Abstract
Activation of translation initiation is essential for the malignant phenotype, and is emerging as a potential therapeutic target. Translation is regulated by the expression of translation initiation factor 4E (eIF4E) as well as the interaction of eIF4E with eIF4E-binding proteins (e.g. 4E-BP1). Rapamycin inhibits translation initiation by decreasing the phosphorylation of 4E-BP1, increasing eIF4E/4E-BP1 interaction. However, rapamycin also inhibits S6K phosphorylation, leading to feedback-loop activation of Akt. We hypothesized that targeting eIF4E directly would inhibit breast cancer cell growth without activating Akt. We demonstrated that eIF4E is ubiquitously expressed in breast cancer cell lines. eIF4E knockdown by siRNA inhibited growth in different breast cancer cell subtypes including triple-negative (ER/PR/HER2-negative) cancer cells. eIF4E knockdown inhibited the growth of cells with varying total and p-4E-BP1 levels, and inhibited rapamycin-insensitive as well as sensitive cell lines. eIF4E knockdown led to a decrease in expression of cyclin D1, Bcl2 and Bcl-xL. eIF4E knockdown did not lead to Akt phosphorylation, but did decrease 4E-BP1 expression. We conclude that eIF4E is a promising target for breast cancer therapy. eIF4E-targeted therapy may be efficacious in a variety of breast cancer subtypes including triple-negative tumors for which currently there are no targeted therapies. Unlike rapamycin and its analogues, eIF4E knockdown is not associated with Akt activation.
Keywords: Translation, targeted therapy, breast cancer, eIF4E, mTOR
Introduction
Translation initiation is deregulated in many human cancers including breast, head and neck, colorectal, bladder, prostate, lung, cervical cancers and lymphomas (1). Translation initiation factor 4E (eIF4E) is rate-limiting for cap-dependent translation (2). It has been proposed that the translational efficiency of mRNA with highly complex 5′ untranslated regions (UTRs) is especially dependent on eIF4E levels (1, 3). eIF4E overexpression leads to selective translation of mRNA such as cyclin D1, Bcl-2, Bcl-xL and VEGF in vitro (4-8). eIF4E expression shows significant correlation with cyclin D1 and VEGF expression in human tumors (9). eIF4E enhances nucleocytoplasmic transport for selected mRNA such as cyclin D1. Thus, eIF4E expression effects the expression of important regulators of cell growth and survival. In vitro eIF4E overexpression mediates growth, proliferative and survival signaling, and has transforming activity in fibroblasts and mammary epithelial cells (10-12). Transgenic eIF4E-expressing mice show a marked increase in tumorigenesis, developing tumors of various histologies (13). Thus, eIF4E also directly acts as an oncogene in vivo.
Activated translation initiation is essential for the malignant breast cancer phenotype (14). Elevated eIF4E levels are critical for angiogenesis and progression of breast carcinomas (15). eIF4E is overexpressed in breast cancer and is a poor prognostic marker in retrospective and prospective studies (16-18). Graff et al have recently shown that eIF4E expression can be downregulated with second generation antisense oligonucleotides, with reduction of in vivo tumor growth in a PC3 prostate cancer model and MDA-MB-231 breast cancer model (19). These data suggest that eIF4E could indeed be pursued as a therapeutic target for breast cancer as well as a variety of other cancers.
The mammalian target of rapamycin (mTOR) kinase in a complex with raptor (mTORC1) leads to the phosphorylation of 4E-BP1 and S6K1, two other key proteins that regulate protein translation (20). Rapamycin and its analogues inhibit mTORC1 signaling. mTOR is being investigated as a potential target in many cancer types and is already a proven therapeutic target for renal cell carcinoma. The rapamycin analogue temsirolimus (Torisel, CCI-779, Wyeth, PA) has been shown to improve overall survival among patients with metastatic renal cell carcinoma (21), leading to its FDA approval. However, eIF4E overexpression partially rescues rapamycin-mediated G1 arrest in vitro, and confers resistance to rapamycin in transgenic lymphoma models in vivo (22, 23). Further, preclinical studies and sequential biopsies in patients treated in a Phase I trial of mTOR inhibitors have determined that mTOR inhibition induces feedback loop activation of Akt, a mechanism that may potentially limit its antitumor activity (24). As this feedback loop activation is thought to be mediated through the inhibition S6K1 by mTOR inhibitors, there is rationale in targeting translation downstream of mTOR, directly at eIF4E.
As activation of translation initiation is a common integral pathway for the malignant phenotype, we hypothesized that eIF4E downregulation would lead to growth inhibition in breast cancer cells of differing tumor subtypes. Further, we hypothesized that targeting eIF4E directly rather than targeting mTORC1 signaling, would not lead to feedback-loop mediated Akt activation.
Materials and Methods
Cell lines and cultures
MDA-MB-468, SK-BR-3, MCF7, MDA-MB-435S, and MDA-MB-231 were obtained from American Type Culture Collection (ATCC, Manassas, VA). MDA-MB-435 was obtained from NCI (Frederick, MD). All cell lines were cultured in DMEM/F12 (Cellgro Mediatech, Inc., Herndon, VA) supplemented with 10% (v/v) fetal bovine serum (SAFC Biosciences, Lenexa, KA) at 37°C and humidified in 5% CO2. For selected experiments, cells were treated with dimethyl sulphoxide (DMSO) (Sigma Chemical Company (St. Louis, MO) or rapamycin (LC laboratories, Woburn, MA).
