Abstract
We previously isolated the SKN7 gene in a screen designed to isolate new components of the G1-S cell cycle transcription machinery in budding yeast. We have now found that Skn7 associates with Mbp1, the DNA-binding component of the G1-S transcription factor DSC1/MBF. SKN7 and MBP1 show several genetic interactions. Skn7 overexpression is lethal and is suppressed by a mutation in MBP1. Similarly, high overexpression of Mbp1 is lethal and can be suppressed by skn7 mutations. SKN7 is also required for MBP1 function in a mutant compromised for G1-specific transcription. Gel-retardation assays indicate that Skn7 is not an integral part of MBF. However, a physical interaction between Skn7 and Mbp1 was detected using two-hybrid assays and GST pulldowns. Thus, Skn7 and Mbp1 seem to form a transcription factor independent of MBF. Genetic data suggest that this new transcription factor could be involved in the bud-emergence process.
INTRODUCTION
In the budding yeast Saccharomyces cerevisiae, entry into the cell cycle is controlled at a point in late G1, called START, when the environmental conditions are assessed. Passage through START commits the cell to a new cell cycle and triggers the initial events, namely DNA replication, spindle pole body duplication, and bud emergence. START depends on the activation of two G1/cdk complexes, Cln1-Cdc28 and Cln2-Cdc28 (reviewed by Nasmyth, 1993). The transcription of the two genes CLN1 and CLN2, and of many other genes acting in late G1, is dependent on the accumulation of Cln3, a third G1 cyclin. When a threshold concentration of the Cln3-Cdc28 kinase complex is reached, a burst of late G1-specific gene transcription occurs, including CLN1 and CLN2 (Dirick et al., 1995; Stuart and Wittenberg, 1995).
The Cln3-Cdc28–dependent transcription of late G1-specific genes is mediated by two related heterodimeric transcription factors, SBF and DSC1/MBF (reviewed by Mendenhall and Hodge, 1998). They each contain a related DNA-binding protein, Swi4 for SBF and Mbp1 for MBF, and the same regulatory protein, Swi6. Although SBF and MBF bind to specific DNA sequences, the SCB element (CACGAAA) and the MCB element (ACGCGT), respectively, their functions are partially redundant (Koch et al., 1993).
In addition to CLN1 and CLN2, it has recently been shown that a large group of genes are expressed in late G1 (Cho et al., 1998; Spellman et al., 1998). These genes code for proteins necessary for the correct execution of START-dependent functions. For example, most DNA replication genes are under the control of MBF (Johnston and Lowndes, 1992). Similarly, the coordinated expression at START, or immediately after, of a number of genes involved in cell wall biosynthesis has been shown to be dependent on SBF (Igual et al., 1996); likewise, genes involved in budding and morphogenesis are also expressed in late G1 (Cho et al., 1998; Spellman et al., 1998). Coordinated cell cycle–dependent transcription through the SCB and MCB elements is thus an important means used by the cell to achieve coordination of the early events of the cell cycle (Johnston, 1992).
Combinations of mutations that inactivate both transcription factors, such as swi4 swi6 and swi4 mbp1, are lethal (Nasmyth and Dirick, 1991; Koch et al., 1993). However, surprisingly, SWI6 is not an essential gene, suggesting additional complexity in the G1 transcriptional machinery. In view of this fact, we screened for additional activators of MCB- and SCB-dependent transcription and identified one gene, SKN7/BRY1 (Morgan et al., 1995b). On a high-copy-number plasmid, SKN7 bypasses the essential requirement for SBF and MBF, restoring CLN1 and CLN2 transcription through their SCB and MCB promoter elements (Morgan et al., 1995b). Skn7, therefore, can activate the expression of late G1-specific genes. However, the means by which it does so is not clear.
The Skn7 protein interacts with the small GTPase Rho1 (Alberts et al., 1998), suggesting that it might be partly controlled by Rho1. In agreement with this idea, mutations in SKN7 and PKC1, one of the known Rho1 effectors (Nonaka et al., 1995), are synthetically lethal (Brown et al., 1994; Morgan et al., 1995b). In budding yeast, the PKC MAPK pathway controls cell wall gene expression (Igual et al., 1996), perhaps through direct regulation of Swi4 (Madden et al., 1997). Given its role in G1 transcription, SKN7 could itself be a transcriptional activator of cell wall genes (Brown et al., 1993, 1994; Morgan et al., 1995b), although direct experimental evidence in support of this idea is lacking. In addition, SKN7 functions in the oxidative stress response (Krems et al., 1996; Morgan et al., 1997), acting as a transcription factor in cooperation with Yap1 in the regulation of at least TRX2 (Morgan et al., 1997). However, any connections between the Skn7-Yap1 interaction, the oxidative stress response, and the G1 transcription program remain obscure.
The SKN7 protein shows in its C-terminal homology to the response regulators of bacterial two-component signal transduction systems (Brown et al., 1993; Morgan et al., 1995b). Skn7 has indeed been shown to form a two-component system in yeast with the histidine kinase Sln1 (Ketala et al., 1998; Li et al., 1998). In prokaryotes, these signaling pathways affect many aspects of cell physiology, including the cell cycle (Quon et al., 1996). They comprise a sensor and a response regulator; stimulation of the sensor leads to a phosphotransfer to a conserved aspartate residue of the response regulator, usually a transcription factor (for reviews, see Bourret et al., 1991; Parkinson, 1993). The SKN7 and SLN1 proteins, therefore, may be part of a two-component system in yeast that regulates late G1 gene expression and cell wall gene expression in response to an as-yet-unidentified signal.
