Abstract
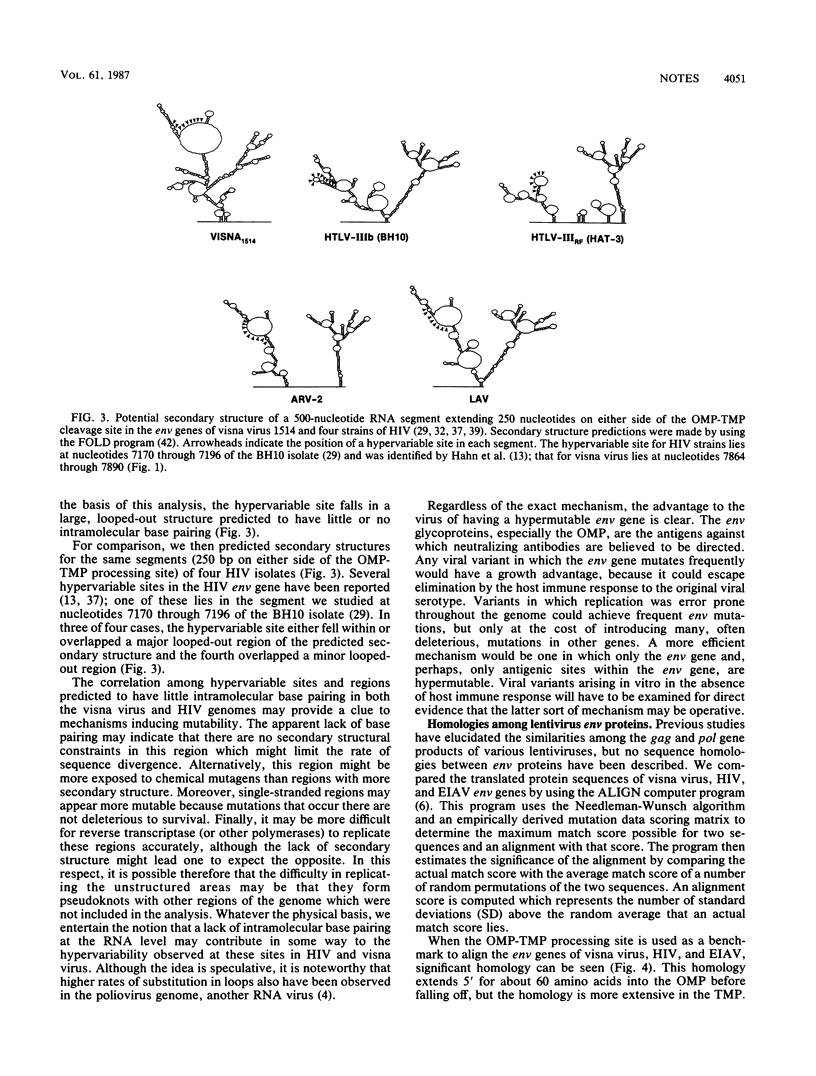
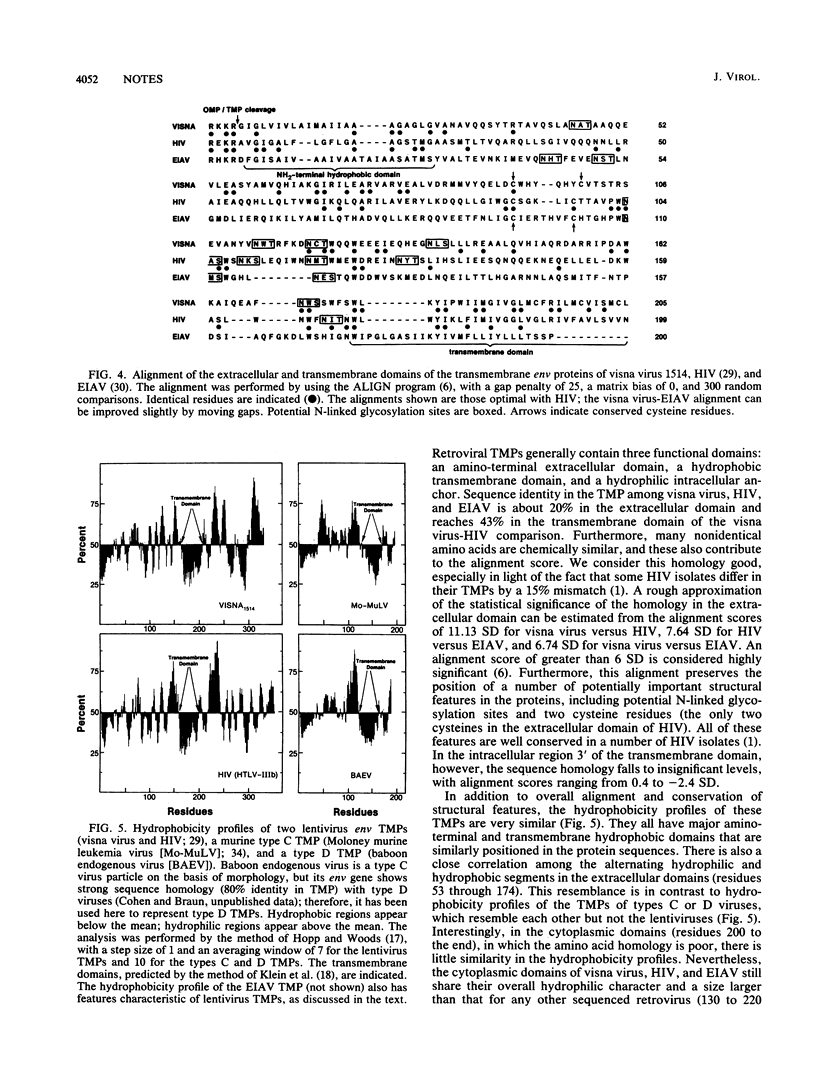
The complete nucleotide sequence of the visna virus 1514 genome was determined. Our sequence confirms the relationship of visna virus and other lentiviruses to human immunodeficiency virus (HIV) both at the level of sequence homology and of genomic organization. Sequence homology is shown to extend to the transmembrane proteins of lentivirus env genes; this homology is strongest in the extracellular domain, suggesting that close structural and functional similarities may also exist among these envelope proteins. Comparison of our data with the sequence of visna virus LV1-1, an antigenic variant derived from strain 1514, demonstrates that the rate of divergence has been about 1.7 x 10(-3) substitutions per nucleotide per year in vivo. This rate is orders of magnitude higher than that for most DNA genomes, but agrees well with estimates of the rate for HIV. A statistically significant cluster of mutations in the env gene appears to represent a hypervariable site and may correspond to the epitope responsible for the antigenic differences between 1514 and LV1-1. Analysis of the potential RNA folding pattern of the visna virus env gene shows that this hypervariable site falls within a region with little potential for intramolecular base pairing. This correlation of hypervariability with lack of RNA secondary structure is strengthened by the fact that it also holds for a hypervariable site in the env gene of HIV.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alizon M., Wain-Hobson S., Montagnier L., Sonigo P. Genetic variability of the AIDS virus: nucleotide sequence analysis of two isolates from African patients. Cell. 1986 Jul 4;46(1):63–74. doi: 10.1016/0092-8674(86)90860-3. [DOI] [PubMed] [Google Scholar]
- Chiu I. M., Yaniv A., Dahlberg J. E., Gazit A., Skuntz S. F., Tronick S. R., Aaronson S. A. Nucleotide sequence evidence for relationship of AIDS retrovirus to lentiviruses. 1985 Sep 26-Oct 2Nature. 317(6035):366–368. doi: 10.1038/317366a0. [DOI] [PubMed] [Google Scholar]
- Clements J. E., D'Antonio N., Narayan O. Genomic changes associated with antigenic variation of visna virus. II. Common nucleotide sequence changes detected in variants from independent isolations. J Mol Biol. 1982 Jul 5;158(3):415–434. doi: 10.1016/0022-2836(82)90207-8. [DOI] [PubMed] [Google Scholar]
- Currey K. M., Peterlin B. M., Maizel J. V., Jr Secondary structure of poliovirus RNA: correlation of computer-predicted with electron microscopically observed structure. Virology. 1986 Jan 15;148(1):33–46. doi: 10.1016/0042-6822(86)90401-0. [DOI] [PubMed] [Google Scholar]
