Summary
Aminoacyl-tRNA synthetases catalyze the attachment of specific amino acids to their cognate tRNAs. Specific aminoacylation is dictated by a set of recognition elements that mark tRNA molecules as substrates for particular synthetases. Escherichia coli prolyl-tRNA synthetase (ProRS) has previously been shown to recognize specific bases of tRNAPro in both the anticodon domain, which mediate initial complex formation, and in the acceptor stem, which is proximal to the site of catalysis. In this work, we unambiguously define the molecular interaction between E. coli ProRS and the acceptor stem of cognate tRNAPro. Oxidative cross-linking studies using 2′-deoxy-8-oxo-7,8-dihydroguanosine-containing proline tRNAs identify a direct interaction between a critical arginine residue (R144) in the active site of E. coli ProRS and the G72 residue in the acceptor stem of tRNAPro. Assays conducted with motif 2 loop variants and tRNA mutants wherein specific atomic groups of G72 were deleted, are consistent with a functionally important hydrogen bonding network between R144 and the major groove of G72. These results taken together with previous studies suggest that breaking this key contact uncouples the allosteric interaction between the anticodon domain and the aminoacylation active site, providing new insights into the communication network that governs the synthetase-tRNA interaction.
Keywords: Aminoacyl-tRNA synthetases, Prolyl-tRNA synthetase, Oxidative cross-linking, 8-Oxo-guanosine, RNA-protein interactions, Coupling network
1. Introduction
Aminoacyl-tRNA synthetases catalyze the attachment of specific amino acids to their cognate tRNAs. This reaction is critical for ensuring high fidelity translation of the genetic code. Specific tRNA binding and aminoacylation are dictated by a set of recognition elements primarily found in the anticodon and acceptor stem of the tRNA [1]. Despite the availability of a large number of X-ray crystal structures of synthetase-tRNA complexes, in most systems, the path of communication between the anticodon recognition event and the site of catalysis, which is located ~70 Å away, is unknown and remains a major open question in the field. Both short- and long-range thermal motions and an allosteric network of functional protein-RNA interactions are likely to play an important role in this process [2–6].
Escherichia coli prolyl-tRNA synthetase (ProRS) is a class II synthetase previously shown to recognize elements in both the anticodon and acceptor stem of tRNAPro [7, 8]. These data are in good agreement with the X-ray crystal structure of Thermus thermophilus ProRS complexed to the anticodon of its cognate tRNA [9]. Although a co-crystal structure of a ProRS with the tRNA acceptor stem bound has not been reported to date, biochemical data have shown that E. coli ProRS recognizes major groove elements of acceptor stem nucleotides A73 and G72 of E. coli tRNAPro [10]. The latter position is unique to tRNAPro isoacceptors in E. coli [7, 11]. Furthermore, residue R144 of E. coli ProRS, which is located in the so-called “motif 2 loop” sequence, 143VRPRF147, is critical for efficient aminoacylation. An R144C substitution completely abolishes tRNA charging, but has no effect on amino acid activation [10]. Cross-linking experiments confirmed the close proximity between the motif 2 loop and the tRNA acceptor stem [10]. Based on these data, a specific interaction between the motif 2 loop and the acceptor stem was proposed [10]. However, due to the length and location of the tether used in the previous cross-linking studies, the exact nature of the interaction could not be elucidated.
A novel method of oxidatively inducing DNA-protein cross-links has been reported using 2′-deoxy-8-oxo-7,8-dihydroguanosine (OG)-substituted DNA [12, 13]. In the presence of Na2IrCl6, OG is readily oxidized to create an electron-deficient center highly susceptible to attack by a nearby nucleophilic protein side chain [12, 14]. Since cross-link formation requires that two functional groups be within hydrogen bonding distance, interactions can be mapped with high precision using this approach. Furthermore, studies are consistent with a mechanism of cross-linking involving nucleophilic attack of an amino acid side chain (e.g., lysine or arginine) at C5 in the major groove of oxidized OG. Although to our knowledge this method has never been applied to the study of RNA-protein interactions, this appeared to be a promising approach in our system for fine structure mapping of the ProRS motif 2 loop-tRNAPro acceptor stem interaction.
