Abstract
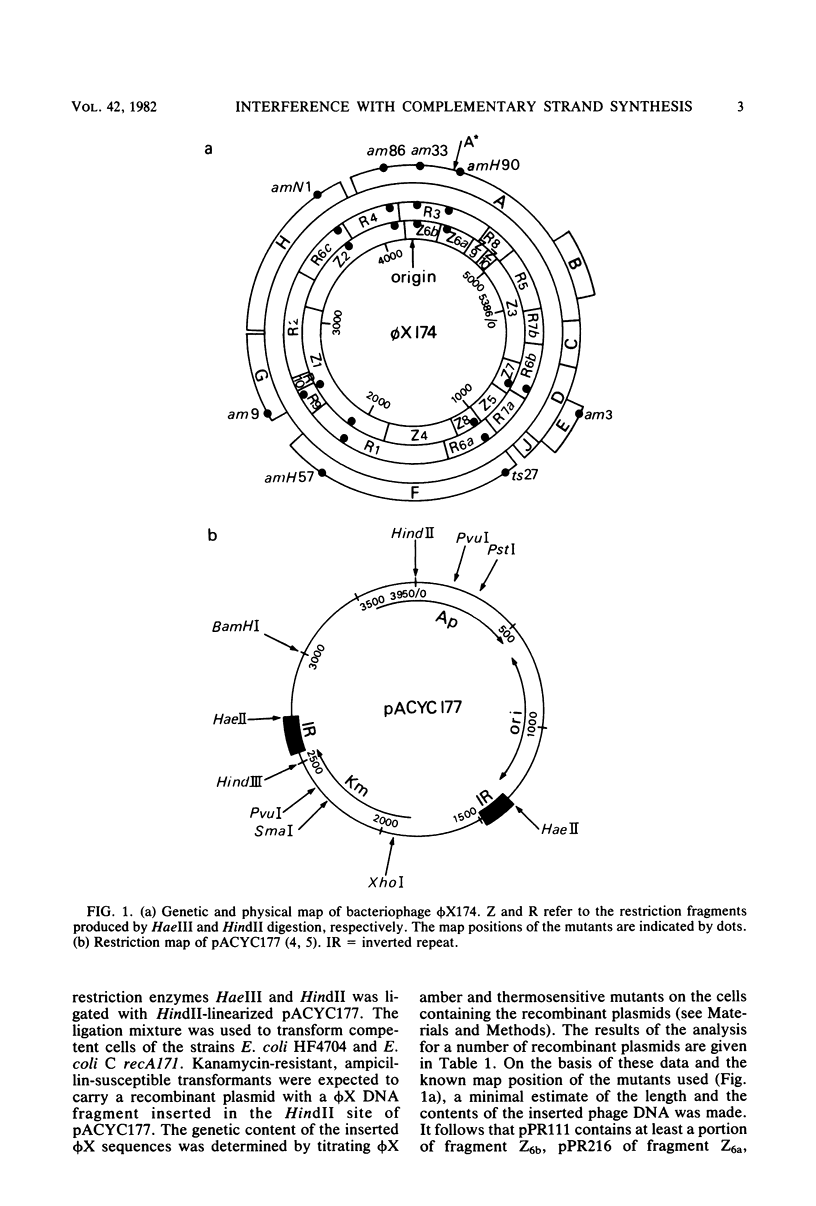
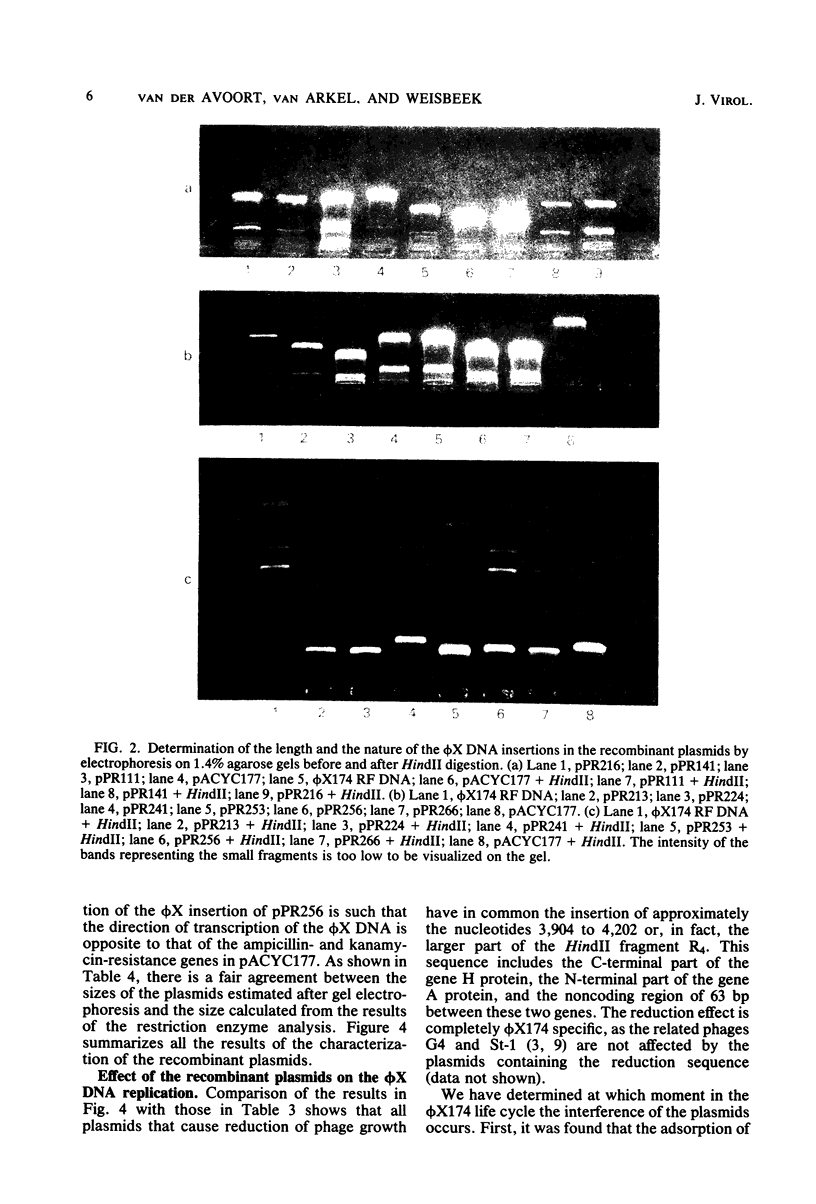
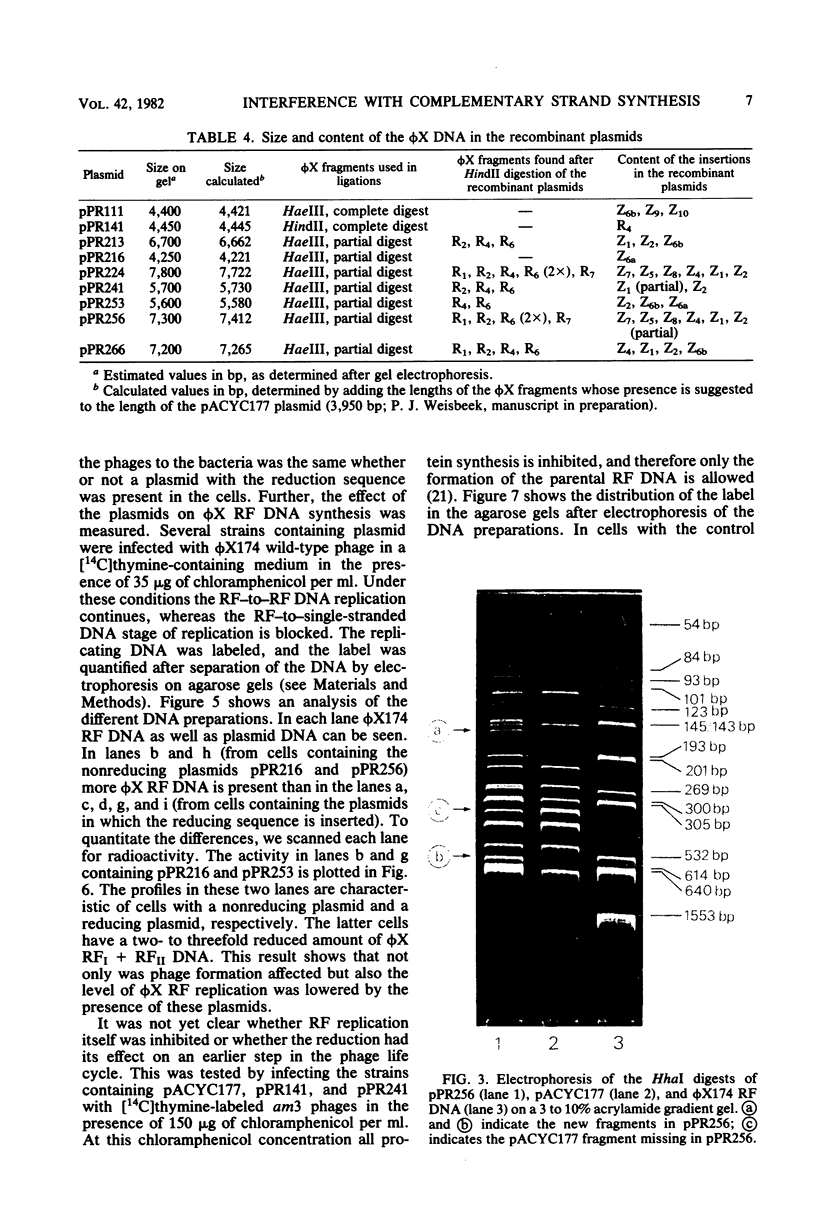
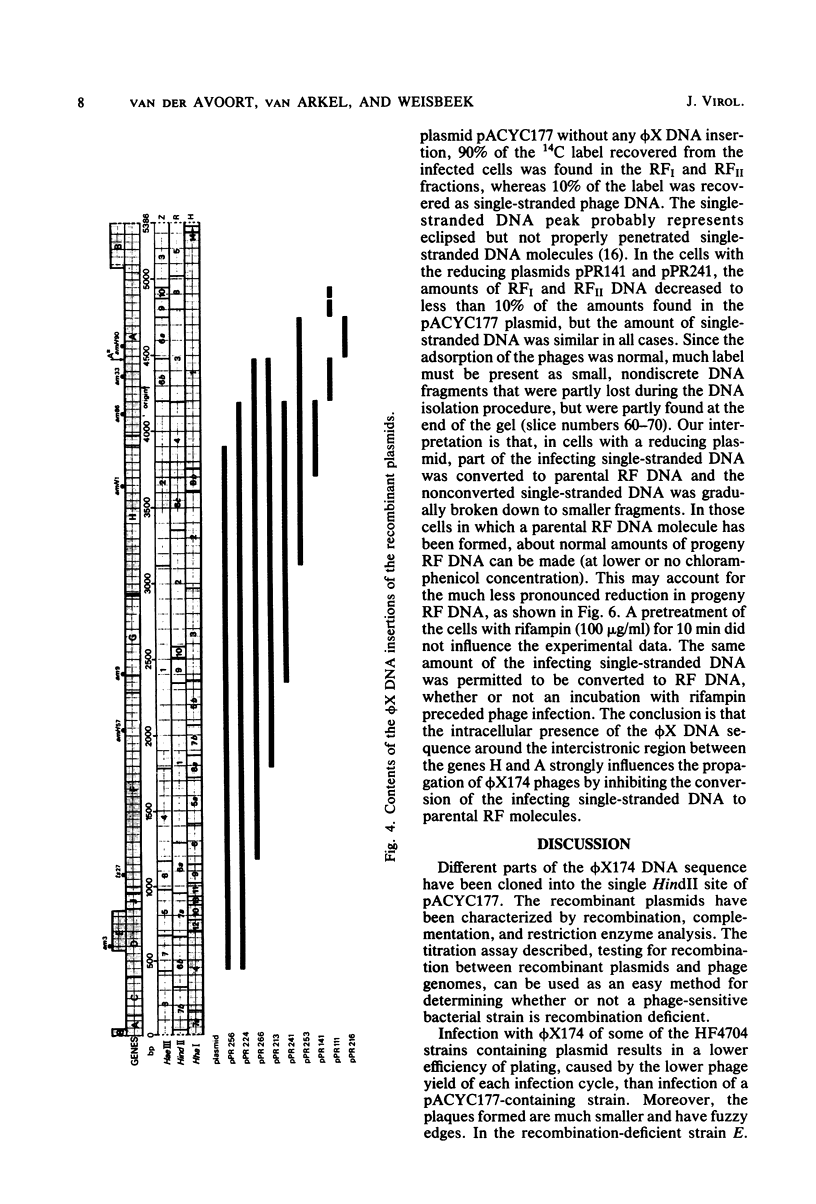
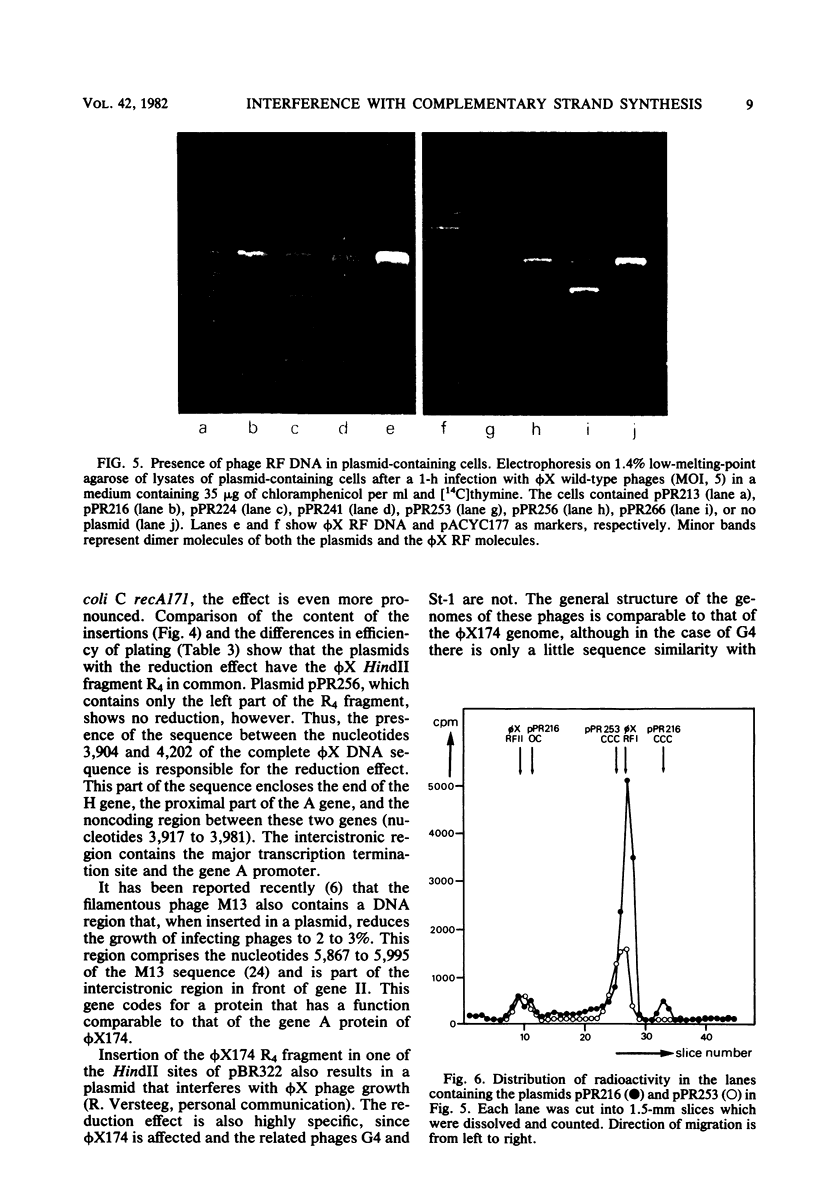
The insertion of a particular phi X DNA sequence in the plasmid pACYC177 strongly decreased the capacity of Escherichia coli cells containing such a plasmid to propagate bacteriophage phi X174. The smallest DNA sequence tested that showed the effect was the HindII fragment R4. This fragment does not code for a complete protein. It contains the sequence specifying the C-terminal part of the gene H protein and the N-terminal part of the gene A protein, as well as the noncoding region between these genes. Analysis of cells that contain plasmids with the "reduction sequence" showed that (i) the adsorption of the phages to the host cells is normal, (ii) in a single infection cycle much less phage is formed, (iii) only 10% of the infecting viral single-stranded DNA is converted to double-stranded replicative-form DNA, and (iv) less progeny replicative form DNA is synthesized. The reduction process is phi X174 specific, since the growth of the related G4 and St-1 phages was not affected in these cells. The effect of the recombinant plasmids on infecting phage DNA shows similarity to the process of superinfection exclusion.