Small interfering RNA (siRNA)
siRNA to eIF4E was custom-ordered duplexes from Dharmacon, Inc. (Lafayette, CO) or Ambion, Inc. (Austin, TX). The four sequences tested were 4E1 (AAGCAAACCUGCGGCUGAUCU) (25), 4E2 (ACAGCAGAGACGAAGUGAC) (25), 4E3 (GGAUGGUAUUGAGCCUAUG) (26), and 4E4 (GGUGGGCACUCUGGUUUUU) (26). Negative control siRNAs were purchased from Ambion, Inc. Transfection was performed using DharmaFECT 1 reagent according to manufacturer's protocol (Dharmacon).
Western blots
Western blotting was perfomed as described elsewhere (27). Membranes were immunoblotted with antibodies to eIF4E, p-eIF4E (S209), Bcl2, Bcl-xL, 4E-BP1, p-4E-BP1 (T70), S6K1, p-S6K1 (T389), Akt, p-Akt (T308), p-Akt (S473) (all from Cell Signaling Technology, Danvers, MA), cyclin D1 (Santa Cruz Biotechnology, Santa Cruz, CA), and actin (Sigma). Membranes were visualized by the Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE) or ECL developer system (GE Healthcare Life Sciences).
Cell Growth and Cell Cycle Analysis
Cell growth was determined by either of two methods: by the relative percentages of metabolically active cells compared with untreated controls on the basis of mitochondrial conversion of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) to formazine or alternately, by the protein content of treated and untreated cells through the sulforhodamine-B (SRB) assay (28). The results were assessed in a 96-well format plate reader performed in triplicates by measuring the absorbance at a wavelength of 570 nm. The IC50 was determined on the basis of the dose-response curves using SRB assay. The commercial software package was obtained from Calcusyn (Biosoft, Cambridge, United Kingdom). Cell cycle was analyzed as described previously (29).
Colony Formation Assay
At 24 h incubation of siRNA, cells were trypsinized, counted and plated at a density of 1-5 × 103 cells/60-mm plate in triplicate for each treatment group. Approximately 2-4 weeks later, plates were fixed, stained with crystal violet, scanned, and colonies were counted using NIH ImageJ v.1.33h software.
Sucrose density gradient
MDA-MB-468 cells were transfected by negative control or 4E3 siRNA, and cultured for 72 h. The sucrose density gradient assay, cycloheximide (Sigma) was added to 100 μg/ml 5 min prior to harvesting. Polysome extracts were lysed in the following buffer: 10 mM Tris-HCl (pH 7.5), 100 mM KCl, 20 mM MgCl2, 0.5% Triton X-100, 0.5% sodium deoxycholate, 6% sucrose, 100 μg/ml cycloheximide, and 1,000 U/ml RNase inhibitor (Roche Diagnostic Corporation, Indianapolis, IN). Lysates were then centrifuged for 5 min at 12,000×g and applied to 15-50% (w/v) sucrose gradient prepared in 10 mM Tris-HCl (pH 7.5), 100 mM KCl, 20 mM MgCl2, and 100 μg/ml cycloheximide, using an ISCO Model 160 Gradient Former (Teledyne Isco, Inc., Lincoln, NE). Sucrose gradients were centrifuged at 38,000×g for 2 h 30 min in a Beckman SW40Ti rotor. After centrifugation, gradients were fractionated by using an ISCO Foxy Jr. fractionator (Teledyne Isco). The absorbance at 254 nm of gradient was analyzed with an ISCO UA-6 UV/VIS detector.
Statistical Analysis
Results were presented as means ± SD. Growth inhibition was normalized to the no treatment group, no treatment growth being set at 100%. For comparison between groups, data were analyzed by Student's t test. Differences between groups were considered statistically significant at P < 0.05.
Results
eIF4E Knockdown Inhibits Cell Growth in Breast Cancer Cell Lines
To determine the potential of eIF4E as a therapeutic target, we assessed the expression of eIF4E on western blotting, and observed that eIF4E is ubiquitously expressed in a panel breast cancer cell lines (Figure 1A). The expression of total 4E-BP1 and p-4E-BP1 was variable in the cell lines. To determine the rapamycin sensitivity of the cell lines, the cell lines were treated with varying concentrations of rapamycin ranging from 0.1 to 1000 nM for 5 days. Figure 1B demonstrates that the panel of cell lines we selected for study varied in their sensitivity to rapamycin's growth-inhibitory effects.
Figure 1.