In this work, the molecular interactions between E. coli ProRS and the acceptor stem of cognate tRNAPro are elucidated using aminoacylation assays and cross-linking with OG-containing tRNAs. The results presented here strongly support a direct hydrogen bonding interaction between a critical arginine residue in the active site of E. coli ProRS and major groove functional groups of G72 in the acceptor stem of tRNAPro. We hypothesize that the R144-G72 interaction constitutes a key component of the network that communicates the anticodon recognition event to the site of catalysis. Thus, more precisely defining this hydrogen-bonding network contributes to our understanding of allosteric coupling junctions influencing long-distance communication in this system.
2. Materials and methods
2.1. Enzyme Purification and Site-Directed Mutagenesis of ProRS
Purification of His-tagged wild-type E. coli ProRS was accomplished as described previously [15]. Site-directed mutagenesis of the motif 2 loop residues was accomplished by overlap extension PCR [16] using DNA primers encoding the desired changes. There is only one cysteine in wild-type E. coli ProRS and this change was previously shown to result in only a minor reduction in aminoacylation efficiency [10]. Due to their availability, three motif 2 mutant proteins (V143C, R144C, and R146C) additionally containing a C443G mutation were also studied in this work. The entire gene was sequenced to verify the specified mutation and to ensure that the mutagenesis procedure did not introduce undesired mutations. E. coli strain SG13009 [pREP4] (Qiagen) was transformed with the mutant plasmids for overexpression and purification, which were performed as described for the wild-type enzyme. ProRS concentrations were based on active-site titrations determined by the adenylate burst assay [17].
2.2. RNA Preparation
Semi-synthetic E. coli ΔC1-tRNAPro was prepared from two fragments as previously described [18]. The omission of C1 facilitates in vitro transcription and results in a tRNA substrate that is ~3-fold more active than a C1-containing tRNAPro transcript. 5′-3/4-length (nucleotides 1–57) fragment of tRNAPro was in vitro transcribed using a DNA template linearized by BstBI digestion of the plasmid used to obtain full-length E. coli ΔC1-tRNAPro as described [8]. The 3′-16-mer fragment was chemically synthesized using the phosphoramidite method on an Expedite™ 8909 Nucleic Acid Synthesis System (PerSeptive Biosystems). The modified deoxynucleotides 8-oxo-7,8-dihydroguanosine (OG), 2-aminopurine (2AP), 7-deaza-A (7A), and 7-deaza-G (7G) (all from Glen Research), were incorporated into position 72 or 73 (tRNA numbering) of the synthetic 3′-16-mer during automated chemical synthesis. Synthetic oligonucleotides were purified on 16% denaturing polyacrylamide gels followed by elution and work-up as described [18]. [32P]-Labeling of the 5′-end of the 16-mer oligonucleotides was carried out following the manufacturer’s protocol using [γ-32P]-ATP (New England Nuclear) and polynucleotide kinase (New England Biolabs). Semi-synthetic tRNA molecules were generated by annealing 1:1 ratios of 5′-3/4-length tRNAs to 3′-16-mer RNA fragments at 60 °C for 3 min, followed by addition of MgCl2 to 10 mM final concentration, cooling at room temperature for 5 min and placement on ice.
2.3. Aminoacylation Assays
Aminoacylation assays were carried out essentially as described previously [15, 19]. Kinetic assays using wild-type, R144K and R144L E. coli ProRS were performed with 50–200 nM enzyme and 2–20 μM E. coli ΔC1-tRNAPro. The kinetic constants were derived from Lineweaver-Burk plots. In the case of semi-synthetic tRNAs containing acceptor stem mutations at position 72 or 73, relative kcat/KM values were determined either from Lineweaver-Burk plots or from the initial rates of aminoacylation, which are proportional to RNA concentration under the conditions used for these experiments. Thus, Vo/[S] is an accurate reflection of kcat/KM.
