of a given protein does not preclude its use as a biomarker, such characterization enriches and informs the development of clinical tools and therapies. Characterization of specific sperm proteins are valuable from a practical standpoint to decide which candidates are the most compelling for further investment of time and resources. A temporal and mechanistic understanding of how a protein plays into a particular developmental or biochemical pathway would allow more refined characterization of sperm dysfunction. For example, inappropriate expression of stage-specific developmental markers in a sperm sample could prompt a diagnosis of aberrant sperm development and may influence decisions about that couple's treatment options. Moreover, molecular characterization of clinical targets can denote those that have the potential to lead to treatments that improve patient fertility outcomes.
How will proteins identified by proteomics be useful in the clinic? One important consideration is that a given clinical biomarker assay should have sufficient sensitivity to identify affected individuals correctly and yet show adequate specificity to avoid false detection of unaffected persons (106). Another consideration for a useful biomarker is that expression changes occur prevalently within a population of infertile patients (107, 108). Some candidate proteins may not meet all criteria. For example, certain biologically relevant markers may be highly sensitive and specific, detecting a defect in only a small subset of the population. Because the etiology of male infertility is complex, the most efficient assays may be those that combine such biomarkers into panels, improving the probability of detecting dysfunction for each patient using one clinical test (109, 110). The continued identification of sperm-specific proteins is essential for the development of a broad range of highly specific markers that can detect infertility arising from numerous possible events.
Finally, clinical tests that indicate sperm dysfunction help clinicians and patients make informed decisions (111–113). 50% of miscarriages have unexplained causes (114). Paternal genomic abnormalities, like sperm DNA damage, are correlated with fertilization success and may also be indicators of post-fertilization outcome, like spontaneous abortion after natural or in vitro fertilization (IVF) (115–117). Tests for the quality of sperm chromatin can be used to inform patients of the statistical likelihood of potential reproductive outcomes, like spontaneous abortion, before they make the decision to proceed with a particular course of ART. Developing reliable and well validated tests will arm both clinician and patient with the necessary information to proceed with the most appropriate treatment options.
Cures for infertility may no longer be just a far off dream. Given the modern revolution in systems-wide approaches like proteomics and genomics, our prospects for understanding this complex syndrome are greatly increased. In addition to proteomic studies of chromatin, proteomic analyses have been performed on seminal fluid and various sperm organelles, including the flagella to define motility and sperm membranes to understand sperm-egg interactions (118–120). Thus there is tremendous opportunity to make important findings and contributions to understand sperm biology, to define causes of infertility, and to bring hope to infertile couples with safe and effective treatment.
Acknowledgments
We thank Bruce Macher for critically reading the manuscript and Mark Sigman for helpful discussions.
Footnotes
Published, MCP Papers in Press, May 25, 2008, DOI 10.1074/mcp.R800005-MCP200
The abbreviations used are: ART, assisted reproductive technology; ICSI, intracytoplasmic sperm injection; SNBP, sperm nuclear basic protein; IVF, in vitro fertilization; 2D-PAGE, two-dimensional poly-acrylamide gel electrophoresis; LC-MS2, liquid chromatography-tandem mass spectrometry; GO, Gene Ontology; RNAi, RNA interference; MudPIT, Multidimensional Protein Identification Technology; TUNEL, terminal deoxynucleotidyl transferase-mediated nick end labeling.