The SKN7 DNA-binding domain shows homology to the heat-shock-factor DNA-binding domain (Brown et al., 1993; Morgan et al., 1995b), and it is thus unlikely that Skn7 binds SCB and MCB elements directly. This suggests that for any G1 transcriptional role, Skn7 would require association with a partner protein. Here we provide genetic and biochemical evidence that the partner for Skn7 in G1-regulated transcription is MBP1, the DNA-binding component of the MBF transcription factor. We show that Skn7 and Mbp1 associate, both in vivo and in vitro, and that in the absence of Swi6, Skn7 is necessary for Mbp1-dependent transcription. Finally, genetic data suggest that the Skn7/Mbp1 complex has a role in bud emergence.
MATERIALS AND METHODS
Strains and Growth Conditions
The haploid strains used in this study were as follows: W303-1a (a ade2-1 trp1-1 can1-100 leu2-3112 his3-11 ura3), K2003 (a ade2 his3 met leu2 trp1 ura3 swi4ts swi6::TRP1), K3294 (a ade2-1 met trp1-1 leu2-3112 can1-100 his3 ura3 ho-lacZ mbp1::URA3), GPY1115 (a, ade2 trp1 leu2 his3 ura3 pkc1::HIS3) (Paravicini et al., 1992), DJTD2-16D (α cdc42-1 leu2 ura3 his4 trp1 gal2), CG379 (α his7-2 leu2-3 trp1-289 ura3-52), and YN166 (trp1 ade2 leu2 ura3 GAL1-URA3 GAL1-lacZ). The skn7Δ, swi6Δ, and swi4Δ strains used in this study are isogenic strains in the W303 background. The mbp1/bor2 mutants used in this study were those isolated in the W303 background.
Yeast Techniques
Cells were grown in YEPD (1% yeast extract, 2% bactopeptone, 2% glucose) or, for diploid or plasmid selection, in synthetic minimal medium (0.67% yeast nitrogen base [YNB], 2% glucose or galactose) supplemented with the appropriate amino acids at 40 μg/ml. The growth temperature was 30°C, unless otherwise stated. Yeast transformations were performed by a modification of the lithium acetate procedure (Gietz and Sugino, 1988).
Assays for β-galactosidase activity were performed on midlog-phase cells as described previously (Guarente, 1983). Activities are given in OD units at 420 nm·min−1·mg−1 protein. Values represent the average of four independent experiments. FACS analysis was carried out as described previously (Igual et al., 1996).
Plasmid Constructs
SKN7 Plasmids
YEp24/SKN7 and pAB36 (pMW20/SKN7) have been described previously (Morgan et al., 1995b), as have pBAM1 (YCplac33/SKN7) and pBAM2 (YCplac33/skn7D427N) (Morgan et al., 1997). pAB56 was created by inserting a 2.9-kilobase PvuII-SphI fragment containing the GAL-SKN7 fusion into YEplac112 (Gietz and Sugino, 1988). pAB52 was created by ligating the coding region of SKN7, with BamHI (5′) and SpeI (3′) linkers added by PCR, into pT7linktag (a kind gift from N. Jones, Imperial Cancer Research Fund, London, United Kingdom) so that SKN7 is under the control of the T7 promoter. pAB53 was constructed by ligating the coding region of SKN7, with BamHI (5′) and SpeI (3′) linkers added by PCR, into pGEX-KG (Pharmacia, Uppsala, Sweden) so that Skn7 can be expressed in Escherichia coli as a fusion with GST. pAB61, pAB63, and pAB64 are deletions of pAB53 in which the fusion protein is truncated after residues 247, 473, and 311, respectively. pAB65 is a fusion between GST and residues 381–623 of SKN7, including the receiver domain. The skn7ΔHR allele, encoding a protein deleted from residues 238–261, was amplified by PCR from the pGADskn7ΔHR plasmid and inserted into the EcoRI site of pGEX-KG in frame with the GST, thus creating pAB81. pAB93 and pAB96 were constructed as follows: the skn7ΔHR allele was cut by PstI and SalI from the pGADskn7ΔHR plasmid and inserted into pB-BRY1/SKN7 (Morgan et al., 1995b) digested with PstI and SalI. A SacI/SalI fragment from the resulting plasmid was then ligated into either YEplac195 or YCplac33, thus placing the skn7ΔHR under the control of the SKN7 promoter, on a 2-μm or a centromeric plasmid to give pAB93 and pAB96, respectively. For pAB97 and pAB98, the SKN7 promoter was amplified by PCR with KpnI and EcoRI linkers added on both ends. The SKN7 ORF was amplified by PCR from residues 151–623 (skn7ΔDBD), i.e., excluding the DNA-binding domain, with an in-frame ATG codon and an EcoRI site added in 5′ and a SpeI site added in 3′. The two PCR fragments were then cloned into Bluescript so that skn7ΔDBD was put under the control of the SKN7 promoter. The pSKN7-skn7ΔDBD construction was then moved as a SacI/KpnI fragment into YEplac195 and YCplac33, creating pAB97 and pAB98, respectively. All four plasmids, i.e., pAB93, pAB96, pAB97, and pAB98, were checked by Western blotting for production of a protein of the expected size in a W303 skn7Δ background.
MBP1 Plasmids
The MBP1 ORF was amplified by PCR, with BamHI sites added at either end of the gene, and then cloned into pEMBLyex4 (Murray, 1987) to create pGAL-MBP1. pAB48 was made by subcloning a SacI/SalI fragment containing the MBP1 gene and its upstream sequences from pCK13 (a kind gift from K. Nasmyth, Institute of Molecular Pathology, Vienna, Austria) into YEplac195. pAB59 was constructed by ligating the coding region of MBP1, with BamHI (5′) and SpeI (3′) linkers added by PCR, into pT7linktag so that MBP1 is under the control of the T7 promoter. For the two-hybrid experiments, an NcoI-BglII fragment, corresponding to residues 215–833, was cloned in frame into pAS1-CYH2 (Harper et al., 1993), thus creating pAB75.