- Daniel M. D., Letvin N. L., King N. W., Kannagi M., Sehgal P. K., Hunt R. D., Kanki P. J., Essex M., Desrosiers R. C. Isolation of T-cell tropic HTLV-III-like retrovirus from macaques. Science. 1985 Jun 7;228(4704):1201–1204. doi: 10.1126/science.3159089. [DOI] [PubMed] [Google Scholar]
- Dayhoff M. O., Barker W. C., Hunt L. T. Establishing homologies in protein sequences. Methods Enzymol. 1983;91:524–545. doi: 10.1016/s0076-6879(83)91049-2. [DOI] [PubMed] [Google Scholar]
- Deininger P. L. Random subcloning of sonicated DNA: application to shotgun DNA sequence analysis. Anal Biochem. 1983 Feb 15;129(1):216–223. doi: 10.1016/0003-2697(83)90072-6. [DOI] [PubMed] [Google Scholar]
- Gojobori T., Li W. H., Graur D. Patterns of nucleotide substitution in pseudogenes and functional genes. J Mol Evol. 1982;18(5):360–369. doi: 10.1007/BF01733904. [DOI] [PubMed] [Google Scholar]
- Gojobori T., Yokoyama S. Rates of evolution of the retroviral oncogene of Moloney murine sarcoma virus and of its cellular homologues. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4198–4201. doi: 10.1073/pnas.82.12.4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonda M. A., Braun M. J., Clements J. E., Pyper J. M., Wong-Staal F., Gallo R. C., Gilden R. V. Human T-cell lymphotropic virus type III shares sequence homology with a family of pathogenic lentiviruses. Proc Natl Acad Sci U S A. 1986 Jun;83(11):4007–4011. doi: 10.1073/pnas.83.11.4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonda M. A., Wong-Staal F., Gallo R. C., Clements J. E., Narayan O., Gilden R. V. Sequence homology and morphologic similarity of HTLV-III and visna virus, a pathogenic lentivirus. Science. 1985 Jan 11;227(4683):173–177. doi: 10.1126/science.2981428. [DOI] [PubMed] [Google Scholar]
- Haase A. T. Pathogenesis of lentivirus infections. Nature. 1986 Jul 10;322(6075):130–136. doi: 10.1038/322130a0. [DOI] [PubMed] [Google Scholar]
- Hahn B. H., Shaw G. M., Taylor M. E., Redfield R. R., Markham P. D., Salahuddin S. Z., Wong-Staal F., Gallo R. C., Parks E. S., Parks W. P. Genetic variation in HTLV-III/LAV over time in patients with AIDS or at risk for AIDS. Science. 1986 Jun 20;232(4757):1548–1553. doi: 10.1126/science.3012778. [DOI] [PubMed] [Google Scholar]
- Harris J. D., Blum H., Scott J., Traynor B., Ventura P., Haase A. Slow virus visna: reproduction in vitro of virus from extrachromosomal DNA. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7212–7215. doi: 10.1073/pnas.81.22.7212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess J. L., Clements J. E., Narayan O. cis- and trans-acting transcriptional regulation of visna virus. Science. 1985 Aug 2;229(4712):482–485. doi: 10.1126/science.2990051. [DOI] [PubMed] [Google Scholar]