This work was supported, in whole or in part, by National Institutes of Health Grant MBRS SCORE S06 GM52588. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Boe-Hansen, G. B., Fedder, J., Ersboll, A. K., and Christensen, P. ( 2006) The sperm chromatin structure assay as a diagnostic tool in the human fertility clinic. Hum. Reprod. 21, 1576–1582 [DOI] [PubMed] [Google Scholar]
- 2.Bungum, M., Humaidan, P., Axmon, A., Spano, M., Bungum, L., Erenpreiss, J., and Giwercman, A. ( 2007) Sperm DNA integrity assessment in prediction of assisted reproduction technology outcome. Hum. Reprod. 22, 174–179 [DOI] [PubMed] [Google Scholar]
- 3.Cebesoy, F. B., Aydos, K., and Unlu, C. ( 2006) Effect of sperm chromatin damage on fertilization ratio and embryo quality post-ICSI. Arch. Androl. 52, 397–402 [DOI] [PubMed] [Google Scholar]
- 4.Andrabi, S. M. ( 2007) Mammalian sperm chromatin structure and assessment of DNA fragmentation. J. Assist. Reprod. Genet. 24, 561–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belokopytova, I. A., Kostyleva, E. I., Tomilin, A. N., and Vorob'ev, V. I. ( 1993) Human male infertility may be due to a decrease of the protamine P2 content in sperm chromatin. Mol. Reprod. Dev. 34, 53–57 [DOI] [PubMed] [Google Scholar]
- 6.Angelopoulou, R., Plastira, K., and Msaouel, P. ( 2007) Spermatozoal sensitive biomarkers to defective protaminosis and fragmented DNA. Reprod. Biol. Endocrinol. 5, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turek, P. J. ( 2005) Practical approaches to the diagnosis and management of male infertility. Nat. Clin. Pract. Urol. 2, 226–238 [DOI] [PubMed] [Google Scholar]
- 8.National Survey of Family Growth ( 2002) cycle 6, Hyattsville, MD, U. S. Dept. of Health and Human Services. (2004) National Center for Health Statistics
- 9.( 2004) Assisted Reproductive Technology (ART) Report. U. S. Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Health Statistics, 2003, Hyattsville, MD
- 10.D'Occhio, M. J., Hengstberger, K. J., and Johnston, S. D. ( 2007) Biology of sperm chromatin structure and relationship to male fertility and embryonic survival. Anim. Reprod. Sci. 101, 1–17 [DOI] [PubMed] [Google Scholar]
- 11.Georgiou, I., Syrrou, M., Pardalidis, N., Karakitsios, K., Mantzavinos, T., Giotitsas, N., Loutradis, D., Dimitriadis, F., Saito, M., Miyagawa, I., Tzoumis, P., Sylakos, A., Kanakas, N., Moustakareas, T., Baltogiannis, D., Touloupides, S., Giannakis, D., Fatouros, M., and Sofikitis, N. ( 2006) Genetic and epigenetic risks of intracytoplasmic sperm injection method. Asian J. Androl. 8, 643–673 [DOI] [PubMed] [Google Scholar]
- 12.Caron, C., Govin, J., Rousseaux, S., and Khochbin, S. ( 2005) How to pack the genome for a safe trip. Prog. Mol. Subcell. Biol. 38, 65–89 [DOI] [PubMed] [Google Scholar]
- 13.Sassone-Corsi, P. ( 2002) Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science 296, 2176–2178 [DOI] [PubMed] [Google Scholar]
- 14.Churikov, D., Zalenskaya, I. A., and Zalensky, A. O. ( 2004) Male germline-specific histones in mouse and man. Cytogenet. Genome Res. 105, 203–214 [DOI] [PubMed] [Google Scholar]
- 15.Dadoune, J. P., Siffroi, J. P., and Alfonsi, M. F. ( 2004) Transcription in haploid male germ cells. Int. Rev. Cytol. 237, 1–56 [DOI] [PubMed] [Google Scholar]
- 16.Swanson, W. J., and Vacquier, V. D. ( 2002) The rapid evolution of reproductive proteins. Nat. Rev. Genet. 3, 137–144 [DOI] [PubMed] [Google Scholar]
- 17.Chu, D. S., Liu, H., Nix, P., Wu, T. F., Ralston, E. J., Yates, J. R., 3rd, and Meyer, B. J. ( 2006) Sperm chromatin proteomics identifies evolutionarily conserved fertility factors. Nature 443, 101–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hackstein, J. H., Hochstenbach, R., and Pearson, P. L. ( 2000) Towards an understanding of the genetics of human male infertility: lessons from flies. Trends Genet. 16, 565–572 [DOI] [PubMed] [Google Scholar]