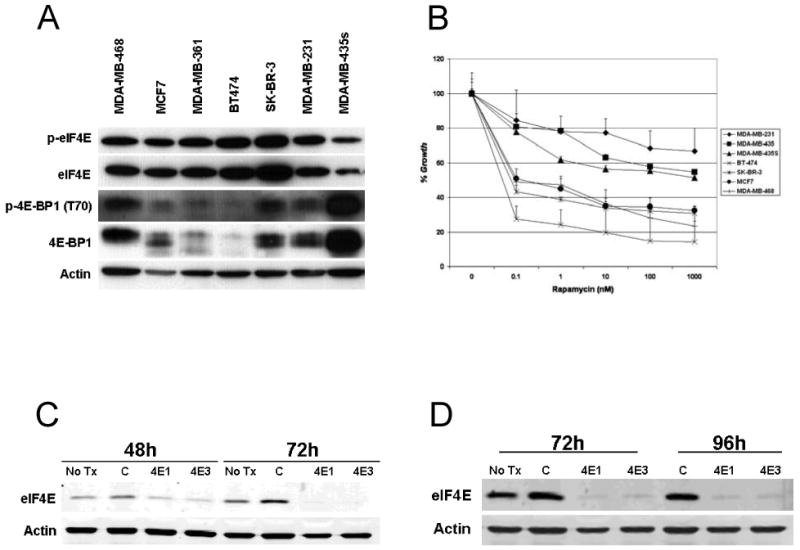
Expression of eIF4E and 4E-BP1 in panel of breast cancer cell lines. A. Cell lysate was harvested from a panel of breast cancer cell lines and separated by SDS-PAGE. Western blotting was performed for p-eIF4E (S209), total eIF4E, p-4E-BP1 (T70), total 4E-BP1 and actin. B. Rapamycin sensitivity of a panel of breast cancer cell lines. Cells plated in 96-well plates in triplicate, and treated with increasing doses of rapamycin. Cell growth was assessed at five days by SRB, and growth was compared to vehicle treated controls (% control). Results represent the average of at least three independent experiments. C. and D. MDA-MB-468 cells were transfected with control siRNA or one of two siRNA sequences for eIF4E (4E1 and 4E3). Cells were lysed at 48, 72 or 96 hours. Western blotting was performed for eIF4E and actin expression.
We tested a siRNA panel of sequences to eIF4E (data not shown) and identified two independent sequences (4E1 and 4E3) that led to marked knockdown of eIF4E expression. Western blotting 48 hours after siRNA transfection demonstrates some decline of eIF4E levels, with further knockdown by 72 hours (Figure 1C) and maintained by 96 hours (Figure 1D).
We next assessed the effect of eIF4E knockdown by transfection of cells with control siRNA or eIF4E siRNA sequences 4E1 or 4E3 (Figure 2A). In each experiment, eIF4E knockdown was confirmed with western blotting for eIF4E and actin at 72 hours (Figure 2A). One day after transfection, cells transfected were trypsinized, counted and plated in 96-well plates in triplicates. After four more days, cell growth was assayed (Figure 2A). In all eight cell lines tested, eIF4E knockdown lead to a statistically significant decrease in growth compared to control siRNA treated cells (p<0.05 for all comparisons of 4E1 or 4E3 to control siRNA). Of note, the cell lines tested represent different subtypes of breast cancer: MCF7 cells being ER+, SKBR3 cells HER2+, and MDA-MB-468, MDA-MB-231, MDA-MB-435 cells being ER/PR/HER2-, or triple negative. Thus eIF4E knockdown inhibited growth in a variety of breast cancer subtypes, including triple negative cells.
Figure 2.
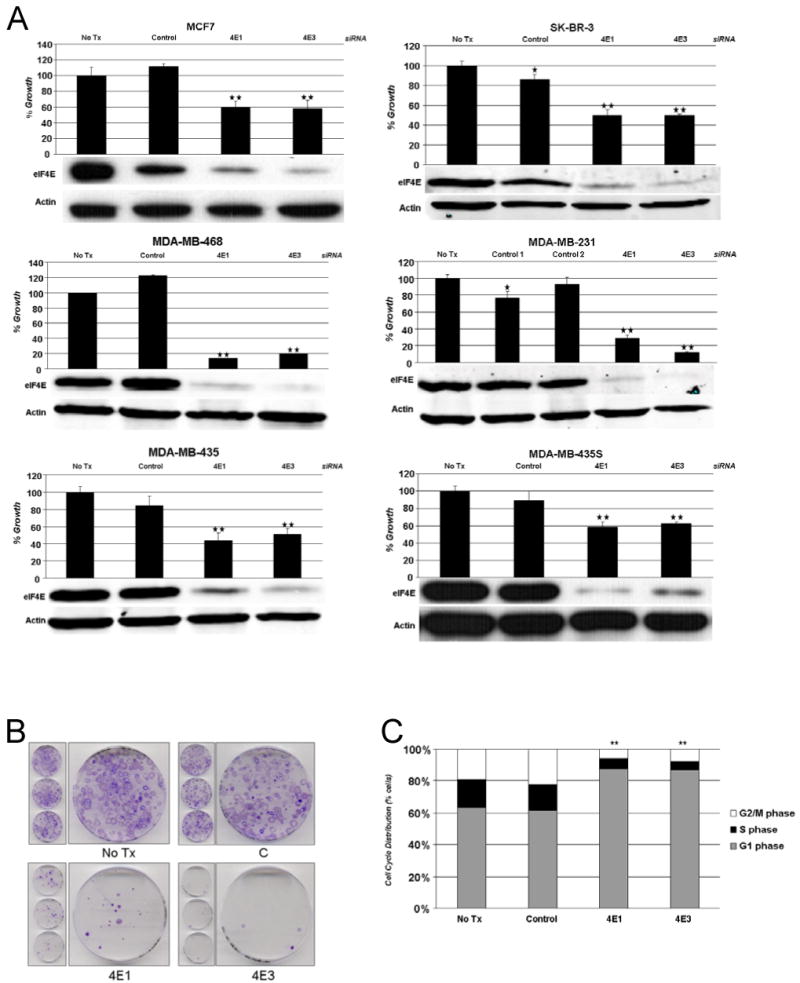
Effect of eIF4E knockdown on cancer cell growth. A. MCF7, SKBR-3 MDA-MB-468, MDA-MB-231, MDA-MB-435, and MDA-MB-435S cells were transfected with control siRNA or one of two eIF4E siRNA sequences (4E1 and 4E3). One day after transfection, the cells were trypsinized, counted and plated in 96 well plates in triplicates. On day 4, cell growth was assayed and cell growth in untreated cells (no tx) was normalized to 100%. Shown are representative experiments (n≥3). Columns, mean; bars, SD. *, P < 0.05, untreated group versus control siRNA group by two tailed