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baas P. D., Jansz H. S. Bacteriophage phiX174 DNA synthesis in a replication-deficient host: determination of the origin of phiX DNA replication. J Mol Biol. 1976 Apr 15;102(3):633–656. doi: 10.1016/0022-2836(76)90339-9. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley D. E. A comparative study of some properties of the phi-X174 type bacteriophages. Can J Microbiol. 1970 Oct;16(10):965–971. doi: 10.1139/m70-165. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary J. M., Ray D. S. Replication of the plasmid pBR322 under the control of a cloned replication origin from the single-stranded DNA phage M13. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4638–4642. doi: 10.1073/pnas.77.8.4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg S., Griffith J., Kornberg A. phiX174 cistron A protein is a multifunctional enzyme in DNA replication. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3198–3202. doi: 10.1073/pnas.74.8.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg S., Scott J. F., Kornberg A. An enzyme system for replication of duplex circular DNA: the replicative form of phage phi X174. Proc Natl Acad Sci U S A. 1976 May;73(5):1594–1597. doi: 10.1073/pnas.73.5.1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godson G. N., Barrell B. G., Staden R., Fiddes J. C. Nucleotide sequence of bacteriophage G4 DNA. Nature. 1978 Nov 16;276(5685):236–247. doi: 10.1038/276236a0. [DOI] [PubMed] [Google Scholar]
- Godson G. N. Evolution of phi-chi 174. Isolation of four new phi-chi-like phages and comparison with phi-chi 174. Virology. 1974 Mar;58(1):272–289. doi: 10.1016/0042-6822(74)90161-5. [DOI] [PubMed] [Google Scholar]
- Humayun M. Z., Chambers R. W. Construction and characterization of an Escherichia coli plasmid bearing a functional gene G of bacteriophage phiX174. Proc Natl Acad Sci U S A. 1978 Feb;75(2):774–778. doi: 10.1073/pnas.75.2.774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison C. A., 3rd, Sinsheimer R. L. Requirement of protein synthesis for bacteriophage phi X174 superinfection exclusion. J Virol. 1971 Jul;8(1):121–124. doi: 10.1128/jvi.8.1.121-124.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langeveld S. A., van Mansfeld A. D., Baas P. D., Jansz H. S., van Arkel G. A., Weisbeek P. J. Nucleotide sequence of the origin of replication in bacteriophage phiX174 RF DNA. Nature. 1978 Feb 2;271(5644):417–420. doi: 10.1038/271417a0. [DOI] [PubMed] [Google Scholar]