- 19.Xu, E. Y., Lee, D. F., Klebes, A., Turek, P. J., Kornberg, T. B., and Reijo Pera, R. A. ( 2003) Human BOULE gene rescues meiotic defects in infertile flies. Hum. Mol. Genet. 12, 169–175 [DOI] [PubMed] [Google Scholar]
- 20.Aitken, R. J., and Baker, M. A. ( 2007) The role of proteomics in understanding sperm cell biology. Int. J. Androl. 30, 1–8 [DOI] [PubMed] [Google Scholar]
- 21.Lefievre, L., Bedu-Addo, K., Conner, S. J., Machado-Oliveira, G. S., Chen, Y., Kirkman-Brown, J. C., Afnan, M. A., Publicover, S. J., Ford, W. C., and Barratt, C. L. ( 2007) Counting sperm does not add up any more: time for a new equation? Reproduction 133, 675–684 [DOI] [PubMed] [Google Scholar]
- 22.Wu, T. F., and Chu, D. S. ( 2008) Epigenetic processes implemented during spermatogenesis distinguish the paternal pronucleus in the embryo. Reprod. Biomed. Online 16, 13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suarez, S. S., Marquez, B., Harris, T. P., and Schimenti, J. C. ( 2007) Different regulatory systems operate in the midpiece and principal piece of the mammalian sperm flagellum. Soc. Reprod. Fertil. Suppl. 65, 331–334 [PubMed] [Google Scholar]
- 24.Suarez, S. S., and Pacey, A. A. ( 2006) Sperm transport in the female reproductive tract. Hum. Reprod. Update 12, 23–37 [DOI] [PubMed] [Google Scholar]
- 25.Brewis, I. A., Moore, H. D., Fraser, L. R., Holt, W. V., Baldi, E., Luconi, M., Gadella, B. M., Ford, W. C., and Harrison, R. A. ( 2005) Molecular mechanisms during sperm capacitation. Hum. Fertil. (Camb.) 8, 253–261 [DOI] [PubMed] [Google Scholar]
- 26.Salicioni, A. M., Platt, M. D., Wertheimer, E. V., Arcelay, E., Allaire, A., Sosnik, J., Visconti, P. E., Brewis, I. A., Moore, H. D., Fraser, L. R., Holt, W. V., Baldi, E., Luconi, M., Gadella, B. M., Ford, W. C., and Harrison, R. A. ( 2007) Signaling pathways involved in sperm capacitation: molecular mechanisms during sperm capacitation. Soc. Reprod. Fertil. Suppl. 65, 245–259 [PubMed] [Google Scholar]
- 27.Tomes, C. N., Salicioni, A. M., Platt, M. D., Wertheimer, E. V., Arcelay, E., Allaire, A., Sosnik, J., Visconti, P. E., Brewis, I. A., Moore, H. D., Fraser, L. R., Holt, W. V., Baldi, E., Luconi, M., Gadella, B. M., Ford, W. C., and Harrison, R. A. ( 2007) Molecular mechanisms of membrane fusion during acrosomal exocytosis signaling pathways involved in sperm capacitation: molecular mechanisms during sperm capacitation. Soc. Reprod. Fertil. Suppl. 65, 275–29117644969 [Google Scholar]
- 28.Chakrabarti, R., Cheng, L., Puri, P., Soler, D., and Vijayaraghavan, S. ( 2007) Protein phosphatase PP1 gamma 2 in sperm morphogenesis and epididymal initiation of sperm motility. Asian J. Androl. 9, 445–452 [DOI] [PubMed] [Google Scholar]
- 29.Chakrabarti, R., Kline, D., Lu, J., Orth, J., Pilder, S., and Vijayaraghavan, S. ( 2007) Analysis of Ppp1cc-null mice suggests a role for PP1 gamma 2 in sperm morphogenesis. Biol. Reprod. 76, 992–1001 [DOI] [PubMed] [Google Scholar]
- 30.Varmuza, S., Jurisicova, A., Okano, K., Hudson, J., Boekelheide, K., and Shipp, E. B. ( 1999) Spermiogenesis is impaired in mice bearing a targeted mutation in the protein phosphatase 1c gamma gene. Dev. Biol. 205, 98–110 [DOI] [PubMed] [Google Scholar]
- 31.Hunt, P., and Hassold, T. ( 2002) Sex matters in meiosis. Science 296, 2181–2183 [DOI] [PubMed] [Google Scholar]
- 32.Allen, M. J., Lee, C., Lee, J. D., Pogany, G. C., Balooch, M., Siekhaus, W. J., and Balhorn, R. ( 1993) Atomic force microscopy of mammalian sperm chromatin. Chromosoma 102, 623–630 [DOI] [PubMed] [Google Scholar]
- 33.Lewis, J. D., Abbott, D. W., and Ausio, J. ( 2003) A haploid affair: core histone transitions during spermatogenesis. Biochem. Cell Biol. 81, 131–140 [DOI] [PubMed] [Google Scholar]
- 34.Lewis, J. D., Song, Y., de Jong, M. E., Bagha, S. M., and Ausio, J. ( 2003) A walk though vertebrate and invertebrate protamines. Chromosoma 111, 473–482 [DOI] [PubMed] [Google Scholar]
- 35.Braun, R. E. ( 2001) Packaging paternal chromosomes with protamine. Nat. Genet. 28, 10–12 [DOI] [PubMed] [Google Scholar]