Student's t-test. **, P < 0.05, control siRNA group versus 4E1 or 4E3 siRNA by two tailed Student's t-test. 72 hours after siRNA transfection, cells were lysed and western blotting was performed for eIF4E and actin expression. Representative western blots demonstrating eIF4E knockdown in each cell line is shown. B. Effect of eIF4E knockdown on anchorage-dependent growth. Cells were transfected with control siRNA or one of two eIF4E siRNA sequences (4E1 and 4E3). One day after transfection, the cells were trypsinized, counted and plated in 60-mm plates (1000 cells/plate). After 2 weeks, colonies were stained with crystal violet. No tx, no treatment. C. Effect eIF4E knockdown on cell cycle. MCF7 cells were transfected with control siRNA or one of two eIF4E siRNA sequences (4E1 and 4E3). Four days after transfection, the attached cells were harvested, and FACS analysis was performed. No tx, no treatment. **, P < 0.05, % cells in G1 in control siRNA group versus 4E1 or 4E3 siRNA by two tailed Student's t-test.
The effect of eIF4E knockdown on anchorage-dependent growth was also assessed by colony formation assay in MDA-MB-435S cells. One day after cells were transfected with control siRNA or one of two eIF4E siRNA sequences, cells were trypsinized, counted and plated in 60-mm plates (1000 cells/plate) and after two weeks, colonies were stained with crystal violet. eIF4E knockdown with either eIF4E siRNA sequence led to a dramatic decline in colony formation ability (Figure 2B).
To elucidate its mechanism of action, we assessed the effect of eIF4E knockdown on the cell cycle with flow cytometry (Figure 2C). MCF7 cells were transfected with control siRNA or one of two eIF4E siRNA sequences (4E1 and 4E3) and four days later cell cycle analysis was performed on attached cells. eIF4E knockdown was associated with a decline in % cells in S phase and a significant increase in % cells in G1 phase (%G1 cells 59% with control siRNA vs. 86% with 4E1, 83% with 4E3, p<0.0001 for both).
eIF4E Knockdown Inhibits Translation and eIF4E's Downstream Targets
We next evaluated the effect of eIF4E knockdown on translation. mRNA that are actively translated usually have multiple ribosomes associated with them, referred to as polysomes, while translationally inactive mRNAs are often sequestered in messenger ribonucleoprotein (mRNP) particles or are associated with a single ribosome (monosome). Polysomal and mRNP particles can be separated by sucrose gradient centrifugation. Therefore, MDA-MB-468 cells were treated with control siRNA versus 4E siRNA and after three days, cell lysates were fractionated on 15-50% sucrose gradients. eIF4E knockdown was associated with a decline of RNA in polysomal fractions and an enrichment in non-polysomal fractions (Figure 3A).
Figure 3.
