Abstract
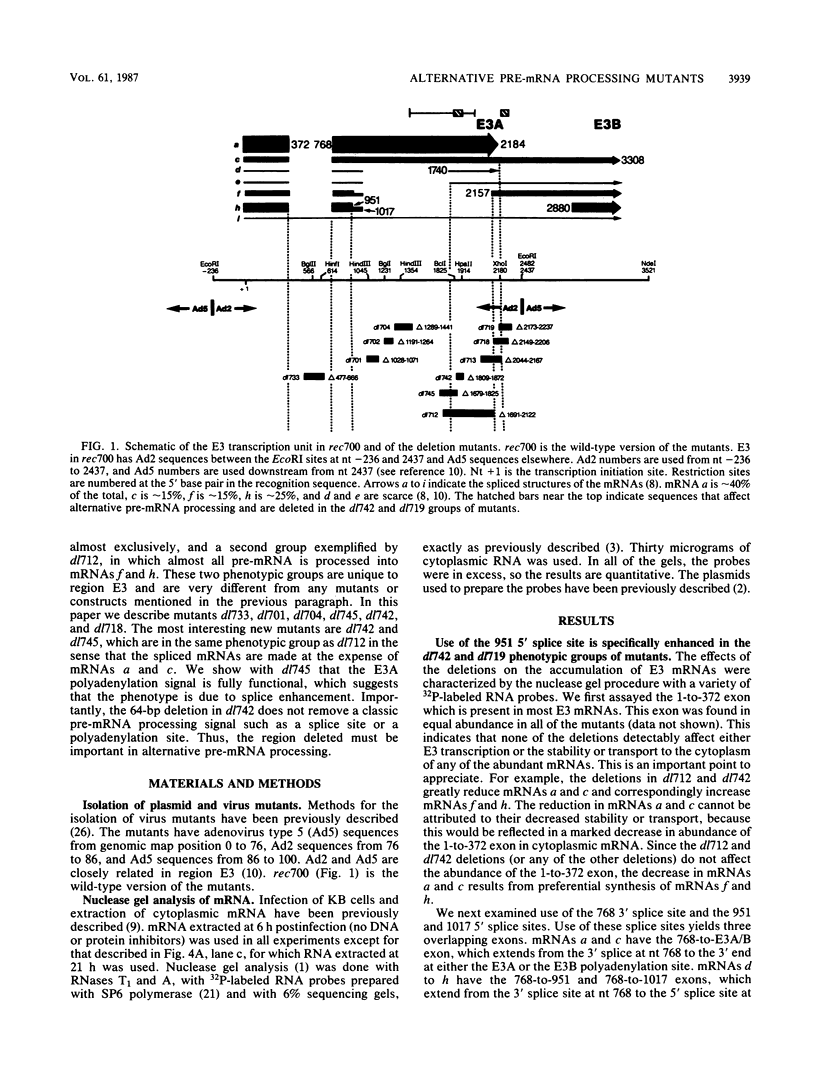
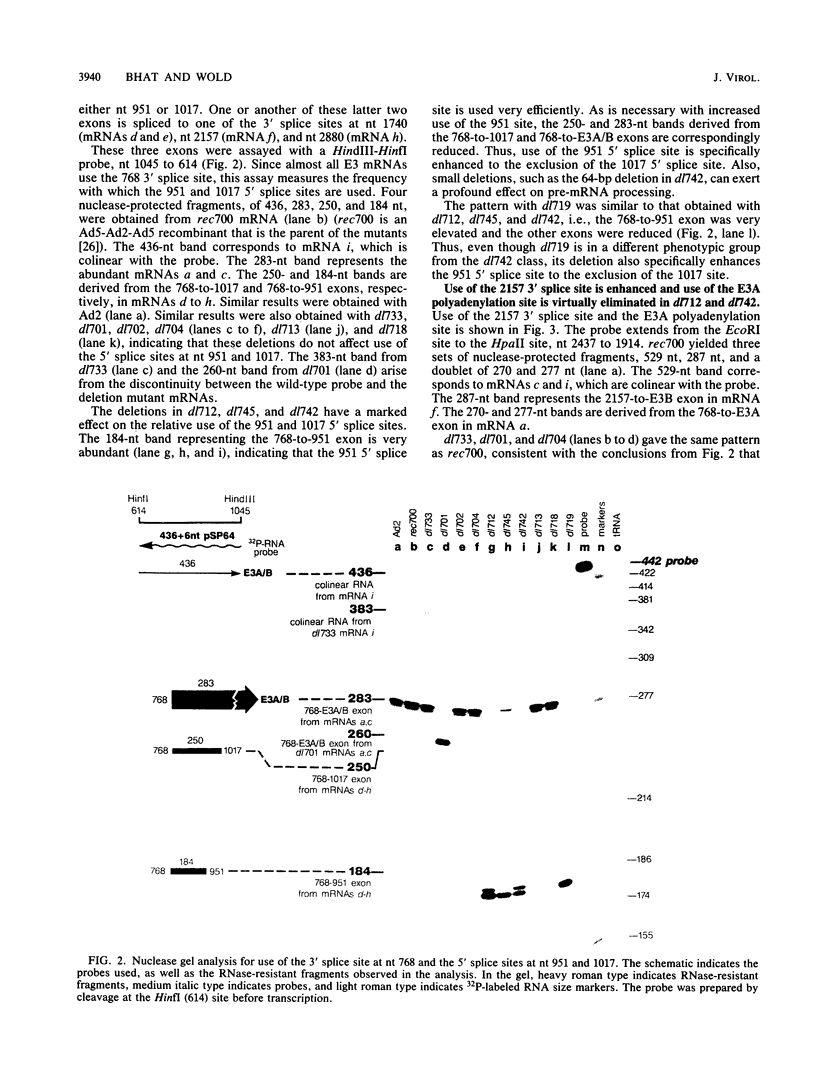
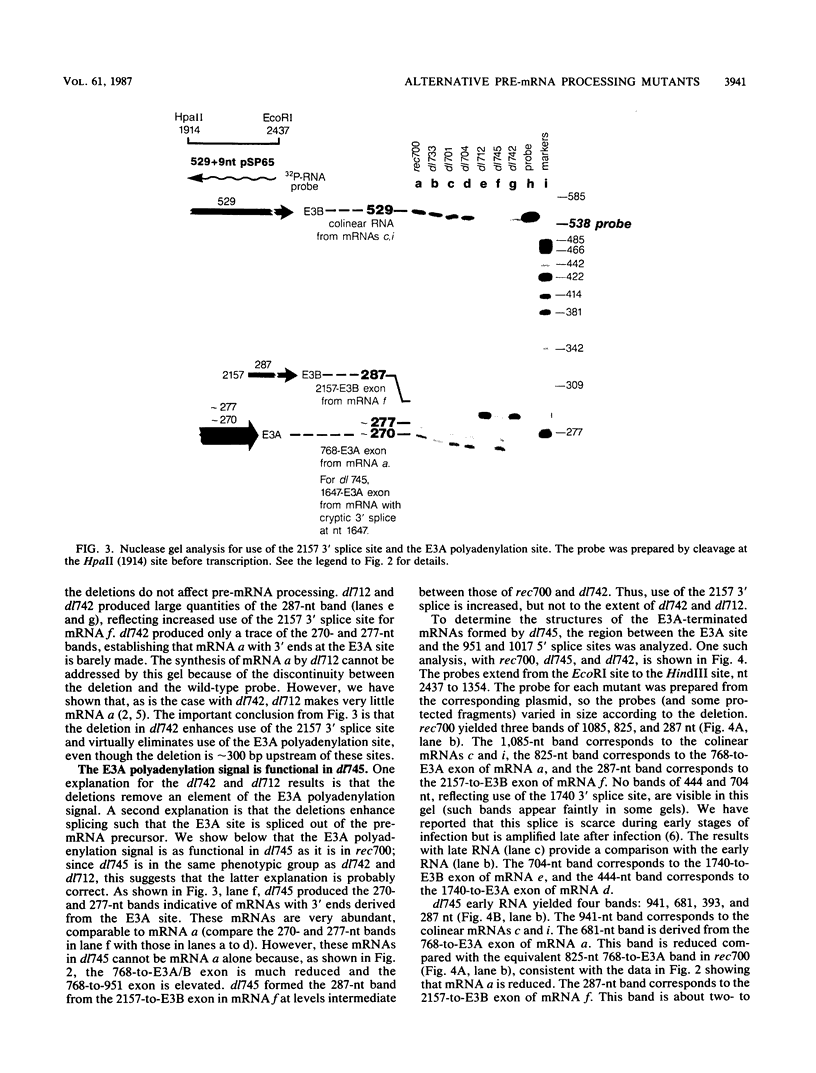
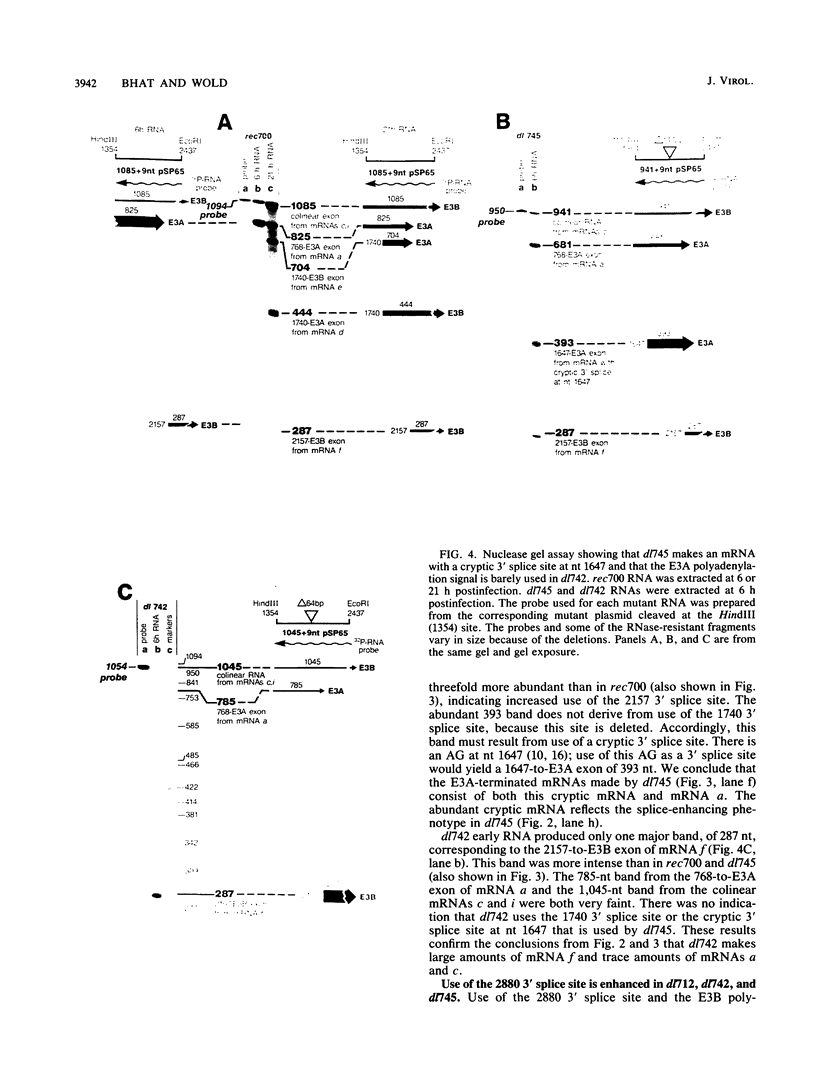
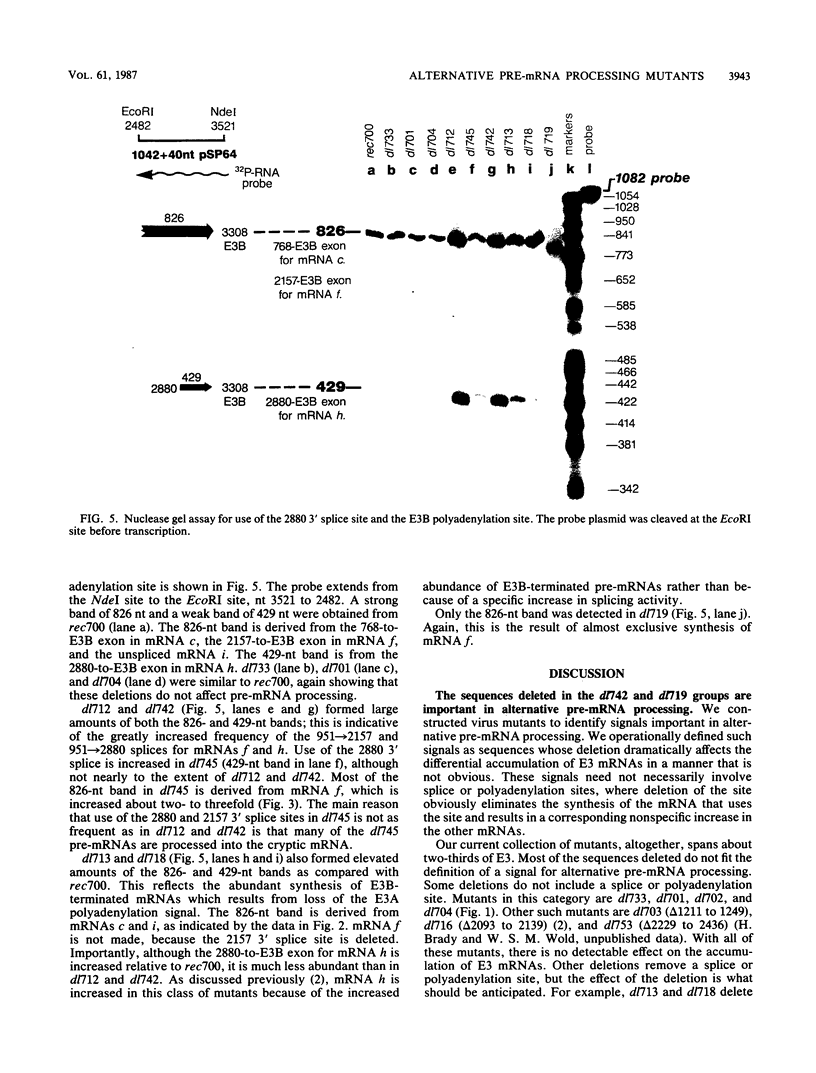
The E3 complex transcription unit of adenovirus encodes overlapping mRNAs (a to i) with different exon structures. The major mRNAs are a (approximately 40% of the total) and c (approximately 15%), which are spliced once, and f (approximately 15%) and h (approximately 25%), which are spliced twice. mRNA a uses the upstream E3A polyadenylation site, and the other mRNAs use the downstream E3B polyadenylation site. We analyzed virus deletion mutants to identify sequences important in alternative pre-mRNA processing in region E3. Our main finding is that a 64-base-pair deletion in dl742 causes mainly mRNAs f and h to be formed. mRNAs a and c are barely made. dl742 does not delete either a splice site or a polyadenylation site. Thus, the sequences deleted must function in alternative pre-mRNA processing independently of the signals at the actual splice and polyadenylation sites. The lack of synthesis of mRNA a by dl742 does not appear to result from a defect in the E3A polyadenylation signal but rather from an increase in splicing activity which results in the synthesis of doubly spliced mRNAs f and h at the expense of singly spliced mRNAs a and c. This suggests, in the wild-type situation, that the frequency of use of the E3A versus the E3B polyadenylation site may be determined by the rate of splicing, as well as, presumably, the rate of cleavage-polyadenylation at the E3A site.