- 36.Cho, C., Willis, W. D., Goulding, E. H., Jung-Ha, H., Choi, Y. C., Hecht, N. B., and Eddy, E. M. ( 2001) Haploinsufficiency of protamine-1 or -2 causes infertility in mice. Nat. Genet. 28, 82–86 [DOI] [PubMed] [Google Scholar]
- 37.Yu, Y. E., Zhang, Y., Unni, E., Shirley, C. R., Deng, J. M., Russell, L. D., Weil, M. M., Behringer, R. R., and Meistrich, M. L. ( 2000) Abnormal spermatogenesis and reduced fertility in transition nuclear protein 1-deficient mice. Proc. Natl. Acad. Sci. U. S. A. 97, 4683–4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao, M., Shirley, C. R., Yu, Y. E., Mohapatra, B., Zhang, Y., Unni, E., Deng, J. M., Arango, N. A., Terry, N. H., Weil, M. M., Russell, L. D., Behringer, R. R., and Meistrich, M. L. ( 2001) Targeted disruption of the transition protein 2 gene affects sperm chromatin structure and reduces fertility in mice. Mol. Cell. Biol. 21, 7243–7255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ooi, S. L., and Henikoff, S. ( 2007) Germline histone dynamics and epigenetics. Curr. Opin. Cell Biol. 19, 1–9 [DOI] [PubMed] [Google Scholar]
- 40.Naaby-Hansen, S., Waterfield, M. D., and Cramer, R. ( 2001) Proteomics–post-genomic cartography to understand gene function. Trends Pharmacol. Sci. 22, 376–384 [DOI] [PubMed] [Google Scholar]
- 41.Martinez-Heredia, J., Estanyol, J. M., Ballesca, J. L., and Oliva, R. ( 2006) Proteomic identification of human sperm proteins. Proteomics 6, 4356–4369 [DOI] [PubMed] [Google Scholar]
- 42.Naaby-Hansen, S., Flickinger, C. J., and Herr, J. C. ( 1997) Two-dimensional gel electrophoretic analysis of vectorially labeled surface proteins of human spermatozoa. Biol. Reprod. 56, 771–787 [DOI] [PubMed] [Google Scholar]
- 43.Li, L. W., Fan, L. Q., Zhu, W. B., Nien, H. C., Sun, B. L., Luo, K. L., Liao, T. T., Tang, L., and Lu, G. X. ( 2007) Establishment of a high-resolution 2-D reference map of human spermatozoal proteins from 12 fertile sperm-bank donors. Asian J. Androl. 9, 321–329 [DOI] [PubMed] [Google Scholar]
- 44.Johnston, D. S., Wooters, J., Kopf, G. S., Qiu, Y., and Roberts, K. P. ( 2005) Analysis of the human sperm proteome. Ann. N. Y. Acad. Sci. 1061, 190–202 [DOI] [PubMed] [Google Scholar]
- 45.Baker, M. A., Reeves, G., Hetherington, L., Muller, J., Baur, I., and Aitken, R. J. ( 2007) Identification of gene products present in Triton X-100 soluble and insoluble fractions of human spermatozoa lysates using LC-MS/MS analysis. Proteomics Clin. Appl. 1, 524–532 [DOI] [PubMed] [Google Scholar]
- 46.Ahn, N. G., Shabb, J. B., Old, W. M., and Resing, K. A. ( 2007) Achieving in-depth proteomics profiling by mass spectrometry. ACS Chem. Biol. 2, 39–52 [DOI] [PubMed] [Google Scholar]
- 47.de Mateo, S., Martinez-Heredia, J., Estanyol, J. M., Domiguez-Fandos, D., Vidal-Taboada, J. M., Ballesca, J. L., and Oliva, R. ( 2007) Marked correlations in protein expression identified by proteomic analysis of human spermatozoa. Proteomics 7, 4264–4277 [DOI] [PubMed] [Google Scholar]
- 48.Lan, N., Montelione, G. T., and Gerstein, M. ( 2003) Ontologies for proteomics: towards a systematic definition of structure and function that scales to the genome level. Curr. Opin. Chem. Biol. 7, 44–54 [DOI] [PubMed] [Google Scholar]
- 49.Dorus, S., Busby, S. A., Gerike, U., Shabanowitz, J., Hunt, D. F., and Karr, T. L. ( 2006) Genomic and functional evolution of the Drosophila melanogaster sperm proteome. Nat. Genet. 38, 1440–1445 [DOI] [PubMed] [Google Scholar]
- 50.Parisi, M., Nuttall, R., Edwards, P., Minor, J., Naiman, D., Lu, J., Doctolero, M., Vainer, M., Chan, C., Malley, J., Eastman, S., and Oliver, B. ( 2004) A survey of ovary-, testis-, and soma-biased gene expression in Drosophila melanogaster adults. Genome Biol. 5, R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O'Bryan, M. K., and de Kretser, D. ( 2006) Mouse models for genes involved in impaired spermatogenesis. Int. J. Androl. 29, 76–89; Discussion 105–108 [DOI] [PubMed] [Google Scholar]
- 52.L'Hernault, S. W. ( 2006) Spermatogenesis. WormBook, 1–14 [DOI] [PMC free article] [PubMed]