2.4. Cross-linking Reactions
RNA-protein cross-linking reactions were carried out using a modification of the procedure described by Burrows and co-workers [12]. Wild-type or mutant ProRS (1–5 μM) and wild-type or OG72-containing semi-synthetic 5′-[32P]- ΔC1-tRNAPro (5–10 μM) were incubated in 50 mM HEPES (pH 7.5) for 5 min at room temperature, followed by addition of 100 μM Na2IrCl6 (Alfa Aesar). In some experiments, proline, ATP, proline + ATP, or 5′-O-[N-(L-prolyl)sulfamoyl]adenosine (Pro-AMS) were added to a final concentration of 20μM prior to addition of Na2IrCl6. After 4 h of incubation at room temperature, reactions were quenched with 50 nM EDTA (pH 8.0). Reaction mixtures were loaded onto 8% sodium dodecyl sulfate-polyacrylamide gels, visualized, and quantified via phosphorimaging analysis of the dried gel.
3. Results and Discussion
We first incorporated amino acid substitutions lysine and leucine at position 144 in the motif 2 loop of E. coli ProRS. Mutation of the positively charged R144 side chain to a neutral leucine residue resulted in an 870-fold reduced aminoacylation catalytic efficiency (Table 1). Even the conservative change to lysine resulted in a large 480-fold decrease in kcat/KM (Table 1). Interestingly, in both cases, the defect was entirely in the kcat parameter, with little effect on the KM. These data confirmed the sensitivity of ProRS to changes at this site and showed that the defect was not at the binding step.
Table 1.
Effect of Single Amino Acid Changes at Position 144 in the Motif 2 Loop of E. coli ProRS on Aminoacylation of E. coli tRNAPro Transcriptsa
| ProRS Mutation | kcat (s−1) | KM (μM) | kcat/KM (s−1.μM −1) | kcat/KM (relative) | x-fold change |
|---|---|---|---|---|---|
| Wild-Type | 0.35 | 18.0 | 1.8×10−2 | 1.0 | 1.0 |
| R144K | 2.5 ×10−4 | 11.0 | 3.7×10−5 | 2.1×10−3 | −480 |
| R144L | 1.0 ×10−4 | 10.3 | 2.1×10−5 | 1.1×10−3 | −870 |
Assays were performed at least twice with a difference of < 2-fold between trials.
Previously, we hypothesized that residues in the motif 2 loop interact closely with acceptor stem residues G72 and A73. If a hydrogen bonding interaction between R144 and G72 and/or A73 occurs, the decrease in aminoacylation caused by mutations at these nucleotides [8] may be completely or partially suppressed by the R144 mutants. We indeed demonstrate in this work that aminoacylation of G72A-tRNAPro is reduced 170-fold compared to wild type tRNAPro when assayed with wild-type ProRS, whereas only a 2.6-fold decrease is observed with R144K ProRS (relative to charging of wild-type tRNAPro by R144K ProRS) (Table 2). Interestingly, R144L ProRS actually prefers G72A-tRNAPro as a substrate; the aminoacylation efficiency measured with this variant is 2.4-fold greater relative to aminoacylation of wild-type tRNAPro (Table 2). These results are consistent with a functional interaction between position 144 of ProRS and G72 of tRNAPro. However, similar results were obtained with an A73G tRNA variant (Table 2). Whereas the kcat/KM of this tRNA is 175-fold reduced in the presence of wild-type ProRS, R144K and R144L ProRS variants are not sensitive to the change of A73 to G.
Table 2.
Ratio of kcat/KM for aminoacylation of wild-type and mutant tRNAPro by wild-type and mutant ProRSa
| G72A | A73G | 2AP72 | 7G72 | 7A73 | |
|---|---|---|---|---|---|
| ProRS (WT) | 170 | 175 | 30 | 52 | 23 |
| R144K (−480) | 2.6 | 1.0 | 2.5 | 2.1 | 7.4 |
| R144L (−870) | 0.42 | 0.91 | 0.43 | 0.06 | 18 |
| V143C (−3) | 230 | 310 | ND | ND | ND |
| R146C (−79) | >106 | >106 | ND | ND | ND |
2AP is 2′-deoxy-2-aminopurine, 7G is 2′-deoxy-7-deaza-G, and 7A is 2′–deoxy-7-deaza A. The 2′–hydroxyl group was previously shown to be dispensable at positions 72 [12] and 73 [5]. The ratios were obtained as follows: (kcat/KM)wild-type tRNA/(kcat/KM)mutant tRNA. ND stands for not determined. The numbers in parenthesis (left column) indicate the fold-decrease in the wild-type (WT) tRNAPro aminoacylation efficiency of the indicated mutant ProRS, relative to WT ProRS (in the case of R144K and R144L) or relative to C443G ProRS (in the case of V143C and R146C, see Materials and Methods). All assays were performed at least twice with a difference of < 2-fold between trials.