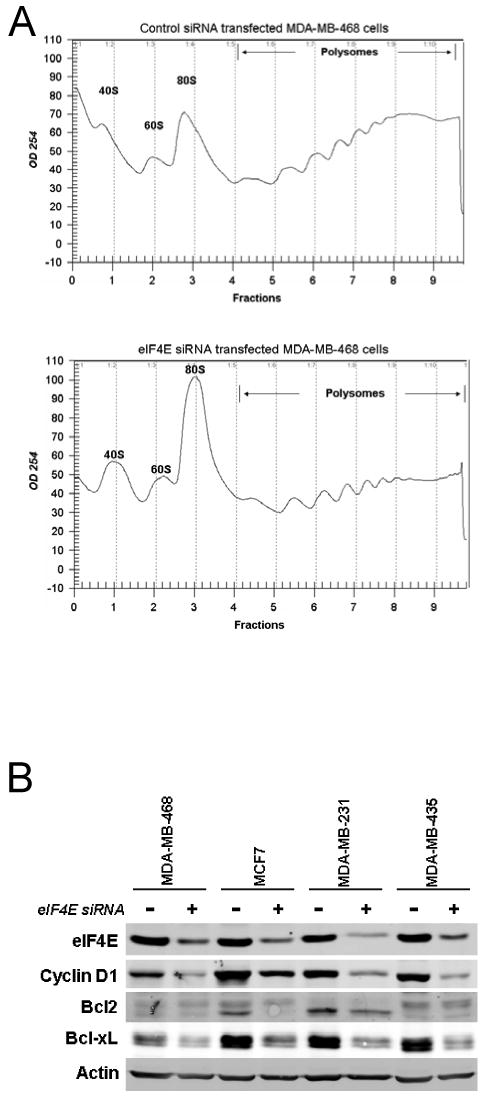
Effect of eIF4E knockdown on translation. A. MDA-MB468 cells were transfected with control siRNA or eIF4E siRNA and were cultured for three days to allow the efficient eIF4E knockdown. Cell lysates were then separated on a 15-50% sucrose gradient. After centrifugation, gradients were fractionated by using an ISCO Foxy Jr. fractionator (Teledyne Isco). The A254 of gradient was analyzed with an ISCO UA-6 UV/VIS detector. B. Effect eIF4E knockdown on eIF4E's translational targets. MDA-MB-468, MCF7, MDA-MB-231 or MDA-MB-435 cells were transfected with control siRNA or eIF4E siRNA (4E3). Three days after transfection, cell lysate was harvested, and western blotting was performed for eIF4E, cyclin D1, Bcl2, Bcl-xL, and actin.
We next evaluated the effect of eIF4E knockdown on the expression of some of the mRNA previously proposed to be selectively regulated by eIF4E, in four different cell lines (Figure 3B). eIF4E knockdown was associated with downregulation of cyclin D1 and Bcl-xL in all four cell lines, and with Bcl2 downregulation in MCF7 and MDA-MB-231 cells.
eIF4E Knockdown Is Not Associated With Activation of Akt but is Associated with a Decrease in 4E-BP1 expression
In previous experiments, inhibition of translation via rapamycin was found to lead to an increase in p-Akt (24, 30). Therefore, we tested the effect of eIF4E knockdown on Akt phosphorylation. We found that eIF4E knockdown did not lead to an increase in Akt phosphorylation in MDA-MB-468, MCF7, MDA-MD-231, and MDA-MB-435 cells. MDA-MB-468 cells, a PTEN mutant cell line with constitutive activation of Akt, had no change in its p-Akt levels, while the latter three cell lines had low p-Akt levels at baseline without activation with eIF4E knockdown (Figure 4A). In contrast, rapamycin treatment enhanced p-Akt levels in MDA-MB-435 cells (Figure 4B). Conditional and modest increases in p-Akt were observed in MCF7 cells, with less robust and less frequent effects noted in MDA-MB-231 and MDA-MB-468 (data not shown). We also evaluated the effect of eIF4E knockdown on the expression of 4E-BP1 and on the expression of S6K1, another mTORC1 target. Of note, western blotting with anti-4E-BP1 antibodies detects multiple bands, representing isoforms generated by differential phosphorylation (29, 31-33), and western blotting with anti-S6K1 antibodies detects multiple bands representing isoforms generated by alternative translation initiation (34) (Kim et al, manuscript in preparation). Interestingly, we found that eIF4E knockdown did decrease the expression of 4E-BP1 expression reproducibly in three of four cell lines, without altering the expression S6K1 (Figure 4C).
Figure 4.
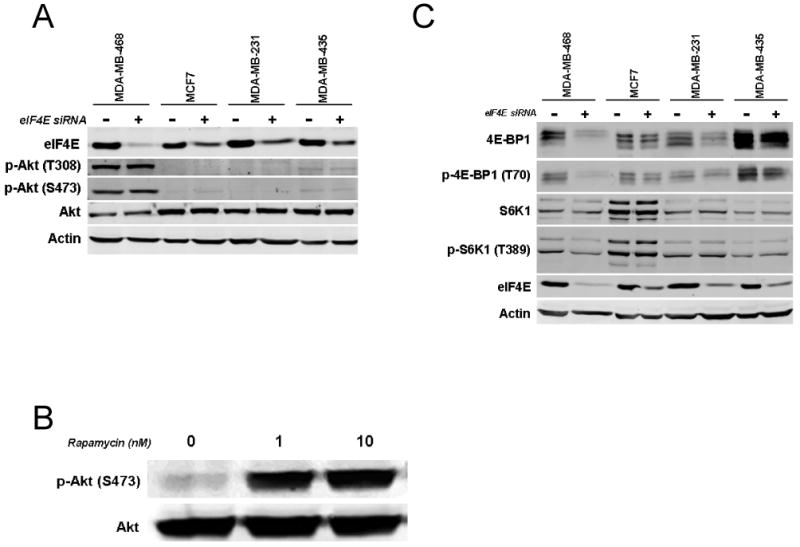
Effect eIF4E knockdown on Akt phosphorylation. A. MDA-MB-468, MCF7, MDA-MB-231 or MDA-MB-435 cells were transfected with control siRNA or eIF4E siRNA (4E3). Three days after transfection, cell lysate was harvested, and western blotting was performed for eIF4E, p-Akt (T308), p-Akt (S473), Akt, and actin. B. MDA-MB-435 cells were treated with vehicle or with 1 or 10nM rapamycin for 24 hours. Western blotting was performed for p-Akt (S473) and Akt. C. MDA-MB-468, MCF7, MDA-MB-231 or MDA-MB-435 cells were transfected with control siRNA or eIF4E siRNA (4E3). Three days after transfection, cell lysate was harvested, and western blotting was performed for 4E-BP1, p-4E-BP1 (T70). S6K1, p-S6K1 (T389), eIF4E, and actin.