The plasmids expressing SWI4 and SWI6 under the control of the T7 promoter have already been described, as has the GAL-CDC42A118 plasmid (Ziman et al., 1991). The 2-μm–based plasmids carrying CLB5 and CLB6 come from laboratory stocks.
Isolation of Mutants Resistant to GAL-SKN7–dependent Lethality
The W303-1A and CG379 strains were transformed with pAB36, a centromeric plasmid carrying GAL-SKN7. Transformants were grown in liquid glucose minimal medium at 25°C until midlog phase (∼5 × 106 cells/ml). They were then plated onto minimal medium, with galactose as the carbon source, and incubated at 25°C. Spontaneous mutants appeared at a frequency of ∼10−7. These were crossed to the wild-type strain of the opposite mating type. If the growth phenotype on galactose was due to a plasmid rearrangement, this would be apparent in the diploid, so only those mutants complemented by the wild type were studied further.
GST-pulldown Assays
GST-pulldown assays were performed essentially as described by Siegmund and Nasmyth (1996), except that after binding of the in vitro synthesized protein to the GST fusion protein, four washes were performed at 500 mM NaCl. Pellets were washed another two times in 20 mM Tris-HCl (pH 8.0), 1 mM EDTA before being boiled in SDS-PAGE sample buffer and loaded on a SDS-polyacrylamide gel. After migration, the gel was fixed, dried, and subjected to autoradiography at −70°C.
Gel Mobility Shift Assays
Protein extracts from log-phase cultures grown at 30°C in YEPD were prepared and gel-retardation assays were performed as previously described (Lowndes et al., 1991). The 3xMCB probe has already been described (Lowndes et al., 1991).
RNA Analysis
Total RNA was isolated from yeast strains grown under the conditions described (Morgan et al., 1995b). Northern hybridization techniques have also been described previously (Morgan et al., 1995b).
RESULTS
Isolation of Mutants Resistant to GAL-SKN7
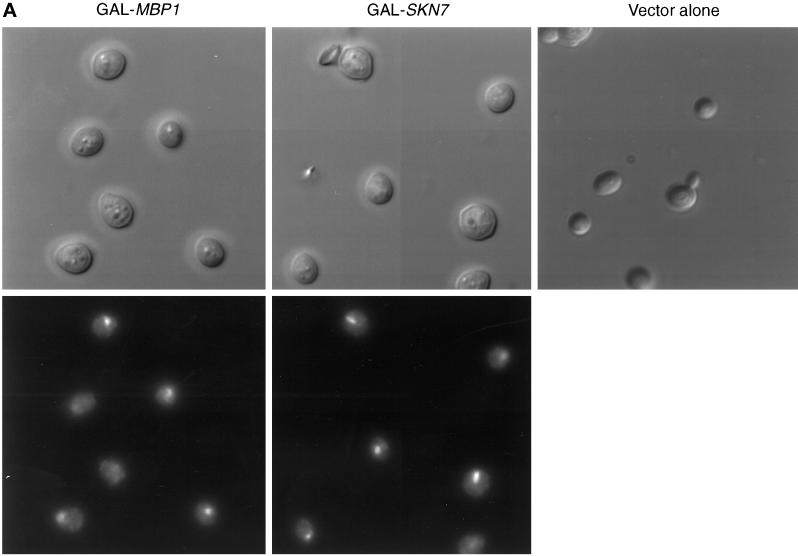
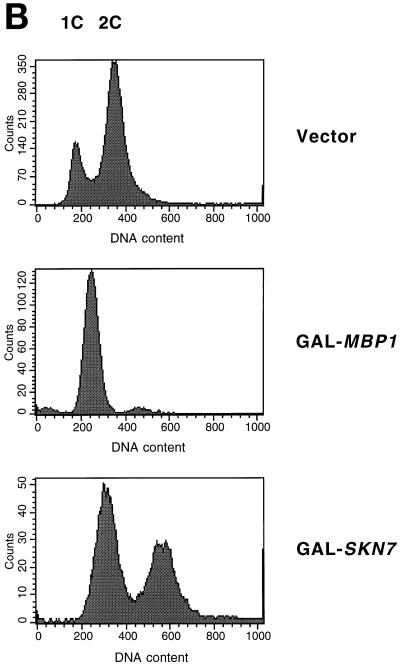
We wished to explore further the interaction of Skn7 with the G1 transcription machinery and to determine its cell cycle role. We previously showed that overexpression of SKN7 from the GAL promoter is lethal (Morgan et al., 1995b) (Figure 1A). Microscopic analysis showed that cultures of cells overexpressing SKN7 accumulate swollen, unbudded cells with a single nucleus (Figure 2A). FACS analysis showed an accumulation of cells with a 1C DNA content, although a significant fraction of the population had initiated S phase and showed a 2C DNA content (Figure 2B).
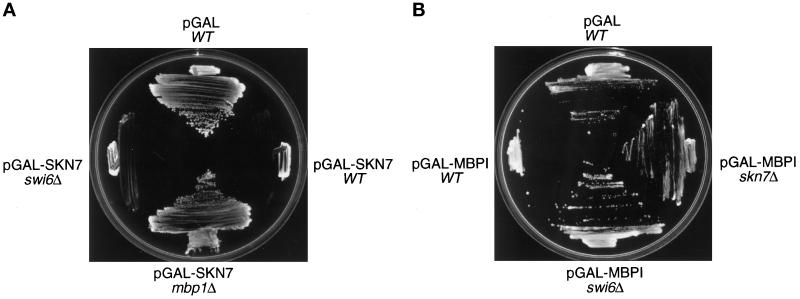
Figure 1.
