Abstract
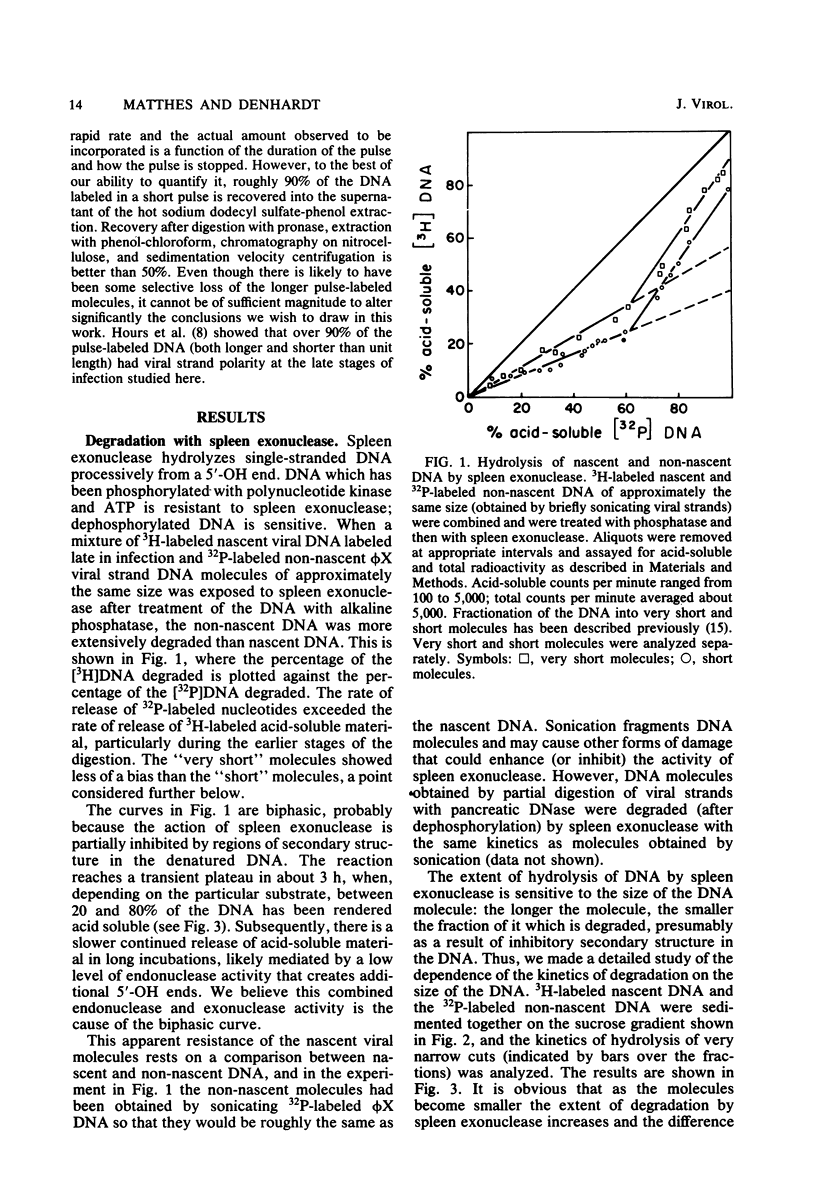
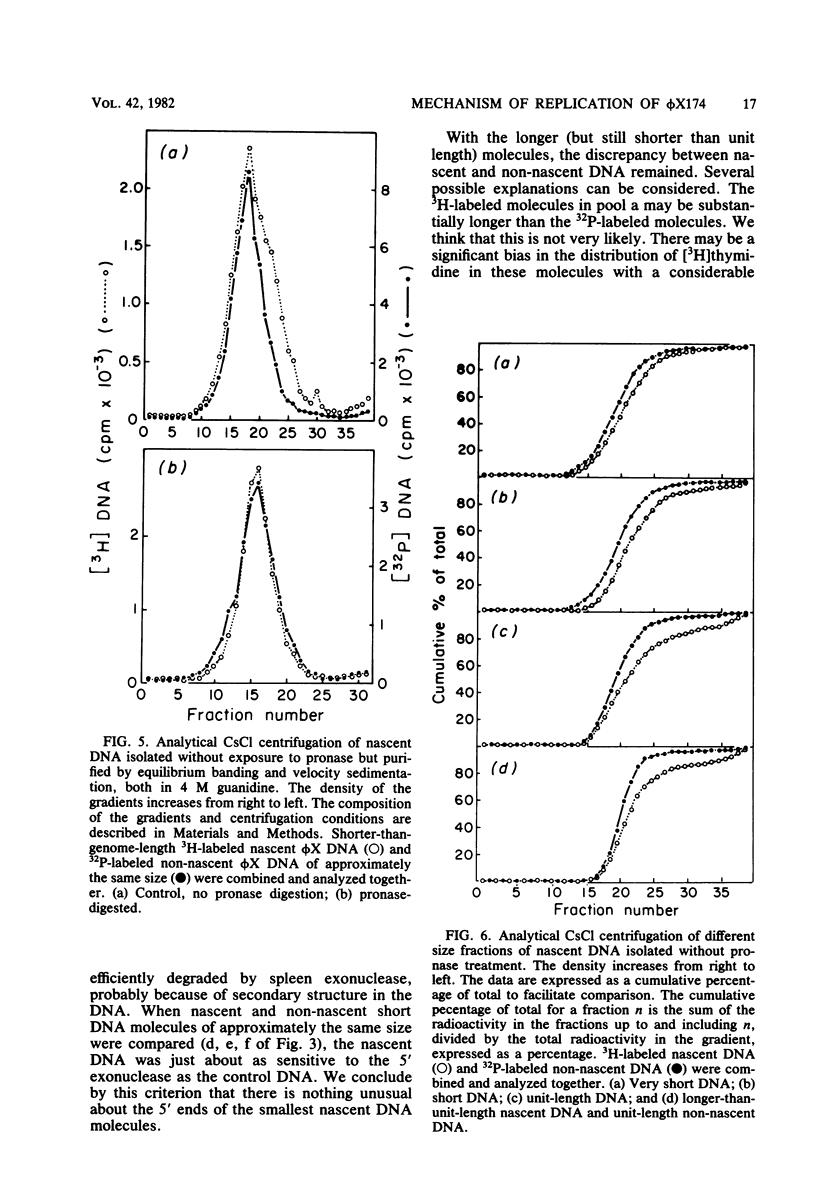
We reported earlier that dephosphorylated nascent phi X174 viral strand DNA molecules were less extensively degraded from the 5' end by spleen exonuclease than were non-nascent molecules. Experiments described here revealed that the insensitivity to the 5'-OH end-specific nuclease was more evident among the longer molecules in the population than among the shorter, all of the molecules being less than unit length in size. The smallest molecules in the population were about as sensitive to the enzyme as the control molecules and hence must possess unblocked 5'-terminal nucleotides. Degradation of the nascent DNA with the 3' end-specific snake venom phosphodiesterase revealed only a small enrichment for [3H]thymidine near the 3' end, seemingly insufficient to account completely for the apparent insensitivity of the longer molecules to spleen exonuclease. When the nascent molecules were isolated without the use of proteolytic enzymes, some pronase-sensitive material was found associated with the DNA, particularly the longer molecules. We suggest that the resistance of the longer nascent (pronase-treated) molecules to spleen exonuclease occurs because they have remnants of the viral gene A or A* protein covalently bound to the 5' end.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Carusi E. A. Evidence for blocked 5'-termini in human adenovirus DNA. Virology. 1977 Jan;76(1):380–394. doi: 10.1016/0042-6822(77)90310-5. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A theory of DNA replication. J Theor Biol. 1972 Mar;34(3):487–508. doi: 10.1016/0022-5193(72)90137-3. [DOI] [PubMed] [Google Scholar]
- Dressler D. H., Denhardt D. T. Mechanism of replication of phi-X-174 single stranded DNA. Nature. 1968 Jul 27;219(5152):346–351. doi: 10.1038/219346a0. [DOI] [PubMed] [Google Scholar]