We also reported that mutations of other positions in the motif 2 loop have significant but much more modest (3- to 80-fold) effects on aminoacylation [10]. To confirm the positional specificity of the G72/A73 interaction within the motif 2 loop, we further tested two additional motif 2 loop mutants (V143C and R146C) that were previously reported to result in 3-fold and 79-fold decreases in kcat/KM, respectively. Here we report that, in contrast to the results obtained with R144 variants, V143C ProRS and R146C ProRS displayed large decreases (230-fold and >106-fold, respectively) in aminoacylation of G72A-tRNAPro relative to charging of wild-type tRNAPro (Table 2). Similar results were obtained with A73G-tRNAPro. These results show that activity changes due to V143C and R146C mutations are not suppressed by mutation of G72A and A73G of tRNAPro, and therefore, they do not support a functional interaction between V143 or R146 and positions 72/73 of the acceptor stem.
Nucleotide base changes such as those described above may also result in indirect effects due to conformational or stacking differences induced by the mutations. More subtle atomic group substitutions or deletions are less likely to have these indirect effects. To further define the nature of the interaction between R144 and G72/A73, atomic group mutants were incorporated into semi-synthetic tRNAPro constructs. Substitution of 2-aminopurine at position 72, which removes the major groove 6-keto oxygen, results in a 30-fold decrease in aminoacylation by wild-type ProRS, but only a 2.5 fold decrease with R144K ProRS (Table 2). Similar to the G72A variant, this mutant displays slightly increased activity with R144L. These data support a direct hydrogen bonding interaction between R144 and the major groove oxygen of G72. Substitution of the N7 position with a carbon by incorporation of 7-deaza G72, results in a 52-fold decrease in charging by wild-type ProRS but only a 2.1-fold decrease in the case of R144K ProRS and a striking 17-fold increase with R144L. The latter may be due to more favorable nonpolar packing between the leucine side chain and the carbon at position 7 (in contrast to the unfavorable polar surface area of N7) [20]. In contrast to the results obtained at position 72, the effects of substituting neighboring base A73 with 7-deaza A are not significantly suppressed (3-fold or less) by changes at R144 (Table 2). In particular, the R144L variant is as sensitive to the 7-deaza A73 change as wild-type ProRS. Taken together, we conclude that R144 interacts preferentially with G72 through two hydrogen-bonding interactions with the 6-keto oxygen and N7 position in the major groove.
We next incorporated OG into the acceptor stem of semi-synthetic E. coli tRNAPro at the critical G72 position. This substitution resulted in only a small (< 2-fold) decrease in aminoacylation by wild-type ProRS (data not shown). Previous work also showed that deoxynucleotide substitution at G72 resulted in a minor (1.2-fold) reduction in kcat/KM [18]. Unmodified and OG-containing tRNAs were incubated with wild-type, R144K, R144L, or R144C E. coli ProRS and Na2IrCl6. As mentioned earlier, these mutations on ProRS did not negatively affect the KM values for tRNAPro (Table 1), suggesting that the defects are not in the binding step. Following the cross-linking reactions, samples were analyzed on denaturing polyacrylamide gels (Fig. 1).
Fig. 1.