The genetic interactions between SKN7 and MBP1. (A) mbp1 mutations confer resistance to pGAL-SKN7–dependent lethality. Derivatives of the wild-type strain W303 (WT) were transformed with either the pMW20 empty vector (pGAL) or the pGAL-SKN7 construct. Transformants were streaked on galactose-containing minimal agar plates and incubated at 30°C. (B) skn7 mutations confer resistance to pGAL-MBP1–dependent lethality. Derivatives of the wild-type strain W303 were transformed with either the pEMBLyex4 empty vector (pGAL) or the pGAL-MBP1 construct. Transformants were streaked on minimal agar plates containing galactose and lacking leucine and incubated at 30°C.
Figure 2.
Overexpression of MBP1 or SKN7 arrests division of cells. W303-1A cells containing pGAL-SKN7 or pGAL-MBP1 were grown at 30°C in YNB medium containing raffinose. Galactose was added, and incubation continued for the equivalent of two generations. (A) Cellular morphology (upper panels) and DAP1 staining (lower panels). (B) FACS analysis. Note that DNA peaks tend to migrate to the right as cells enlarge during cell cycle arrest.
Because mutations conferring resistance to GAL-SKN7 expression might affect proteins interacting in some way with SKN7, spontaneous mutants resistant to SKN7 overexpression were isolated (see MATERIALS AND METHODS). Twenty-two recessive mutants were thus identified, and these fell into two complementation groups, bor1 and bor2 (for BRY1/SKN7 overexpression resistant). We recovered 12 bor1 mutants and 10 bor2 mutants, which suggests that our screen was saturated.
At this time, we became aware that high overexpression of MBP1 was also lethal (see below). Because both MBP1 and SKN7 act through SCB and MCB elements (see INTRODUCTION), we tested whether the MBP1 gene was required for GAL-SKN7–dependent lethality. Indeed, mbp1Δ cells containing pMW20-SKN7 (pGAL-SKN7) (Morgan et al., 1995b) were able to grow in the presence of galactose (Figure 1A). Reintroduction of MBP1 into the mbp1Δ pGAL-SKN7 strain restored sensitivity to galactose (our unpublished results), confirming that resistance to GAL-SKN7 is conferred by the mbp1 mutation. We checked by Northern blotting that SKN7 transcription is not regulated by MBP1 and that MBP1 expression is not under the control of SKN7 (our unpublished results).
We then determined whether the mbp1Δ mutation belonged to either the bor1 or the bor2 complementation group. Diploids of mbp1Δ with mutants from the bor2 complementation group retained resistance to GAL-SKN7 expression. Moreover, when a mbp1Δ/bor2 diploid was sporulated, all of the spores were resistant to GAL-SKN7; hence, MBP1 and BOR2 are allelic. We also showed that BOR1 was allelic to GAL3, so the bor1 mutants were not studied further.
The Mbp1 protein interacts with Swi6 in the MBF transcription factor, and it is also homologous to Swi4 (Koch et al., 1993). Therefore, we tested whether swi4 and swi6 mutants would be resistant to SKN7 overexpression. Importantly, neither a swi6Δ mutant (Figure 1A) nor a swi4Δ mutant (our unpublished results) containing the GAL-SKN7 plasmid was able to grow on galactose. The genetic interaction of Skn7 with components of the G1 transcriptional machinery was specific for MBP1.
Overexpression of MBP1 Is Lethal and Is Suppressed by skn7Δ
The MBP1 gene was inserted into the pEMBLyex4 2-μm–based vector under control of the GAL promoter. This plasmid also bears the leu2-d allele, so that its copy number can be boosted by selecting for leucine, thus achieving even higher levels of expression. Wild-type W303 containing the pEMBLyex4/MBP1 plasmid (pGAL-MBP1) does not grow on minimal medium containing galactose but lacking leucine (Figure 1B). High overexpression of MBP1 is thus toxic in yeast. Under similar conditions, neither SWI4 nor SWI6 overexpression is lethal (our unpublished results). As with Skn7 overexpression, cells overexpressing MBP1 arrested as large, round, unbudded cells with a single nucleus (Figure 2A). However, in contrast to SKN7 overexpression, these cells showed little evidence of any initiation of S phase, but they arrested with a clear 1C peak of DNA (Figure 2B).
Given the interactions between MBP1 and SWI4 and SWI6, we tested whether swi4Δ and swi6Δ mutants, as well as skn7Δ, would be resistant to GAL-MBP1. These three mutants containing pGAL-MBP1 were tested for growth on minimal medium containing galactose and lacking leucine. The swi4Δ pGAL-MBP1 did not grow on galactose (our unpublished results), whereas both swi6Δ pGAL-MBP1 and skn7Δ pGAL-MBP1 grew under these conditions (Figure 1B). The swi6Δ and skn7Δ mutations, therefore, are able to confer resistance to MBP1 overexpression, indicating that both Swi6 and Skn7 participate in this Mbp1-induced lethality.