Discussion
Activation of translation initiation is critical for cancer cell growth, survival and tumor progression. Thus translation is a rational target for novel cancer therapeutics (35). Here we demonstrate that eIF4E knockdown inhibited growth in different breast cancer cell subtypes including triple-negative tumors for which currently there are no targeted therapies. Further eIF4E knockdown inhibited rapamycin-insensitive as well as sensitive cell lines. eIF4E knockdown decreased polysomal recruitment, and the expression of cyclin D1, Bcl2 and Bcl-xL. eIF4E knockdown did not lead to Akt phosphorylation. These data suggest that eIF4E is a promising target for breast cancer therapy. eIF4E-targeted therapies may have wide efficacy, in tumors driven by different oncogenic stimuli, and may be effective in rapamycin-resistant tumors.
When targeting a critical process like translation, a major question is whether the drug will have significant effects on normal cellular processes, leading to limiting toxicities and too narrow a therapeutic window. In order to address these issues, attempted in vivo siRNA delivery in two independent experiments using neutral liposomal siRNA delivery technologies that have been previously described (36). Unfortunately although we were able to deliver a fluorescent control siRNA into tumor xenografts and normal tissues, we did not observe knockdown of eIF4E at the RNA or protein level with two different 4E siRNA (data not shown). Thus we were unable to address the question of in vivo efficacy or toxicity in our studies. However, recently Graff et al did demonstrate that eIF4E knockdown can be achieved using antisense eIF4E oligonucleotides in a prostate cancer and a breast cancer xenograft model (19). They demonstrated that eIF4E knockdown inhibits in vivo growth, and although 80% eIF4E knockdown was observed in essential organs, no toxicity was observed in these mice models (19). These data provide in vivo evidence that cancers may be more susceptible to eIF4E inhibition than normal tissue and have provided the foundation for bringing eIF4E-specific antisense molecule LY2275796 into Phase 1 clinical studies (37).
Another potential approach to inhibit translation is by interfering with its binding to the 7-methyl guanosine cap, or by interfering with its binding to the multidomain adaptor protein eIF4G, thus interfering with assembly of the translation initiation complex eIF4F. Kentsis et al. reported that the antiviral guanosine analogue ribovirin binds to eIF4E at the site used by the 7-methyl guanosine cap, competing with eIF4E binding and disrupting the transport and translation of mRNAs regulated by eIF4E (38). eIF4E-binding motif peptides can also interfere with eIF4E-eIF4G binding, translation initiation, cell cycle, and survival, providing proof of concept that eIF4E-binding small molecule inhibitors may have utility in cancer therapy (39), (40). Recently, Moerke et al. performed a high-throughput screen for inhibitors of the eIF4E/eIF4G interaction (41). The most potent compound exhibited in vitro activity against multiple cancer cell lines, and appeared to have preferential effect on transformed cells (41).
Cancer therapy remains challenging due to survival pathways that emerge once a promising cancer target is inhibited. Our data suggests that eIF4E knockdown, unlike rapamycin–mediated translational inhibition, is not associated with feedback loop activation of Akt. However, eIF4E knockdown was associated with a decline in 4E-BP1 levels. Larsson et al recently reported that eIF4E overexpression in human mammary epithelial cells led to an increased translational efficiency of 4E-BP1 transcripts (12). This suggests that 4E-BP1 is under eIF4E-mediated translational control, thus eIF4E knockdown-induced decline in 4E-BP1 levels may at least in part attributable to a decrease in its translation. As activation of translation initiation is critical to the malignant phenotype, the cell thus appears to respond to a decrease in eIF4E levels by trying to increase available eIF4E by decreasing its inhibitory protein, a potential compensatory mechanism to maintain active translation.
In conclusion, our results add to the rapidly increasing body of literature demonstrating the critical role of activated translation initiation for cancer cell growth. Our data suggest that eIF4E is a promising therapeutic target in a variety of breast cancer subtypes, including triple negative tumors for which no targeted therapies are available at this time. Strategies to decrease eIF4E expression such as antisense oligonucleotides or novel siRNA delivery systems, as well as novel small molecular inhibitors of eIF4F complex formation may be pursued as potential anti-tumor strategies.
Acknowledgments
We thank Toi Clayton Soh for her assistance with manuscript preparation.
Grant support: This work was supported by the Elsa Pardee Foundation, NIH1 R01 CA112199, U.T. M.D. Anderson Physician-Scientist Award (to F.M.-B.), 1K23CA121994-01 (to A.G.-A.), NIH 5 T32 CA09599 (to A.S. and D. L.) and the U.T. M. D. Anderson Cancer Center Support Core Grant (CA 16672).