- Lindqvist B. H., Sinsheimer R. L. The process of infection with bacteriophage phi-X174. XV. Bacteriophage DNA synthesis in abortive infections with a set of conditional lethal mutants. J Mol Biol. 1967 Nov 28;30(1):69–80. doi: 10.1016/0022-2836(67)90244-6. [DOI] [PubMed] [Google Scholar]
- Murray N. E., Bruce S. A., Murray K. Molecular cloning of the DNA ligase gene from bacteriophage T4. II. Amplification and preparation of the gene product. J Mol Biol. 1979 Aug 15;132(3):493–505. doi: 10.1016/0022-2836(79)90271-7. [DOI] [PubMed] [Google Scholar]
- Newbold J. E., Sinsheimer R. L. The process of infection with bacteriophage phiX174. XXXII. Early steps in the infection process: attachment, eclipse and DNA penetration. J Mol Biol. 1970 Apr 14;49(1):49–66. doi: 10.1016/0022-2836(70)90375-x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Friedmann T., Air G. M., Barrell B. G., Brown N. L., Fiddes J. C., Hutchison C. A., 3rd, Slocombe P. M., Smith M. The nucleotide sequence of bacteriophage phiX174. J Mol Biol. 1978 Oct 25;125(2):225–246. doi: 10.1016/0022-2836(78)90346-7. [DOI] [PubMed] [Google Scholar]