SKN7 Is Necessary for Suppression of a swi4ts swi6Δ Strain by MBP1
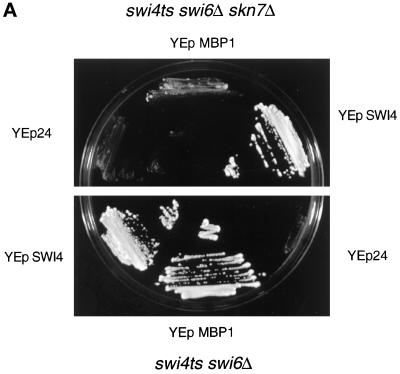
The above data suggest that Mbp1 and Skn7 might interact. Because Mbp1 is part of the MBF transcription factor, we explored whether Skn7 could be part of MBF. Initially, we addressed this by a genetic experiment. In a swi6Δ mutant, no MBF activity can be detected, suggesting that Mbp1 requires Swi6 to bind DNA, at least in vitro (Dirick et al., 1992; Lowndes et al., 1992). However, a high-copy-number plasmid carrying MBP1 is able to rescue the temperature sensitivity of the swi4ts swi6Δ mutant (Figure 3A). Mbp1, therefore, might associate with a transcriptional activator other than Swi6 to suppress the double mutant. To test whether this is Skn7, we transformed MBP1 on a high-copy-number plasmid into an isogenic swi4ts swi6Δ skn7Δ strain. The MBP1 plasmid is totally unable to suppress the temperature sensitivity of this strain (Figure 3A). Reintroduction of a centromeric plasmid carrying SKN7 restores the ability to grow at 37°C, but the skn7ΔDBD allele, which lacks the DNA-binding domain, is unable to do so (our unpublished results). Thus, Skn7 function is required for MBP1 to suppress swi4ts swi6Δ, consistent with a Skn7 and Mbp1 association.
Figure 3.
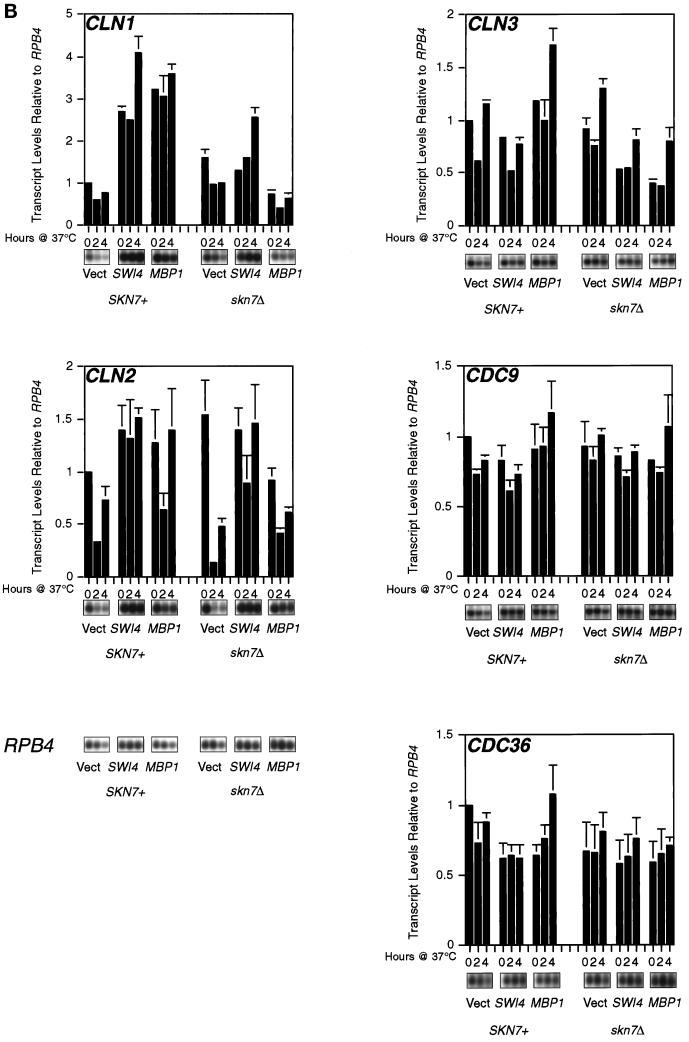
SKN7 function is required for MBP1 suppression of the swi4ts swi6Δ mutant. (A) Skn7 is necessary for suppression of swi4ts swi6Δ by high-copy-number MBP1. The swi4ts swi6Δ and swi4ts swi6Δ skn7Δ strains were transformed with YEp24 or a multicopy plasmid carrying either SWI4 or MBP1 and streaked on YEPD agar plates at 37°C. (B) Skn7 is required for increased cyclin levels in Mbp1 suppression of a swi4ts swi6Δ strain. The appropriate strains (see text) were grown to midlog phase in YNB glucose at 25°C, transferred to 37°C for the times shown, and sampled, and total RNA was extracted and a Northern blot was prepared (Morgan et al., 1995b). Probes were internal fragments of the genes concerned, and RPB4 was used as a loading control. This experiment was repeated, and an average of the data points is presented above with error bars, but raw data are shown for only one experiment. For the two sets of data to be directly comparable, levels are presented relative to the level at 0 time of strain K2003 containing vector alone. This value was arbitrarily set at 1. Note that where no error bar is shown, the difference between the two samples concerned was insignificant and the graphing package used was unable to draw the error bars. Quantitation was by phosphorimager.