- Dubeau L., Denhardt D. T. The mechanism of replication of phi X 174. XVIII. Gene A and A* proteins of phi X 174 bind tightly to phi X 174 replicative form DNA. Biochim Biophys Acta. 1981 Mar 26;653(1):52–60. doi: 10.1016/0005-2787(81)90103-9. [DOI] [PubMed] [Google Scholar]
- Eisenberg S., Griffith J., Kornberg A. phiX174 cistron A protein is a multifunctional enzyme in DNA replication. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3198–3202. doi: 10.1073/pnas.74.8.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W., Dressler D. DNA replication: the rolling circle model. Cold Spring Harb Symp Quant Biol. 1968;33:473–484. doi: 10.1101/sqb.1968.033.01.055. [DOI] [PubMed] [Google Scholar]
- Ikeda J. E., Yudelevich A., Shimamoto N., Hurwitz J. Role of polymeric forms of the bacteriophage phi X174 coded gene A protein in phi XRFI DNA cleavage. J Biol Chem. 1979 Oct 10;254(19):9416–9428. [PubMed] [Google Scholar]
- Ito J. Bacteriophage phi29 terminal protein: its association with the 5' termini of the phi29 genome. J Virol. 1978 Dec;28(3):895–904. doi: 10.1128/jvi.28.3.895-904.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosawa Y., Ogawa T., Hirose S., Okazaki T., Okazaki R. Mechanism of DNA chain growth. XV. RNA-linked nascent DNA pieces in Escherichia coli strains assayed with spleen exonuclease. J Mol Biol. 1975 Aug 25;96(4):653–664. doi: 10.1016/0022-2836(75)90144-8. [DOI] [PubMed] [Google Scholar]
- Langeveld S. A., van Mansfeld A. D., de Winter J. M., Weisbeek P. J. Cleavage of single-stranded DNA by the A and A* proteins of bacteriophage phi X174. Nucleic Acids Res. 1979 Dec 20;7(8):2177–2188. doi: 10.1093/nar/7.8.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langeveld S. A., van Mansfeld A. D., van der Ende A., van de Pol J. H., van Arkel G. A., Weisbeek P. J. The nuclease specificity of the bacteriophage phi X174 A* protein. Nucleic Acids Res. 1981 Feb 11;9(3):545–562. doi: 10.1093/nar/9.