Abbreviations used
- eIF4E
eukaryotic translation initiation factor
- 4E-BP1
eIF4E-binding proteins
- 5′ UTR
5′ untranslated region
- mRNP
messenger ribonucleoprotein, mTOR, mammalian target of rapamycin, ER, estrogen receptor
- PR
progesterone receptor
Footnotes
Disclosure of Potential Conflicts of Interest: FMB is a collaborator on Eli Lilly & Co. Clinical Trial (MDACC 2005-0770). The other authors disclosed no potential conflicts of interest.
References
- 1.De Benedetti A, Graff JR. eIF-4E expression and its role in malignancies and metastases. Oncogene. 2004;23:3189–99. doi: 10.1038/sj.onc.1207545. [DOI] [PubMed] [Google Scholar]
- 2.Zimmer SG, DeBenedetti A, Graff JR. Translational control of malignancy: the mRNA cap-binding protein, eIF-4E, as a central regulator of tumor formation, growth, invasion and metastasis. Anticancer Res. 2000;20:1343–51. [PubMed] [Google Scholar]
- 3.Koromilas AE, Lazaris-Karatzas A, Sonenberg N. mRNAs containing extensive secondary structure in their 5′ non-coding region translate efficiently in cells overexpressing initiation factor eIF-4E. Embo J. 1992;11:4153–8. doi: 10.1002/j.1460-2075.1992.tb05508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenwald IB, Kaspar R, Rousseau D, et al. Eukaryotic translation initiation factor 4E regulates expression of cyclin D1 at transcriptional and post-transcriptional levels. J Biol Chem. 1995;270:21176–80. doi: 10.1074/jbc.270.36.21176. [DOI] [PubMed] [Google Scholar]
- 5.Rhoads RE. Regulation of eukaryotic protein synthesis by initiation factors. J Biol Chem. 1993;268:3017–20. [PubMed] [Google Scholar]
- 6.Pelletier J, Sonenberg N. Insertion mutagenesis to increase secondary structure within the 5′ noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell. 1985;40:515–26. doi: 10.1016/0092-8674(85)90200-4. [DOI] [PubMed] [Google Scholar]
- 7.Kozak M. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc Natl Acad Sci U S A. 1986;83:2850–4. doi: 10.1073/pnas.83.9.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li S, Takasu T, Perlman DM, et al. Translation factor eIF4E rescues cells from Myc-dependent apoptosis by inhibiting cytochrome c release. J Biol Chem. 2003;278:3015–22. doi: 10.1074/jbc.M208821200. [DOI] [PubMed] [Google Scholar]
- 9.Yang SX, Hewitt SM, Steinberg SM, Liewehr DJ, Swain SM. Expression levels of eIF4E, VEGF, and cyclin D1, and correlation of eIF4E with VEGF and cyclin D1 in multi-tumor tissue microarray. Oncol Rep. 2007;17:281–7. [PubMed] [Google Scholar]
- 10.De Benedetti A, Rhoads RE. Overexpression of eukaryotic protein synthesis initiation factor 4E in HeLa cells results in aberrant growth and morphology. Proc Natl Acad Sci U S A. 1990;87:8212–6. doi: 10.1073/pnas.87.21.8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fingar DC, Salama S, Tsou C, Harlow E, Blenis J. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 2002;16:1472–87. doi: 10.1101/gad.995802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larsson O, Li S, Issaenko OA, et al. Eukaryotic translation initiation factor 4E induced progression of primary human mammary epithelial cells along the cancer pathway is associated with targeted translational deregulation of oncogenic drivers and inhibitors. Cancer Res. 2007;67:6814–24. doi: 10.1158/0008-5472.CAN-07-0752. [DOI] [PubMed] [Google Scholar]
- 13.Ruggero D, Montanaro L, Ma L, et al. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat Med. 2004;10:484–6. doi: 10.1038/nm1042. [DOI] [PubMed] [Google Scholar]
- 14.Avdulov S, Li S, Michalek V, et al. Activation of translation complex eIF4F is essential for the genesis and maintenance of the malignant phenotype in human mammary epithelial cells. Cancer Cell. 2004;5:553–63. doi: 10.1016/j.ccr.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 15.Nathan CA, Carter P, Liu L, et al. Elevated expression of eIF4E and FGF-2 isoforms during vascularization of breast carcinomas. Oncogene. 1997;15:1087–94. doi: 10.1038/sj.onc.1201272. [DOI] [PubMed] [Google Scholar]
- 16.Li BD, McDonald JC, Nassar R, De Benedetti A. Clinical outcome in stage I to III breast carcinoma and eIF4E overexpression. Ann Surg. 1998;227:756–61. doi: 10.1097/00000658-199805000-00016. discussion 61-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Byrnes K, White S, Chu Q, et al. High eIF4E, VEGF, and microvessel density in stage I to III breast cancer. Ann Surg. 2006;243:684–90. doi: 10.1097/01.sla.0000216770.23642.d8. discussion 91-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li BD, Liu L, Dawson M, De Benedetti A. Overexpression of eukaryotic initiation factor 4E (eIF4E) in breast carcinoma. Cancer. 1997;79:2385–90. [PubMed] [Google Scholar]
- 19.Graff JR, Konicek BW, Vincent TM, et al. Therapeutic suppression of translation initiation factor eIF4E expression reduces tumor growth without toxicity. J Clin Invest. 2007;117:2638–48. doi: 10.1172/JCI32044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dancey JE. Therapeutic targets: MTOR and related pathways. Cancer Biol Ther. 2006;5:1065–73. doi: 10.4161/cbt.5.9.3175. [DOI] [PubMed] [Google Scholar]
- 21.Hudes GR. mTOR as a target for therapy of renal cancer. Clin Adv Hematol Oncol. 2007;5:772–4. [PubMed] [Google Scholar]
- 22.Fingar DC, Richardson CJ, Tee AR, Cheatham L, Tsou C, Blenis J. mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/eukaryotic translation initiation factor 4E. Mol Cell Biol. 2004;24:200–16. doi: 10.1128/MCB.24.1.200-216.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wendel HG, De Stanchina E, Fridman JS, et al. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;428:332–7. doi: 10.1038/nature02369. [DOI] [PubMed] [Google Scholar]
- 24.O'Reilly KE, Rojo F, She QB, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–8. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sachidanandam R. RNAi Codex. 2007 11/20/2007 [cited; Available from: http://codex.cshl.edu/scripts/newmain.pl?content=protocols.html.