The suppression of the swi4ts swi6Δ strain by MBPI must entail increased cyclin expression. This, of course, is also the basis of suppression of this strain by high-copy-number SKN7 (Morgan et al., 1995b). The Skn7 requirement for Mbp1 suppression, therefore, should be reflected in G1 cyclin levels, and Northern hybridization confirmed this (Figure 3B). We introduced high-copy-number MBP1 into the isogenic swi4ts swi6Δ and swi4ts swi6Δ skn7Δ strain mentioned above. As controls, we also used high-copy-number SWI4 and the empty vector. The resulting strains were grown to midlog phase and transferred to 37°C, and the transcript levels of CLN1 and CLN2 were examined. In the presence of SKN7, both SWI4 and MBP1 led to abundant CLN1 expression at 37°C. In the absence of SKN7, high-copy-number SW14 still stimulated CLN1 levels but, importantly, high-copy-number MBP1 failed to stimulate CLN1 expression. Smaller but reproducible effects on CLN2 expression were also seen (Figure 3B).
As control transcripts, we examined CDC9 and CDC36, which were unaffected by the skn7Δ mutation. However, there was a slight effect on CLN3 levels. This was not apparent when MET4 was used as a loading control, which had no effect on relative levels of other transcripts (our unpublished results). Therefore, the apparent effect on CLN3 in Figure 3B may not be significant. However, it is intriguing that Skn7 affects the function of the Mcm1 transcription factor (Yu et al., 1995) and, in turn, Mcm1 functions in the regulation of CLN3 expression (McInerny et al., 1997).
In summary, the physiological consequences of the Skn7-Mbp1 interaction can be visualized. Ablation of SKN7 results in a marked decrease in CLN1 transcript levels in the presence of high-copy-number MBP1. This is sufficient to explain the SKN7 requirement for the MBP1 suppression of a swi4ts swi6Δ strain. The minor effects on CLN2 may also contribute to this effect.
Skn7 Is Not Part of the Core MBF
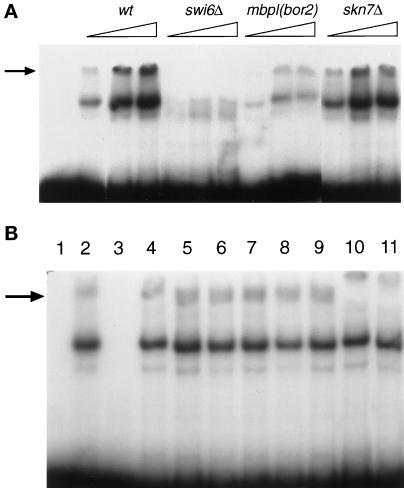
As currently characterized, MBF consists of Mbp1 and Swi6 (see INTRODUCTION). To investigate whether Skn7 is part of MBF, we performed gel-retardation experiments using as a probe a synthetic oligonucleotide containing 3xMCB sites (Lowndes et al., 1991). In cell extracts from wild-type cells, a retarded complex could be seen (Lowndes et al., 1991), which, as expected for MBF, was absent in a swi6Δ strain, was much reduced in mbp1 (bor2) (Figure 4A), and was competed away by an excess of cold MCB probe (Figure 4B, lanes 2 and 3). It was also supershifted by antibodies against Swi6 (Figure 4B, lane 10). However, this complex was clearly not supershifted by the addition of antibodies directed against Skn7 (Figure 4B, lanes 7–9). It is important to note that the lowest concentration of Skn7 antibodies used in this experiment has previously been shown to supershift a promoter complex containing Skn7 (Morgan et al., 1997). The retarded band was also still present in a skn7Δ strain and was of the usual mobility (Figure 4A). Moreover, the MBF in the skn7Δ extracts could still be supershifted by Swi6 antibodies (Figure 4B, lane11). Our results thus strongly suggest that Skn7 is not part of the core MBF that binds simple MCB repeats and that MBF is also clearly not dependent on Skn7.
Figure 4.
Skn7 is not part of the core MBF. MBF was analyzed by gel retardation with a radioactively labeled 3xMCB probe (Lowndes et al., 1991). (A) Deletion of SKN7 does not affect MBF. Increasing amounts of crude extract were assayed from the strains shown. Note that the mbp1 mutation used is a point mutation (an allele of bor2) rather than a deletion. (B) Antibodies to Skn7 do not supershift MBF. Lane 1, free probe; lanes 2–10, wild-type (W303) crude extract; lane 2, no competitor; lane 3, competition with a 100-fold excess of the cold probe; lanes 4–6, preimmune serum (1:100, 1:200, 1:500); lanes 7–9, anti-Skn7 antibodies (1:100, 1:200, 1:500); lane 10, anti-Swi6 antibodies (1:200); lane 11, W303 skn7Δ crude extracts with anti-Swi6 antibodies (1:200).
Two-Hybrid Interaction between MBP1 and SKN7
The genetic data described above suggest a direct interaction between Skn7 and Mbp1. To address this, the MBP1 gene was cloned in frame with the GAL4 DNA-binding domain and tested for interaction in the two-hybrid system with SKN7 (Table 1). As expected, MBP1 interacted strongly with the control SWI6. Significantly, MBP1 also interacted strongly with SKN7. Because both proteins are transcriptional activators, the entire SWI6 and SKN7 genes, rather than fusions to the GAL4 activation domain, were used in this study. On the other hand, there was no interaction between MBP1 and an unrelated fission yeast gene, psh1+ (Millar, personal communication); similarly, there was no interaction between SKN7 and SWI6 (our unpublished results) or the Schizosaccharomyces pombe gene crk1+ (Table 1). These data clearly support some form of physical interaction between Skn7 and Mbp1.
Table 1.
SKN7 and MBP1 interact in a two-hybrid assay
| MBP1 | crk1+ | |
|---|---|---|
| SWI6 | 4.62 ± 0.23 | 0.23 ± 0.01 |
| SKN7 | 4.39 ± 0.99 | 0.15 ± 0.06 |
| skn7ΔHR | 0.49 ± 0.1 | 0.19 ± 0.1 |
| psh1+ | 0.31 ± 0.01 | NTa |
β-Galactosidase activities are given in OD units at 420 nm·min−1·mg−1 protein. Values represent the average of four independent experiments.