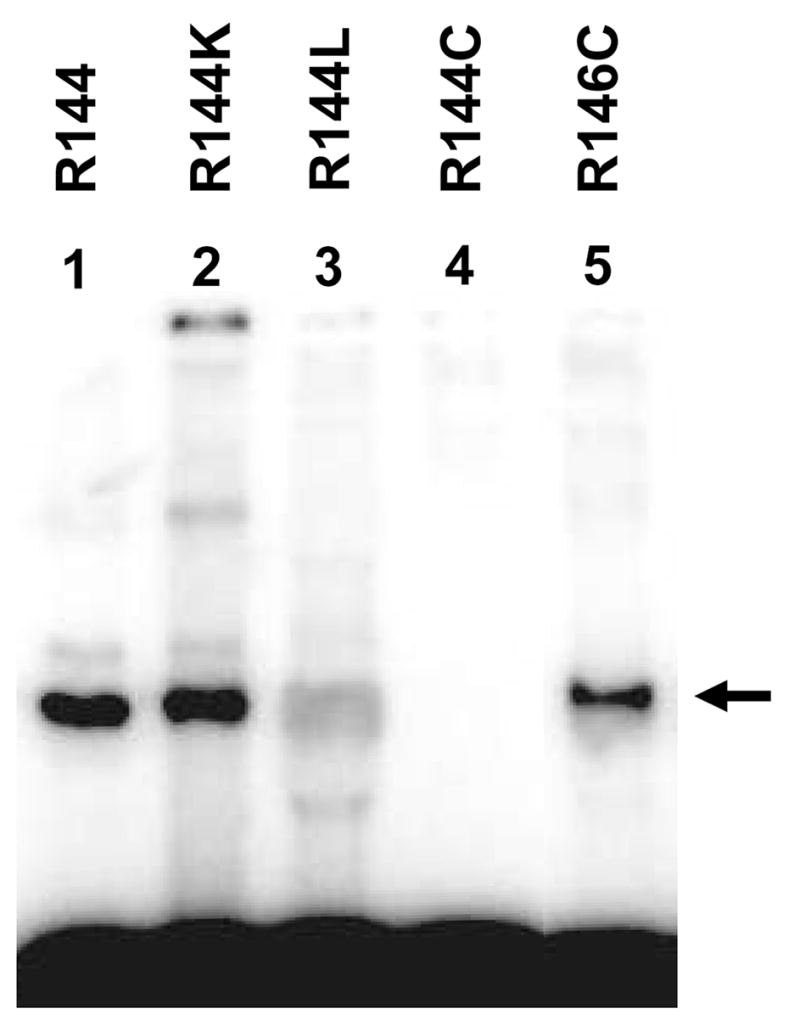
Denaturing polyacrylamide gel (8%) showing the results of oxidative cross-linking of semi-synthetic OG72-tRNAPro to wild-type (lane 1) and mutated E. coli ProRS (lane 2–5). As discussed in the Materials and Methods, R144C (lane 4) and R146C (lane 5) ProRS variants contain an additional C443G mutation. The cross-linked adduct is indicated by an arrow.
As shown in Fig. 1, a cross-linked adduct is detected when wild-type and R144K ProRS are incubated with OG72-tRNA under oxidative conditions (lanes 1 and 2). Cross-linking is dependent upon the presence of an arginine or lysine residue at position 144, as R144L ProRS shows only slight non-specific (see below) cross-linking (lane 3) and no cross-link is detected with R144C ProRS (lane 4). Cross-linking is not observed in the control reactions using unmodified tRNAPro (data not shown). These results support the conclusion that residue 144 is in close proximity (~1.5 Å) to G72 and that both arginine and lysine side chains can react with oxidized OG, as previously reported [13, 14]. The lack of cross-link formation with R144C and R144L ProRS suggests that the nearby motif 2 loop residue, R146, is not involved in close interaction with G72 and cannot substitute for the function of R144. This result is consistent with the kinetic data for R146C ProRS, which also suggested that R146 is not functionally involved in interaction with G72/A73 (Table 2). To confirm this hypothesis further, we showed that a R146C ProRS variant cross-links to OG72-tRNA, albeit with an ~1.3-fold reduced yield relative to wild-type or R144K ProRS (Fig. 1, lane 5), which may be due to non-optimal positioning of the R144 side chain with respect to the major groove of OG72. Taken together, these data support the role of R144 in specific cross-link formation to OG72.