- 26.Lynch M, Chen L, Ravitz MJ, et al. hnRNP K binds a core polypyrimidine element in the eukaryotic translation initiation factor 4E (eIF4E) promoter, and its regulation of eIF4E contributes to neoplastic transformation. Mol Cell Biol. 2005;25:6436–53. doi: 10.1128/MCB.25.15.6436-6453.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mondesire WH, Jian W, Zhang H, et al. Targeting mammalian target of rapamycin synergistically enhances chemotherapy-induced cytotoxicity in breast cancer cells. Clin Cancer Res. 2004;10:7031–42. doi: 10.1158/1078-0432.CCR-04-0361. [DOI] [PubMed] [Google Scholar]
- 28.Voigt W. Sulforhodamine B assay and chemosensitivity. Methods Mol Med. 2005;110:39–48. doi: 10.1385/1-59259-869-2:039. [DOI] [PubMed] [Google Scholar]
- 29.Noh WC, Mondesire WH, Peng J, et al. Determinants of rapamycin sensitivity in breast cancer cells. Clin Cancer Res. 2004;10:1013–23. doi: 10.1158/1078-0432.ccr-03-0043. [DOI] [PubMed] [Google Scholar]
- 30.Akcakanat A, Singh G, Hung MC, Meric-Bernstam F. Rapamycin regulates the phosphorylation of rictor. Biochem Biophys Res Commun. 2007;362:330–3. doi: 10.1016/j.bbrc.2007.07.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beretta L, Gingras AC, Svitkin YV, Hall MN, Sonenberg N. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. Embo J. 1996;15:658–64. [PMC free article] [PubMed] [Google Scholar]
- 32.Gingras AC, Kennedy SG, O'Leary MA, Sonenberg N, Hay N. 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt(PKB) signaling pathway. Genes Dev. 1998;12:502–13. doi: 10.1101/gad.12.4.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gingras AC, Raught B, Gygi SP, et al. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 2001;15:2852–64. doi: 10.1101/gad.912401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grove JR, Banerjee P, Balasubramanyam A, et al. Cloning and expression of two human p70 S6 kinase polypeptides differing only at their amino termini. Mol Cell Biol. 1991;11:5541–50. doi: 10.1128/mcb.11.11.5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meric F, Hunt KK. Translation initiation in cancer: a novel target for therapy. Mol Cancer Ther. 2002;1:971–9. [PubMed] [Google Scholar]
- 36.Landen CN, Jr, Chavez-Reyes A, Bucana C, et al. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res. 2005;65:6910–8. doi: 10.1158/0008-5472.CAN-05-0530. [DOI] [PubMed] [Google Scholar]
- 37.Pharmaceuticals I. Featured Research - Journal of Clinical Investigation. 2007 [cited 11/12/2007]; Available from: http://www.isispharm.com/featured_research_JCI_2007-09-05.html.
- 38.Kentsis A, Topisirovic I, Culjkovic B, Shao L, Borden KL. Ribavirin suppresses eIF4E-mediated oncogenic transformation by physical mimicry of the 7-methyl guanosine mRNA cap. Proc Natl Acad Sci U S A. 2004;101:18105–10. doi: 10.1073/pnas.0406927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salaun P, Boulben S, Mulner-Lorillon O, et al. Embryonic-stage-dependent changes in the level of eIF4E-binding proteins during early development of sea urchin embryos. J Cell Sci. 2005;118:1385–94. doi: 10.1242/jcs.01716. [DOI] [PubMed] [Google Scholar]
- 40.Herbert TP, Fahraeus R, Prescott A, Lane DP, Proud CG. Rapid induction of apoptosis mediated by peptides that bind initiation factor eIF4E. Curr Biol. 2000;10:793–6. doi: 10.1016/s0960-9822(00)00567-4. [DOI] [PubMed] [Google Scholar]
- 41.Moerke NJ, Aktas H, Chen H, et al. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell. 2007;128:257–67. doi: 10.1016/j.cell.2006.11.046. [DOI] [PubMed] [Google Scholar]