NT, not tested.
In Vitro Interaction between Mbp1 and Skn7
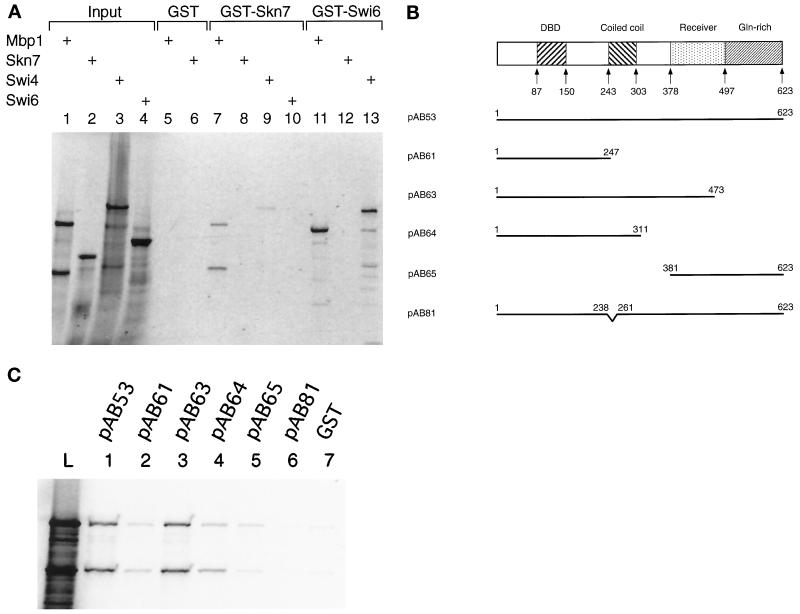
We used GST-pulldown experiments to determine whether the interaction between MBP1 and SKN7 was direct or whether it required ancillary proteins. Once again, we used Swi6 as a control and, as expected, in vitro synthesized Mbp1 was retained by a GST-Swi6 fusion protein but not by GST alone (Figure 5A, lanes 11 and 5). More importantly, Mbp1 also clearly bound to a GST-Skn7 fusion protein (Figure 5A, lane 7). Note that in vitro synthesized Mbp1 runs as two bands, with the faster-migrating band being most probably a C-terminal truncation, because it is not retained by GST-Swi6 (Figure 5A, compare lanes 1 and 11). Both forms, however, are bound by GST-Skn7. The Mbp1-Skn7 association is a strong interaction, for it is stable in up to 1 M salt, like the Mbp1/Swi6 complex (our unpublished results). On the other hand, Swi4 shows only a weak interaction with GST-Skn7, although it is strongly retained by GST-Swi6 (Figure 5A, lanes 9 and 13). Emphasizing the specificity of the Mbp1-Skn7 association, no interaction could be detected between in vitro synthesized Skn7 and GST-Swi6 (Figure 5A, lane 12). In the reverse experiment, in vitro synthesized Swi6 also was not retained by GST-Skn7 (Figure 5A, lane 10). The Skn7 protein thus interacts directly with Mbp1 but not with Swi6 and only weakly with Swi4 (see DISCUSSION).
Figure 5.
In vitro interaction between Skn7 and Mbp1. (A) The interaction between Skn7 and Mbp1 in GST-pulldown experiments. The fusion proteins GST-Skn7 (lanes 7–10) and GST-Swi6 (lanes 11–13) and the negative control GST alone (lanes 5 and 6) were purified on glutathione beads and incubated with in vitro translated, radioactively labeled Mbp1 (lanes 5, 7, and 11), Skn7 (lanes 6, 8, and 12), Swi4 (lanes 9 and 13), or Swi6 (lane 10). The bound proteins were separated by SDS-PAGE and visualized by autoradiography. The total amount of radioactive protein added to each binding reaction is shown in lanes 1 (Mbp1), 2 (Skn7), 3 (Swi4), and 4 (Swi6). (B) Schematic representation of the mutant derivatives of GST-Skn7 used to identify the regions of interaction of Skn7 with Mbp1. The different domains of Skn7 are illustrated, and the amino acid positions of the truncations and internal deletions are indicated. DBD, DNA-binding domain; pAB81, skn7ΔHR (see text). (C) The different derivatives of the GST-Skn7 fusion protein were assayed for their ability to bind in vitro translated, radioactively labeled Mbp1 in GST-pulldown experiments. The total amount of radioactive Mbp1 added to each binding reaction is shown (lane L). GST (lane 7) was used as a negative control.
Attempts at coimmunoprecipitation were only partially successful, i.e., a weak but irreproducible signal was detected (our unpublished results). Possibly the proportion of Skn7 complexed with Mbp1 at any one time is too low for such experiments (see DISCUSSION).
The Receiver Domain and the HR Region in the Skn7 Protein Are Required for the Interaction with Mbp1
In a preliminary attempt to identify the region(s) of the Skn7 protein necessary for the interaction with Mbp1, deletions were made in the GST-Skn7 fusion protein (Figure 5B). These deletions were then tested for their ability to bind Mbp1 (Figure 5C). A deletion starting from the C terminus of the protein, removing the glutamine-rich region and a small part of the receiver domain (pAB63), was still able to bind Mbp1 to the same extent as the full protein (Figure 5C, compare lanes 3 and 1). On the other hand, when the receiver domain was completely deleted (pAB64), the interaction was substantially reduced (Figure 5C, lane 4). It was further reduced when the coiled-coil region was deleted (Figure 5C, lane 2). However, a fusion between GST and the receiver domain (residues 381–623, pAB65) only bound Mbp1 very weakly, if at all (Figure 5C, lane 5). Thus, although the receiver domain is involved in the interaction, other parts of the protein are involved as well, probably including the coiled-coil domain.