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berk A. J., Sharp P. A. Structure of the adenovirus 2 early mRNAs. Cell. 1978 Jul;14(3):695–711. doi: 10.1016/0092-8674(78)90252-0. [DOI] [PubMed] [Google Scholar]
- Bhat B. M., Brady H. A., Pursley M. H., Wold W. S. Deletion mutants that alter differential RNA processing in the E3 complex transcription unit of adenovirus. J Mol Biol. 1986 Aug 20;190(4):543–557. doi: 10.1016/0022-2836(86)90240-8. [DOI] [PubMed] [Google Scholar]
- Bhat B. M., Brady H. A., Wold W. S. Virus deletion mutants that affect a 3' splice site in the E3 transcription unit of adenovirus 2. Mol Cell Biol. 1985 Sep;5(9):2405–2413. doi: 10.1128/mcb.5.9.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat B. M., Wold W. S. ATTAAA as well as downstream sequences are required for RNA 3'-end formation in the E3 complex transcription unit of adenovirus. Mol Cell Biol. 1985 Nov;5(11):3183–3193. doi: 10.1128/mcb.5.11.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat B. M., Wold W. S. Adenovirus mutants with splice-enhancing mutations in the E3 complex transcription unit are also defective in E3A RNA 3'-end formation. J Virol. 1986 Mar;57(3):1155–1158. doi: 10.1128/jvi.57.3.1155-1158.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat B. M., Wold W. S. Genetic analysis of mRNA synthesis in adenovirus region E3 at different stages of productive infection by RNA-processing mutants. J Virol. 1986 Oct;60(1):54–63. doi: 10.1128/jvi.60.1.54-63.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Chow L. T., Broker T. R., Lewis J. B. Complex splicing patterns of RNAs from the early regions of adenovirus-2. J Mol Biol. 1979 Oct 25;134(2):265–303. doi: 10.1016/0022-2836(79)90036-6. [DOI] [PubMed] [Google Scholar]