- Shlomai J., Kornberg A. An Escherichia coli replication protein that recognizes a unique sequence within a hairpin region in phi X174 DNA. Proc Natl Acad Sci U S A. 1980 Feb;77(2):799–803. doi: 10.1073/pnas.77.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Tessman E. S., Borrás M. T., Sun I. L. Superinfection in bacteriophage S13 and determination of the number of bacteriophage particles which can function in an infected cell. J Virol. 1971 Jul;8(1):111–120. doi: 10.1128/jvi.8.1.111-120.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessman E. S. Mutants of bacteriophage S13 blocked in infectious DNA synthesis. J Mol Biol. 1966 May;17(1):218–236. doi: 10.1016/s0022-2836(66)80104-3. [DOI] [PubMed] [Google Scholar]
- Yarus M. J., Sinsheimer R. L. The process of infection with bacteriophage phiX174. 8. Evidence for an essential bacterial "site". J Virol. 1967 Feb;1(1):135–144. doi: 10.1128/jvi.1.1.135-144.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wezenbeek P. M., Hulsebos T. J., Schoenmakers J. G. Nucleotide sequence of the filamentous bacteriophage M13 DNA genome: comparison with phage fd. Gene. 1980 Oct;11(1-2):129–148. doi: 10.1016/0378-1119(80)90093-1. [DOI] [PubMed] [Google Scholar]