- 53.Williamson, A., and Lehmann, R. ( 1996) Germ cell development in Drosophila. Annu. Rev. Cell Dev. Biol. 12, 365–391 [DOI] [PubMed] [Google Scholar]
- 54.Cooke, H. J., and Saunders, P. T. ( 2002) Mouse models of male infertility. Nat. Rev. Genet. 3, 790–801 [DOI] [PubMed] [Google Scholar]
- 55.Matzuk, M. M., and Lamb, D. J. ( 2002) Genetic dissection of mammalian fertility pathways. Nat. Cell Biol. 4, (suppl.) S41–S49 [DOI] [PubMed] [Google Scholar]
- 56.Bult, C. J., Eppig, J. T., Kadin, J. A., Richardson, J. E., and Blake, J. A. ( 2008) The Mouse Genome Database (MGD): mouse biology and model systems. Nucleic Acids Res. 36, D724–D728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rogers, A., Antoshechkin, I., Bieri, T., Blasiar, D., Bastiani, C., Canaran, P., Chan, J., Chen, W. J., Davis, P., Fernandes, J., Fiedler, T. J., Han, M., Harris, T. W., Kishore, R., Lee, R., McKay, S., Muller, H. M., Nakamura, C., Ozersky, P., Petcherski, A., Schindelman, G., Schwarz, E. M., Spooner, W., Tuli, M. A., Van Auken, K., Wang, D., Wang, X., Williams, G., Yook, K., Durbin, R., Stein, L. D., Spieth, J., and Sternberg, P. W. ( 2008) WormBase 2007. Nucleic Acids Res. 36, D612–D617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilson, R. J., Goodman, J. L., and Strelets, V. B. ( 2008) FlyBase: integration and improvements to query tools. Nucleic Acids Res. 36, D588–D593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Andersen, J. S., and Mann, M. ( 2006) Organellar proteomics: turning inventories into insights. EMBO Rep. 7, 874–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu, H., Sadygov, R. G., and Yates, J. R., 3rd ( 2004) A model for random sampling and estimation of relative protein abundance. Anal. Chem. 76, 4193–4201 [DOI] [PubMed] [Google Scholar]
- 61.Graumann, J., Hubner, N. C., Kim, J. B., Ko, K., Moser, M., Kumar, C., Cox, J., Scholer, H., and Mann, M. ( 2008) Stable isotope labeling by amino acids in cell culture (SILAC) and proteome quantitation of mouse embryonic stem cells to a depth of 5,111 proteins. Mol. Cell. Proteomics 7, 672–683 [DOI] [PubMed] [Google Scholar]
- 62.Oh, P., Li, Y., Yu, J., Durr, E., Krasinska, K. M., Carver, L. A., Testa, J. E., and Schnitzer, J. E. ( 2004) Subtractive proteomic mapping of the endothelial surface in lung and solid tumours for tissue-specific therapy. Nature 429, 629–635 [DOI] [PubMed] [Google Scholar]
- 63.Schirmer, E. C., Florens, L., Guan, T., Yates, J. R., 3rd, and Gerace, L. ( 2003) Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science 301, 1380–1382 [DOI] [PubMed] [Google Scholar]
- 64.Liu, H., Lin, D., and Yates, J. R., 3rd ( 2002) Multidimensional separations for protein/peptide analysis in the post-genomic era. BioTechniques 32, 898–911 [DOI] [PubMed] [Google Scholar]
- 65.Tabb, D. L., McDonald, W. H., and Yates, J. R., 3rd ( 2002) DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1, 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gatewood, J. M., Cook, G. R., Balhorn, R., Bradbury, E. M., and Schmid, C. W. ( 1987) Sequence-specific packaging of DNA in human sperm chromatin. Science 236, 962–964 [DOI] [PubMed] [Google Scholar]
- 67.Wykes, S. M., and Krawetz, S. A. ( 2003) The structural organization of sperm chromatin. J. Biol. Chem. 278, 29471–29477 [DOI] [PubMed] [Google Scholar]
- 68.Govin, J., Escoffier, E., Rousseaux, S., Kuhn, L., Ferro, M., Thevenon, J., Catena, R., Davidson, I., Garin, J., Khochbin, S., and Caron, C. ( 2007) Pericentric heterochromatin reprogramming by new histone variants during mouse spermiogenesis. J. Cell Biol. 176, 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grewal, S. I., and Jia, S. ( 2007) Heterochromatin revisited. Nat. Rev. Genet 8, 35–46 [DOI] [PubMed] [Google Scholar]
- 70.Govin, J., Caron, C., Lestrat, C., Rousseaux, S., and Khochbin, S. ( 2004) The role of histones in chromatin remodelling during mammalian spermiogenesis. Eur. J. Biochem. 271, 3459–3469 [DOI] [PubMed] [Google Scholar]