The crystal structure of T. thermophilus ProRS bound to prolyl-adenylate shows a close interaction between the adenylate and the motif 2 loop, as well as an ordering and movement of the loop upon substrate binding [21]. To gain further support for the specific involvement of E. coli ProRS motif 2 loop residues in cross-link formation, we performed cross-linking studies in the presence of proline, ATP, proline and ATP, or a non-hydrolysable sulfamoyl analogue of prolyl-adenylate, Pro-AMS.
The efficiency of OG72-tRNAPro cross-linking to R144K ProRS is suppressed only slightly in the presence of proline alone (−1.4-fold) or ATP alone (−2.8-fold), but is more significantly reduced in the presence of Pro-AMS (−4.5-fold) or both proline and ATP (−5.9-fold) (Fig. 2). These results support the proximity of the observed cross-linked adduct to the aminoacylation active site, and suggest either that conformational changes upon adenylate formation result in the reduced level of cross-link formation, or that the adenylate sterically blocks interaction of G72 with active site residues. In contrast, the slight cross-linking observed with R144L ProRS (Fig. 1, lane 3) is not affected by the presence of substrates (data not shown), indicating that the cross-linking observed with this variant is likely due to non-specific interactions outside the active site.
Fig. 2.
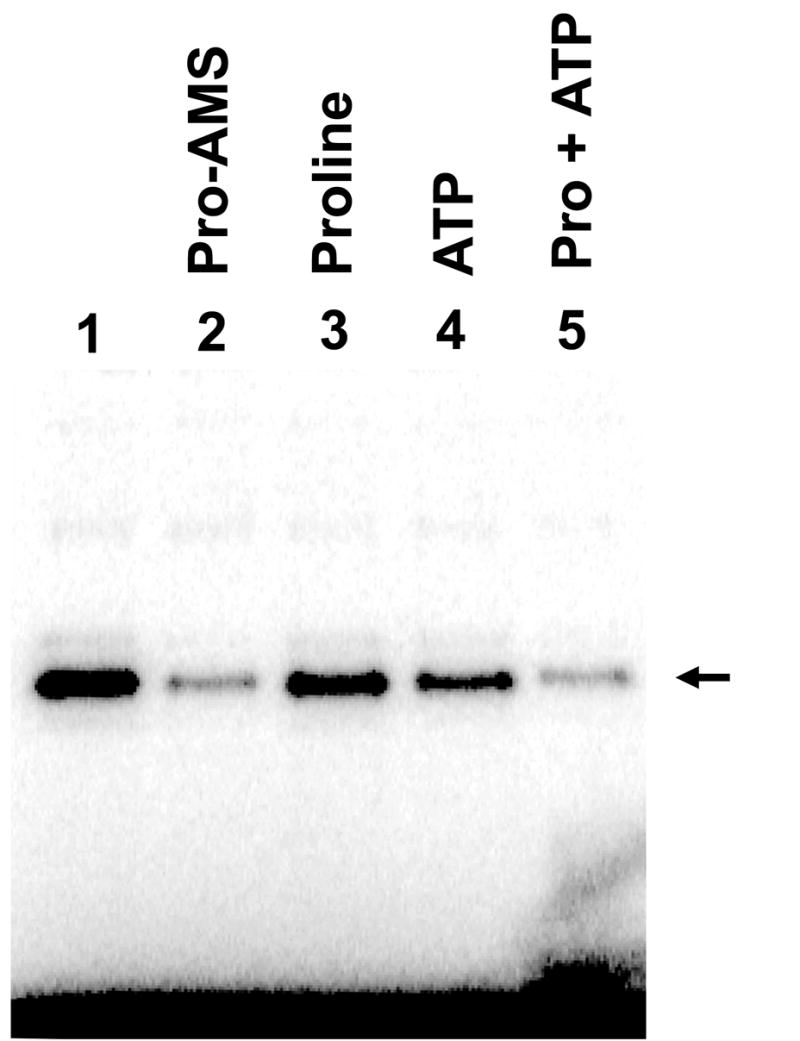
Denaturing polyacrylamide gel (8%) showing the results of oxidative cross-linking of semi-synthetic OG72-tRNAPro to R144K E. coli ProRS in the absence (lane 1) and presence of the indicated substrates (lanes 3–5) or the Pro-AMS adenylate analogue (lane 4). The cross-linked adduct is indicated by an arrow.