- Cladaras C., Bhat B., Wold W. S. Mapping the 5' ends, 3' ends, and splice sites of mRNAs from the early E3 transcription unit of adenovirus 5. Virology. 1985 Jan 15;140(1):44–54. doi: 10.1016/0042-6822(85)90444-1. [DOI] [PubMed] [Google Scholar]
- Cladaras C., Wold W. S. DNA sequence of the early E3 transcription unit of adenovirus 5. Virology. 1985 Jan 15;140(1):28–43. doi: 10.1016/0042-6822(85)90443-x. [DOI] [PubMed] [Google Scholar]
- Danner D., Leder P. Role of an RNA cleavage/poly(A) addition site in the production of membrane-bound and secreted IgM mRNA. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8658–8662. doi: 10.1073/pnas.82.24.8658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell J. E., Jr Variety in the level of gene control in eukaryotic cells. Nature. 1982 Jun 3;297(5865):365–371. doi: 10.1038/297365a0. [DOI] [PubMed] [Google Scholar]
- Deutscher S. L., Bhat B. M., Pursley M. H., Cladaras C., Wold W. S. Novel deletion mutants that enhance a distant upstream 5' splice in the E3 transcription unit of adenovirus 2. Nucleic Acids Res. 1985 Aug 26;13(16):5771–5788. doi: 10.1093/nar/13.16.5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallinaro H., Sittler A., Jacob M. In vivo splicing of the premRNAs from early region 3 of adenovirus-2: association of precursors, intermediates and products with hnRNP. Nucleic Acids Res. 1986 May 27;14(10):4171–4185. doi: 10.1093/nar/14.10.