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida Y., Okazaki T., Okazaki R. Discontinuous replication of replicative form DNA from bacteriophage phiX174. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2776–2779. doi: 10.1073/pnas.74.7.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthes M., Denhardt D. T. The mechanism of replication of phi x174 DNA. XVI. Evidence that the phi x174 viral strand is synthesized discontinuously. J Mol Biol. 1980 Jan 5;136(1):45–63. doi: 10.1016/0022-2836(80)90365-4. [DOI] [PubMed] [Google Scholar]
- Matthes M., Weisbeek P. J., Denhardt D. T. Mechanism of replication of bacteriophage phi X174 XIX. Initiation of phi X174 viral strand DNA synthesis at internal sites on the genome. J Virol. 1982 Apr;42(1):301–305. doi: 10.1128/jvi.42.1.301-305.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabin E. Z., Preiss B., Fraser M. J. A nuclease from Neurospora crassa conidia specific for single-stranded nucleic acids. Prep Biochem. 1971;1(4):283–307. doi: 10.1080/00327487108081946. [DOI] [PubMed] [Google Scholar]
- Siegmann D. W., Werner R. Novel structure at 5'-ends of nascent DNA chains. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3438–3442. doi: 10.1073/pnas.73.10.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinsheimer R. L., Knippers R., Komano T. Stages in the replication of bacteriophage phi X174 DNA in vivo. Cold Spring Harb Symp Quant Biol. 1968;33:443–447. doi: 10.1101/sqb.1968.033.01.051. [DOI] [PubMed] [Google Scholar]
- Sumida-Yasumoto C., Ikeda J. E., Benz E., Marians K. J., Vicuna R., Sugrue S., Zipursky S. L., Hurwitz J. Replication of phiX174 DNA: in vitro synthesis of phiX RFI DNA and circular, single-stranded DNA. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):311–329. doi: 10.1101/sqb.1979.043.01.038. [DOI] [PubMed] [Google Scholar]