- 71.Govin, J., Caron, C., Escoffier, E., Ferro, M., Kuhn, L., Rousseaux, S., Eddy, E. M., Garin, J., and Khochbin, S. ( 2006) Post-meiotic shifts in HSPA2/HSP70.2 chaperone activity during mouse spermatogenesis. J. Biol. Chem. 281, 37888–37892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huszar, G., Vigue, L., and Morshedi, M. ( 1992) Sperm creatine phosphokinase M-isoform ratios and fertilizing potential of men: a blinded study of 84 couples treated with in vitro fertilization. Fertil. Steril. 57, 882–888 [PubMed] [Google Scholar]
- 73.Huszar, G., Vigue, L., and Oehninger, S. ( 1994) Creatine kinase immunocytochemistry of human sperm-hemizona complexes: selective binding of sperm with mature creatine kinase-staining pattern. Fertil. Steril. 61, 136–142 [DOI] [PubMed] [Google Scholar]
- 74.Dix, D. J., Allen, J. W., Collins, B. W., Poorman-Allen, P., Mori, C., Blizard, D. R., Brown, P. R., Goulding, E. H., Strong, B. D., and Eddy, E. M. ( 1997) HSP70–2 is required for desynapsis of synaptonemal complexes during meiotic prophase in juvenile and adult mouse spermatocytes. Development 124, 4595–4603 [DOI] [PubMed] [Google Scholar]
- 75.Eddy, E. M. ( 1999) Role of heat shock protein HSP70–2 in spermatogenesis. Rev. Reprod. 4, 23–30 [DOI] [PubMed] [Google Scholar]
- 76.Allen, J. W., Dix, D. J., Collins, B. W., Merrick, B. A., He, C., Selkirk, J. K., Poorman-Allen, P., Dresser, M. E., and Eddy, E. M. ( 1996) HSP70–2 is part of the synaptonemal complex in mouse and hamster spermatocytes. Chromosoma 104, 414–421 [DOI] [PubMed] [Google Scholar]
- 77.Dix, D. J., Allen, J. W., Collins, B. W., Mori, C., Nakamura, N., Poorman-Allen, P., Goulding, E. H., and Eddy, E. M. ( 1996) Targeted gene disruption of Hsp70–2 results in failed meiosis, germ cell apoptosis, and male infertility. Proc. Natl. Acad. Sci. U. S. A. 93, 3264–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ergur, A. R., Dokras, A., Giraldo, J. L., Habana, A., Kovanci, E., and Huszar, G. ( 2002) Sperm maturity and treatment choice of in vitro fertilization (IVF) or intracytoplasmic sperm injection: diminished sperm HspA2 chaperone levels predict IVF failure. Fertil. Steril. 77, 910–918 [DOI] [PubMed] [Google Scholar]
- 79.Huszar, G., Stone, K., Dix, D., and Vigue, L. ( 2000) Putative creatine kinase M-isoform in human sperm is identified as the 70-kilodalton heat shock protein HspA2. Biol. Reprod. 63, 925–932 [DOI] [PubMed] [Google Scholar]
- 80.Martinez-Heredia, J., de Mateo, S., Vidal-Taboada, J. M., Ballesca, J. L., and Oliva, R. ( 2008) Identification of proteomic differences in asthenozoospermic sperm samples. Hum. Reprod. 23, 783–791 [DOI] [PubMed] [Google Scholar]
- 81.Huszar, G., Ozkavukcu, S., Jakab, A., Celik-Ozenci, C., Sati, G. L., and Cayli, S. ( 2006) Hyaluronic acid binding ability of human sperm reflects cellular maturity and fertilizing potential: selection of sperm for intracytoplasmic sperm injection. Curr. Opin. Obstet. Gynecol. 18, 260–267 [DOI] [PubMed] [Google Scholar]
- 82.Cayli, S., Jakab, A., Ovari, L., Delpiano, E., Celik-Ozenci, C., Sakkas, D., Ward, D., and Huszar, G. ( 2003) Biochemical markers of sperm function: male fertility and sperm selection for ICSI. Reprod. Biomed. Online 7, 462–468 [DOI] [PubMed] [Google Scholar]
- 83.Huszar, G., Jakab, A., Sakkas, D., Ozenci, C. C., Cayli, S., Delpiano, E., and Ozkavukcu, S. ( 2007) Fertility testing and ICSI sperm selection by hyaluronic acid binding: clinical and genetic aspects. Reprod. Biomed. Online 14, 650–663 [DOI] [PubMed] [Google Scholar]
- 84.Huszar, G., Ozenci, C. C., Cayli, S., Zavaczki, Z., Hansch, E., and Vigue, L. ( 2003) Hyaluronic acid binding by human sperm indicates cellular maturity, viability, and unreacted acrosomal status. Fertil. Steril. 79, Suppl. 3, 1616–1624 [DOI] [PubMed] [Google Scholar]
- 85.Zhao, C., Huo, R., Wang, F. Q., Lin, M., Zhou, Z. M., and Sha, J. H. ( 2007) Identification of several proteins involved in regulation of sperm motility by proteomic analysis. Fertil. Steril. 87, 436–438 [DOI] [PubMed] [Google Scholar]