- Holland J., Spindler K., Horodyski F., Grabau E., Nichol S., VandePol S. Rapid evolution of RNA genomes. Science. 1982 Mar 26;215(4540):1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- Hopp T. P., Woods K. R. Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein P., Kanehisa M., DeLisi C. The detection and classification of membrane-spanning proteins. Biochim Biophys Acta. 1985 May 28;815(3):468–476. doi: 10.1016/0005-2736(85)90375-x. [DOI] [PubMed] [Google Scholar]
- Li W. H., Gojobori T., Nei M. Pseudogenes as a paradigm of neutral evolution. Nature. 1981 Jul 16;292(5820):237–239. doi: 10.1038/292237a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Molineaux S., Clements J. E. Molecular cloning of unintegrated visna viral DNA and characterization of frequent deletions in the 3' terminus. Gene. 1983 Aug;23(2):137–148. doi: 10.1016/0378-1119(83)90045-8. [DOI] [PubMed] [Google Scholar]
- Narayan O., Griffin D. E., Chase J. Antigenic shift of visna virus in persistently infected sheep. Science. 1977 Jul 22;197(4301):376–378. doi: 10.1126/science.195339. [DOI] [PubMed] [Google Scholar]
- Narayan O., Griffin D. E., Clements J. E. Virus mutation during 'slow infection': temporal development and characterization of mutants of visna virus recovered from sheep. J Gen Virol. 1978 Nov;41(2):343–352. doi: 10.1099/0022-1317-41-2-343. [DOI] [PubMed] [Google Scholar]
- Narayan O., Kennedy-Stoskopf S., Sheffer D., Griffin D. E., Clements J. E. Activation of caprine arthritis-encephalitis virus expression during maturation of monocytes to macrophages. Infect Immun. 1983 Jul;41(1):67–73. doi: 10.1128/iai.41.1.67-73.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan O., Sheffer D., Griffin D. E., Clements J., Hess J. Lack of neutralizing antibodies to caprine arthritis-encephalitis lentivirus in persistently infected goats can be overcome by immunization with inactivated Mycobacterium tuberculosis. J Virol. 1984 Feb;49(2):349–355. doi: 10.1128/jvi.49.2.349-355.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Payne S., Parekh B., Montelaro R. C., Issel C. J. Genomic alterations associated with persistent infections by equine infectious anaemia virus, a retrovirus. J Gen Virol. 1984 Aug;65(Pt 8):1395–1399. doi: 10.1099/0022-1317-65-8-1395. [DOI] [PubMed] [Google Scholar]
- Pedersen N. C., Ho E. W., Brown M. L., Yamamoto J. K. Isolation of a T-lymphotropic virus from domestic cats with an immunodeficiency-like syndrome. Science. 1987 Feb 13;235(4790):790–793. doi: 10.1126/science.3643650. [DOI] [PubMed] [Google Scholar]
- Ratner L., Haseltine W., Patarca R., Livak K. J., Starcich B., Josephs S. F., Doran E. R., Rafalski J. A., Whitehorn E. A., Baumeister K. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985 Jan 24;313(6000):277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- Rushlow K., Olsen K., Stiegler G., Payne S. L., Montelaro R. C., Issel C. J. Lentivirus genomic organization: the complete nucleotide sequence of the env gene region of equine infectious anemia virus. Virology. 1986 Dec;155(2):309–321. doi: 10.1016/0042-6822(86)90195-9. [DOI] [PubMed] [Google Scholar]