In summary, mutagenesis and kinetic data, together with a novel cross-linking approach were used to identify a specific hydrogen bonding interaction between the arginine side chain at position 144 of the motif 2 loop of E. coli ProRS and the major groove functional groups of G72 in the tRNAPro acceptor stem. Changes at R144 do not substantially alter the Michaelis constant for tRNA, but significantly affect the kcat parameter. Transfer RNA aminoacylation has been described as a multistep process involving initial formation of an “encounter complex”, which generally depends on specific anticodon interactions, followed by an “accommodation” process that involves conformational changes in both partners resulting in correct positioning of the CCA end in the active site [22]. We propose that the R144-G72 contact plays a critical role in the latter process, in effect coupling the anticodon recognition/binding event to catalysis by optimally positioning the CCA-3′ end into the active site. The results also indicate that A73 is likely to be involved in a hydrogen-bonding network involving nearby motif 2 loop residues, although the exact nature of this interaction is not yet clear.
Based on deposited structures in the Protein Data Bank (http://www.pdb.org), arginine-guanosine interactions are the most abundant contacts found in the amino acid-nucleotide interaction database (AANT; http://aant.icmb.utexas.edu/) [23]. Thus, the biochemical approaches described here should be widely applicable to the identification of nucleic acid-protein interactions for which high resolution structural information obtained by NMR or X-ray crystallography is not available.
Acknowledgments
We thank Drs. Penny J. Beuning and Abbey E. Rosen for synthesizing the synthetic oligonucleotides used in this study. We would also like to thank Ms. Carmen Silvers for assistance with purification of ProRS mutants. This work was funded by National Institutes of Health grant GM49928 (K.M.-F.) and T32 GM08277 (B.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Giege R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ghosh A, Vishveshwara S. A study of communication pathways in methionyl-tRNA synthetase by molecular dynamics simulations and structure network analysis. Proc Natl Acad Sci USA. 2007;104:15711–15716. doi: 10.1073/pnas.0704459104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hammes-Schiffer S, Benkovic SJ. Relating protein motion to catalysis. Annu Rev Biochem. 2006;75:519–541. doi: 10.1146/annurev.biochem.75.103004.142800. [DOI] [PubMed] [Google Scholar]
- 4.Budiman ME, Knaggs MH, Fetrow JS, Alexander RW. Using molecular dynamics to map interaction networks in an aminoacyl-tRNA synthetase. Proteins. 2007;68:670–689. doi: 10.1002/prot.21426. [DOI] [PubMed] [Google Scholar]
- 5.Perona JJ, Hou YM. Indirect readout of tRNA for aminoacylation. Biochemistry. 2007;46:10419–10432. doi: 10.1021/bi7014647. [DOI] [PubMed] [Google Scholar]
- 6.First EA, Fersht AR. Analysis of the role of the KMSKS loop in the catalytic mechanism of the tyrosyl-tRNA synthetase using multimutant cycles. Biochemistry. 1995;34:5030–5043. doi: 10.1021/bi00015a014. [DOI] [PubMed] [Google Scholar]