- 86.Pixton, K. L., Deeks, E. D., Flesch, F. M., Moseley, F. L., Bjorndahl, L., Ashton, P. R., Barratt, C. L., and Brewis, I. A. ( 2004) Sperm proteome mapping of a patient who experienced failed fertilization at IVF reveals altered expression of at least 20 proteins compared with fertile donors: case report. Hum. Reprod. 19, 1438–1447 [DOI] [PubMed] [Google Scholar]
- 87.Weber, R. F., Dohle, G. R., and Romijn, J. C. ( 2005) Clinical laboratory evaluation of male subfertility. Adv. Clin. Chem. 40, 317–364 [DOI] [PubMed] [Google Scholar]
- 88.Oliva, R. ( 2006) Protamines and male infertility. Hum. Reprod. Update 12, 417–435 [DOI] [PubMed] [Google Scholar]
- 89.Torregrosa, N., Dominguez-Fandos, D., Camejo, M. I., Shirley, C. R., Meistrich, M. L., Ballesca, J. L., and Oliva, R. ( 2006) Protamine 2 precursors, protamine 1/protamine 2 ratio, DNA integrity and other sperm parameters in infertile patients. Hum. Reprod. 21, 2084–2089 [DOI] [PubMed] [Google Scholar]
- 90.Aoki, V. W., Liu, L., Jones, K. P., Hatasaka, H. H., Gibson, M., Peterson, C. M., and Carrell, D. T. ( 2006) Sperm protamine 1/protamine 2 ratios are related to in vitro fertilization pregnancy rates and predictive of fertilization ability. Fertil. Steril. 86, 1408–1415 [DOI] [PubMed] [Google Scholar]
- 91.Balhorn, R., Reed, S., and Tanphaichitr, N. ( 1988) Aberrant protamine 1/protamine 2 ratios in sperm of infertile human males. Experientia 44, 52–55 [DOI] [PubMed] [Google Scholar]
- 92.de Yebra, L., Ballesca, J. L., Vanrell, J. A., Bassas, L., and Oliva, R. ( 1993) Complete selective absence of protamine P2 in humans. J. Biol. Chem. 268, 10553–10557 [PubMed] [Google Scholar]
- 93.Mengual, L., Ballesca, J. L., Ascaso, C., and Oliva, R. ( 2003) Marked differences in protamine content and P1/P2 ratios in sperm cells from percoll fractions between patients and controls. J. Androl. 24, 438–447 [DOI] [PubMed] [Google Scholar]
- 94.Shifman, M. A., Li, Y., Colangelo, C. M., Stone, K. L., Wu, T. L., Cheung, K. H., Miller, P. L., and Williams, K. R. ( 2007) YPED: a web-accessible database system for protein expression analysis. J. Proteome Res. 6, 4019–4024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Taylor, C. F., Paton, N. W., Lilley, K. S., Binz, P. A., Julian, R. K., Jr., Jones, A. R., Zhu, W., Apweiler, R., Aebersold, R., Deutsch, E. W., Dunn, M. J., Heck, A. J., Leitner, A., Macht, M., Mann, M., Martens, L., Neubert, T. A., Patterson, S. D., Ping, P., Seymour, S. L., Souda, P., Tsugita, A., Vandekerckhove, J., Vondriska, T. M., Whitelegge, J. P., Wilkins, M. R., Xenarios, I., Yates, J. R., 3rd, and Hermjakob, H. ( 2007) The minimum information about a proteomics experiment (MIAPE). Nat. Biotechnol. 25, 887–893 [DOI] [PubMed] [Google Scholar]
- 96.Vizcaino, J. A., Martens, L., Hermjakob, H., Julian, R. K., and Paton, N. W. ( 2007) The PSI formal document process and its implementation on the PSI website. Proteomics 7, 2355–2357 [DOI] [PubMed] [Google Scholar]
- 97.Siuti, N., and Kelleher, N. L. ( 2007) Decoding protein modifications using top-down mass spectrometry. Nat. Methods 4, 817–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Witze, E. S., Old, W. M., Resing, K. A., and Ahn, N. G. ( 2007) Mapping protein post-translational modifications with mass spectrometry. Nat. Methods 4, 798–806 [DOI] [PubMed] [Google Scholar]
- 99.Collins, M. O., Yu, L., and Choudhary, J. S. ( 2007) Analysis of protein phosphorylation on a proteome-scale. Proteomics 7, 2751–2768 [DOI] [PubMed] [Google Scholar]
- 100.Ding, S. J., Qian, W. J., and Smith, R. D. ( 2007) Quantitative proteomic approaches for studying phosphotyrosine signaling. Expert Rev. Proteomics 4, 13–23 [DOI] [PubMed] [Google Scholar]
- 101.Goshe, M. B. ( 2006) Characterizing phosphoproteins and phosphoproteomes using mass spectrometry. Brief Funct. Genomic Proteomic 4, 363–376 [DOI] [PubMed] [Google Scholar]
- 102.Meinnel, T., and Giglione, C. ( 2008) Tools for analyzing and predicting N-terminal protein modifications. Proteomics 8, 626–649 [DOI] [PubMed] [Google Scholar]