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. R. Pre-mRNA splicing. Annu Rev Genet. 1986;20:671–708. doi: 10.1146/annurev.ge.20.120186.003323. [DOI] [PubMed] [Google Scholar]
- Hwang L. S., Park J., Gilboa E. Role of intron-contained sequences in formation of moloney murine leukemia virus env mRNA. Mol Cell Biol. 1984 Nov;4(11):2289–2297. doi: 10.1128/mcb.4.11.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hérissé J., Courtois G., Galibert F. Nucleotide sequence of the EcoRI D fragment of adenovirus 2 genome. Nucleic Acids Res. 1980 May 24;8(10):2173–2192. doi: 10.1093/nar/8.10.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury G., Gruss P., Dhar R., Lai C. J. Processing and expression of early SV40 mRNA: a role for RNA conformation in splicing. Cell. 1979 Sep;18(1):85–92. doi: 10.1016/0092-8674(79)90356-8. [DOI] [PubMed] [Google Scholar]
- Kobrin B. J., Milcarek C., Morrison S. L. Sequences near the 3' secretion-specific polyadenylation site influence levels of secretion-specific and membrane-specific IgG2b mRNA in myeloma cells. Mol Cell Biol. 1986 May;6(5):1687–1697. doi: 10.1128/mcb.6.5.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leff S. E., Rosenfeld M. G., Evans R. M. Complex transcriptional units: diversity in gene expression by alternative RNA processing. Annu Rev Biochem. 1986;55:1091–1117. doi: 10.1146/annurev.bi.55.070186.005303. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K., Vuocolo G. A. Synthesis of two mRNAs by utilization of alternate polyadenylation sites: expression of SV40-mouse immunoglobulin mu chain gene recombinants in Cos monkey cells. EMBO J. 1984 Apr;3(4):689–699. doi: 10.1002/j.1460-2075.1984.tb01871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson M. L., Perry R. P. Regulated production of mu m and mu s mRNA requires linkage of the poly(A) addition sites and is dependent on the length of the mu s-mu m intron. Proc Natl Acad Sci U S A. 1986 Dec;83(23):8883–8887. doi: 10.1073/pnas.83.23.8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somasekhar M. B., Mertz J. E. Exon mutations that affect the choice of splice sites used in processing the SV40 late transcripts. Nucleic Acids Res. 1985 Aug 12;13(15):5591–5609. doi: 10.1093/nar/13.15.5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulfendahl P. J., Pettersson U., Akusjärvi G. Splicing of the adenovirus-2 E1A 13S mRNA requires a minimal intron length and specific intron signals. Nucleic Acids Res. 1985 Sep 11;13(17):6299–6315. doi: 10.1093/nar/13.17.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold W. S., Deutscher S. L., Takemori N., Bhat B. M., Magie S. C. Evidence that AGUAUAUGA and CCAAGAUGA initiate translation in the same mRNA region E3 of adenovirus. Virology. 1986 Jan 15;148(1):168–180. doi: 10.1016/0042-6822(86)90412-5. [DOI] [PubMed] [Google Scholar]
- Zeevi M., Nevins J. R., Darnell J. E., Jr Nuclear RNA is spliced in the absence of poly(A) addition. Cell. 1981 Oct;26(1 Pt 1):39–46. doi: 10.1016/0092-8674(81)90031-3. [DOI] [PubMed] [Google Scholar]








