Abstract
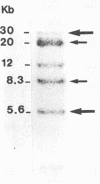
fim-3 is a new proviral integration region involved in 23% (16 of 68) of Friend murine leukemia virus-induced myeloblastic leukemias. This region is distinct from 20 oncogenes and from putative oncogenes tested so far. Proviruses are integrated in a 16-kilobase region, always in the same orientation. No RNA expression of fim-3 was detected.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cuypers H. T., Selten G. C., Zijlstra M., de Goede R. E., Melief C. J., Berns A. J. Tumor progression in murine leukemia virus-induced T-cell lymphomas: monitoring clonal selections with viral and cellular probes. J Virol. 1986 Oct;60(1):230–241. doi: 10.1128/jvi.60.1.230-241.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisselbrecht S., Fichelson S., Sola B., Bordereaux D., Hampe A., André C., Galibert F., Tambourin P. Frequent c-fms activation by proviral insertion in mouse myeloblastic leukaemias. Nature. 1987 Sep 17;329(6136):259–261. doi: 10.1038/329259a0. [DOI] [PubMed] [Google Scholar]
- Graham M., Adams J. M., Cory S. Murine T lymphomas with retroviral inserts in the chromosomal 15 locus for plasmacytoma variant translocations. 1985 Apr 25-May 1Nature. 314(6013):740–743. doi: 10.1038/314740a0. [DOI] [PubMed] [Google Scholar]
- Moreau-Gachelin F., Robert-Lezenes J., Mathieu-Mahul D., Gisselbrecht S., Larsen C. J. Isolation and characterization of a gp 70+ non producer Friend tumor cell clone. Biochimie. 1983 Apr-May;65(4-5):259–266. doi: 10.1016/s0300-9084(83)80277-6. [DOI] [PubMed] [Google Scholar]
- Selten G., Cuypers H. T., Berns A. Proviral activation of the putative oncogene Pim-1 in MuLV induced T-cell lymphomas. EMBO J. 1985 Jul;4(7):1793–1798. doi: 10.1002/j.1460-2075.1985.tb03852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selten G., Cuypers H. T., Zijlstra M., Melief C., Berns A. Involvement of c-myc in MuLV-induced T cell lymphomas in mice: frequency and mechanisms of activation. EMBO J. 1984 Dec 20;3(13):3215–3222. doi: 10.1002/j.1460-2075.1984.tb02281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya T., Mak T. W. Host control of susceptibility to erythroleukemia and to the types of leukemia induced by Friend murine leukemia virus: initial and late stages. Cell. 1982 Dec;31(2 Pt 1):483–493. doi: 10.1016/0092-8674(82)90141-6. [DOI] [PubMed] [Google Scholar]
- Silver J. E., Fredrickson T. N. A new gene that controls the type of leukemia induced by Friend murine leukemia virus. J Exp Med. 1983 Aug 1;158(2):493–505. doi: 10.1084/jem.158.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola B., Fichelson S., Bordereaux D., Tambourin P. E., Gisselbrecht S. fim-1 and fim-2: two new integration regions of Friend murine leukemia virus in myeloblastic leukemias. J Virol. 1986 Nov;60(2):718–725. doi: 10.1128/jvi.60.2.718-725.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsichlis P. N., Strauss P. G., Lohse M. A. Concerted DNA rearrangements in Moloney murine leukemia virus-induced thymomas: a potential synergistic relationship in oncogenesis. J Virol. 1985 Oct;56(1):258–267. doi: 10.1128/jvi.56.1.258-267.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]