- 103.Fraser, L., Wysocki, P., Ciereszko, A., Plucienniczak, G., Kotlowska, M., Kordan, W., Wojtczak, M., Dietrich, G., and Strzezek, J. ( 2006) Application of biochemical markers for identification of biological properties of animal semen. Reprod. Biol. 6, Suppl. 1, 5–20 [PubMed] [Google Scholar]
- 104.Fouchecourt, S., Metayer, S., Locatelli, A., Dacheux, F., and Dacheux, J. L. ( 2000) Stallion epididymal fluid proteome: qualitative and quantitative characterization; secretion and dynamic changes of major proteins. Biol. Reprod. 62, 1790–1803 [DOI] [PubMed] [Google Scholar]
- 105.Peddinti, D., Nanduri, B., Kaya, A., Feugang, J. M., Burgess, S. C., and Memili, E. ( 2008) Comprehensive proteomic analysis of bovine spermatozoa of varying fertility rates and identification of biomarkers associated with fertility. BMC Syst. Biol. 2, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kohn, E. C., Azad, N., Annunziata, C., Dhamoon, A. S., and Whiteley, G. ( 2007) Proteomics as a tool for biomarker discovery. Dis. Markers 23, 411–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Frank, R., and Hargreaves, R. ( 2003) Clinical biomarkers in drug discovery and development. Nat. Rev. Drug Discov. 2, 566–580 [DOI] [PubMed] [Google Scholar]
- 108.Colburn, W. A., and Lee, J. W. ( 2003) Biomarkers, validation and pharmacokinetic-pharmacodynamic modelling. Clin. Pharmacokinet. 42, 997–1022 [DOI] [PubMed] [Google Scholar]
- 109.Xiao, Z., Prieto, D., Conrads, T. P., Veenstra, T. D., and Issaq, H. J. ( 2005) Proteomic patterns: their potential for disease diagnosis. Mol. Cell Endocrinol. 230, 95–106 [DOI] [PubMed] [Google Scholar]
- 110.Lesko, L. J., and Atkinson, A. J., Jr. ( 2001) Use of biomarkers and surrogate endpoints in drug development and regulatory decision making: criteria, validation, strategies. Annu. Rev. Pharmacol. Toxicol. 41, 347–366 [DOI] [PubMed] [Google Scholar]
- 111.Peters, K., Jackson, D., and Rudge, T. ( 2007) Failures of reproduction: problematising ‘success' in assisted reproductive technology. Nurs. Inq. 14, 125–131 [DOI] [PubMed] [Google Scholar]
- 112.Aittomaki, K., Wennerholm, U. B., Bergh, C., Selbing, A., Hazekamp, J., and Nygren, K. G. ( 2004) Safety issues in assisted reproduction technology: should ICSI patients have genetic testing before treatment? A practical proposition to help patient information. Hum. Reprod. 19, 472–476 [DOI] [PubMed] [Google Scholar]
- 113.Raeburn, S. ( 1998) Genetic counselling issues and male infertility. Hum. Fertil. (Camb.) 1, 44–47 [DOI] [PubMed] [Google Scholar]
- 114.Li, T. C. ( 1998) Recurrent miscarriage: principles of management. Hum. Reprod. 13, 478–482 [DOI] [PubMed] [Google Scholar]
- 115.Lin, M. H., Kuo-Kuang Lee, R., Li, S. H., Lu, C. H., Sun, F. J., and Hwu, Y. M. ( 2007) Sperm chromatin structure assay parameters are not related to fertilization rates, embryo quality, and pregnancy rates in in vitro fertilization and intracytoplasmic sperm injection, but might be related to spontaneous abortion rates. Fertil. Steril. Epub ahead of print [DOI] [PubMed]
- 116.Virro, M. R., Larson-Cook, K. L., and Evenson, D. P. ( 2004) Sperm chromatin structure assay (SCSA) parameters are related to fertilization, blastocyst development, and ongoing pregnancy in in vitro fertilization and intracytoplasmic sperm injection cycles. Fertil. Steril. 81, 1289–1295 [DOI] [PubMed] [Google Scholar]
- 117.Zini, A., Meriano, J., Kader, K., Jarvi, K., Laskin, C. A., and Cadesky, K. ( 2005) Potential adverse effect of sperm DNA damage on embryo quality after ICSI. Hum. Reprod. 20, 3476–3480 [DOI] [PubMed] [Google Scholar]
- 118.Pilch, B., and Mann, M. ( 2006) Large-scale and high-confidence proteomic analysis of human seminal plasma. Genome Biol. 7, R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cao, W., Gerton, G. L., and Moss, S. B. ( 2006) Proteomic profiling of accessory structures from the mouse sperm flagellum. Mol. Cell. Proteomics 5, 801–810 [DOI] [PubMed] [Google Scholar]
- 120.Stein, K. K., Go, J. C., Lane, W. S., Primakoff, P., and Myles, D. G. ( 2006) Proteomic analysis of sperm regions that mediate sperm-egg interactions. Proteomics 6, 3533–3543 [DOI] [PubMed] [Google Scholar]