- Salinovich O., Payne S. L., Montelaro R. C., Hussain K. A., Issel C. J., Schnorr K. L. Rapid emergence of novel antigenic and genetic variants of equine infectious anemia virus during persistent infection. J Virol. 1986 Jan;57(1):71–80. doi: 10.1128/jvi.57.1.71-80.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Pescador R., Power M. D., Barr P. J., Steimer K. S., Stempien M. M., Brown-Shimer S. L., Gee W. W., Renard A., Randolph A., Levy J. A. Nucleotide sequence and expression of an AIDS-associated retrovirus (ARV-2). Science. 1985 Feb 1;227(4686):484–492. doi: 10.1126/science.2578227. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinnick T. M., Lerner R. A., Sutcliffe J. G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981 Oct 15;293(5833):543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- Sonigo P., Alizon M., Staskus K., Klatzmann D., Cole S., Danos O., Retzel E., Tiollais P., Haase A., Wain-Hobson S. Nucleotide sequence of the visna lentivirus: relationship to the AIDS virus. Cell. 1985 Aug;42(1):369–382. doi: 10.1016/s0092-8674(85)80132-x. [DOI] [PubMed] [Google Scholar]
- Staden R. Automation of the computer handling of gel reading data produced by the shotgun method of DNA sequencing. Nucleic Acids Res. 1982 Aug 11;10(15):4731–4751. doi: 10.1093/nar/10.15.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starcich B. R., Hahn B. H., Shaw G. M., McNeely P. D., Modrow S., Wolf H., Parks E. S., Parks W. P., Josephs S. F., Gallo R. C. Identification and characterization of conserved and variable regions in the envelope gene of HTLV-III/LAV, the retrovirus of AIDS. Cell. 1986 Jun 6;45(5):637–648. doi: 10.1016/0092-8674(86)90778-6. [DOI] [PubMed] [Google Scholar]
- Stephens R. M., Casey J. W., Rice N. R. Equine infectious anemia virus gag and pol genes: relatedness to visna and AIDS virus. Science. 1986 Feb 7;231(4738):589–594. doi: 10.1126/science.3003905. [DOI] [PubMed] [Google Scholar]
- Wain-Hobson S., Sonigo P., Danos O., Cole S., Alizon M. Nucleotide sequence of the AIDS virus, LAV. Cell. 1985 Jan;40(1):9–17. doi: 10.1016/0092-8674(85)90303-4. [DOI] [PubMed] [Google Scholar]
- Weiss R. A., Clapham P. R., Cheingsong-Popov R., Dalgleish A. G., Carne C. A., Weller I. V., Tedder R. S. Neutralization of human T-lymphotropic virus type III by sera of AIDS and AIDS-risk patients. Nature. 1985 Jul 4;316(6023):69–72. doi: 10.1038/316069a0. [DOI] [PubMed] [Google Scholar]
- Wong-Staal F., Gallo R. C. Human T-lymphotropic retroviruses. Nature. 1985 Oct 3;317(6036):395–403. doi: 10.1038/317395a0. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]