- 7.McClain WH, Schneider J, Gabriel K. Distinctive acceptor-end structure and other determinants of Escherichia coli tRNA(Pro) identity. Nucleic Acids Res. 1994;22:522–529. doi: 10.1093/nar/22.3.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu H, Peterson R, Kessler J, Musier-Forsyth K. Molecular recognition of tRNA(Pro) by Escherichia coli proline tRNA synthetase in vitro. Nucleic Acids Res. 1995;23:165–169. doi: 10.1093/nar/23.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yaremchuk A, Cusack S, Tukalo M. Crystal structure of a eukaryote/archaeon-like protyl-tRNA synthetase and its complex with tRNAPro(CGG) EMBO J. 2000;19:4745–4758. doi: 10.1093/emboj/19.17.4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burke B, Yang F, Chen F, Stehlin C, Chan B, Musier-Forsyth K. Evolutionary coadaptation of the motif 2--acceptor stem interaction in the class II prolyl-tRNA synthetase system. Biochemistry. 2000;39:15540–15547. doi: 10.1021/bi001835p. [DOI] [PubMed] [Google Scholar]
- 11.Sprinzl M, Vassilenko KS. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 2005;33:D139–140. doi: 10.1093/nar/gki012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hickerson RP, Chepanoske CL, Williams SD, David SS, Burrows CJ. Mechanism-Based DNA-Protein Cross-Linking of MutY via Oxidation of 8-Oxoguanosine. J Am Chem Soc. 1999;121:9901–9902. [Google Scholar]
- 13.Johansen ME, Muller JG, Xu X, Burrows CJ. Oxidatively induced DNA-protein cross-linking between single-stranded binding protein and oligodeoxynucleotides containing 8-oxo-7,8-dihydro-2′-deoxyguanosine. Biochemistry. 2005;44:5660–5671. doi: 10.1021/bi047580n. [DOI] [PubMed] [Google Scholar]
- 14.Muller JG, Duarte V, Hickerson RP, Burrows CJ. Gel electrophoretic detection of 7,8-dihydro-8-oxoguanine and 7, 8-dihydro-8-oxoadenine via oxidation by Ir (IV) Nucleic Acids Res. 1998;26:2247–2249. doi: 10.1093/nar/26.9.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heacock D, Forsyth CJ, Shiba K, Musier-Forsyth K. Synthesis and Aminoacyl-tRNA Synthetase Inhibitory Activity of Prolyl Adenylate Analogs. Bioorganic Chemistry. 1996;24:273–289. [Google Scholar]
- 16.Horton RM, Pease LR. In: Directed Mutagenesis. McPherson MJ, editor. IRL press; New York: 1991. pp. 217–247. [Google Scholar]
- 17.Fersht AR, Ashford JS, Bruton CJ, Jakes R, Koch GL, Hartley BS. Active site titration and aminoacyl adenylate binding stoichiometry of aminoacyl-tRNA synthetases. Biochemistry. 1975;14:1–4. doi: 10.1021/bi00672a001. [DOI] [PubMed] [Google Scholar]
- 18.Yap LP, Musier-Forsyth K. Transfer RNA aminoacylation: identification of a critical ribose 2′-hydroxyl-base interaction. RNA. 1995;1:418–424. [PMC free article] [PubMed] [Google Scholar]
- 19.Stehlin C, Heacock DH, Liu H, Musier-Forsyth K. Chemical modification and site-directed mutagenesis of the single cysteine in motif 3 of class II Escherichia coli prolyl-tRNA synthetase. Biochemistry. 1997;36:2932–2938. doi: 10.1021/bi962295s. [DOI] [PubMed] [Google Scholar]
- 20.Ashworth J, Havranek JJ, Duarte CM, Sussman D, Monnat RJ, Jr, Stoddard BL, Baker D. Computational redesign of endonuclease DNA binding and cleavage specificity. Nature. 2006;441:656–659. doi: 10.1038/nature04818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yaremchuk A, Tukalo M, Grotli M, Cusack S. A succession of substrate induced conformational changes ensures the amino acid specificity of Thermus thermophilus prolyl-tRNA synthetase: comparison with histidyl-tRNA synthetase. J Mol Biol. 2001;309:989–1002. doi: 10.1006/jmbi.2001.4712. [DOI] [PubMed] [Google Scholar]
- 22.Guth EC, Francklyn CS. Kinetic discrimination of tRNA identity by the conserved motif 2 loop of a class II aminoacyl-tRNA synthetase. Mol Cell. 2007;25:531–542. doi: 10.1016/j.molcel.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffman MM, Khrapov MA, Cox JC, Yao J, Tong L, Ellington AD. AANT: the Amino Acid-Nucleotide Interaction Database. Nucleic Acids Res. 2004;32:D174–181. doi: 10.1093/nar/gkh128. [DOI] [PMC free article] [PubMed] [Google Scholar]