A short region in the Skn7 protein, designated HR and encompassing part of the coiled-coil domain, has been shown to mediate interactions between Skn7 and Rho1 (Alberts et al., 1998). The HR region lies between residues 238 and 261 at the beginning of the coiled-coil domain (Figure 5B), and this might account for the apparent role of the coiled-coil domain in the interaction with Mbp1. Indeed, deletion of the HR sequences almost abolished the binding to Mbp1 (Figure 5C, lane 6). In addition, a construct lacking the HR region (a kind gift from R. Treisman) was almost inert in a two-hybrid interaction with Mbp1 (Table 1). Therefore, at least the HR region of the coiled-coil domain and the receiver domain are required for interaction of Skn7 with Mbp1. Consistent with this finding, the HR region and the receiver domain are required for the in vivo function of Skn7. A swi4ts swi6Δ was not suppressed by SKN7 lacking either the HR region or the receiver domain (Alberts et al., 1998).
SKN7 and MBP1 Are Involved in Bud Emergence
The genetic data described above regarding rescue of the double mutant swi4ts swi6Δ imply a role for Skn7 and Mbp1 in cyclin expression, although it is far from clear whether this is of physiological relevance in a normal cell cycle (see DISCUSSION). Therefore, we investigated other events of G1 in which Skn7 and Mbp1 could participate. Some cell wall genes have SCB and MCB elements in their promoters and are under cell cycle control (Igual et al., 1996). Previous data, especially the finding that skn7Δ and pkc1Δ are synthetically lethal, suggested that SKN7 may have a role in cell wall metabolism (Brown et al., 1993, 1994; Morgan et al., 1995b; see INTRODUCTION). Similarly, even though mbp1 and pkc1 mutations are not synthetically lethal (Igual et al., 1996), we found that strains deleted for both genes display a synthetic enhancement of spore lethality: 77% of the double-mutant spores were dead compared with 30 and 7% for pkc1Δ and mbp1Δ single-mutant spores, respectively. This finding might suggest a role for Mbp1 in cell wall metabolism, but we could find no obvious cell wall defects in mbp1/bor2 mutants or in skn7Δ strains, and we could find no effect of SKN7 and MBP1 on expression of the cell wall genes that we examined (our unpublished results).
Another key event in G1 is bud emergence. Significantly, the GAL-SKN7–induced lethality can be rescued by a 2-μm plasmid carrying either CLN1 or CLN2 but not by high-copy-number CLB5 or CLB6 (Figure 6A). This suggests that SKN7 and MBP1 could be involved in the budding process, because CLN1 and CLN2 promote bud emergence, partly through actin cytoskeleton reorganization (Benton et al., 1993; Cvrckova and Nasmyth, 1993; Lew and Reed, 1993). Therefore, we looked for genetic interactions between SKN7 and genes involved in bud emergence, particularly CDC42, which encodes a small GTPase that plays a central role in actin polarization (Pringle et al., 1995). When a 2-μm–based plasmid carrying SKN7 was introduced into the cdc42-1 mutant, the restrictive temperature of this mutant was decreased from 37 to 34°C (Figure 6B). The skn7ΔHR and skn7D427N alleles showed the same effect (Alberts et al., 1998). On the other hand, cdc42-1 was not affected by the skn7ΔDBD allele (our unpublished results), indicating that the effect of high-copy-number SKN7 on cdc42-1 growth is likely to be transcriptional. Note that the Skn7 DNA-binding domain is known to be functional (Morgan et al., 1997). However, neither CDC42 nor CDC24 expression is controlled by SKN7 (our unpublished results). As with cdc42-1, we found that the restrictive temperature of a bem1Δ mutant, another mutant defective in actin polarization, is also decreased by a high-copy-number plasmid carrying SKN7 (our unpublished results). When a dominant-negative allele of CDC42, CDC42A118, is expressed from the GAL promoter, growth of wild-type strains is almost abolished on galactose (Ziman et al., 1991). Mutations in SKN7 or MBP1 suppressed the CDC42A118 lethality on galactose, whereas swi6Δ had no effect (Figure 6C). Moreover, introducing 2-μm–based SKN7 or MBP1 plasmids into W303 carrying GAL-CDC42A118 completely eliminated any residual growth on galactose. These results strongly suggest that the G1 function of SKN7 and MBP1 lies in bud emergence.
Figure 6.
Genetic interactions between SKN7, MBP1, and genes involved in bud emergence. (A) pGAL-SKN7–dependent lethality is suppressed by high-copy-number CLN1. W303-1A containing pAB56 (2-μm, TRP1, GAL-SKN7) was transformed with YEp24 or a multicopy plasmid carrying CLN1, CLB5, or CLB6. Transformants were streaked on galactose-containing minimal agar plates and incubated at 30°C. (B) The temperature sensitivity of a cdc42-1 mutant is enhanced by high-copy-number SKN7. A cdc42-1 strain was transformed with the plasmids shown and streaked onto YEPD agar plates at 34°C. (C) pGAL-CDC42A118–dependent lethality is suppressed by skn7 and mbp1 mutations. The pGAL-CDC42A118 plasmid was transformed into W303, W303 swi6Δ, W303 skn7Δ, and W303 mbp1/bor2. The transformants were streaked onto galactose-containing minimal plates at 30°C.
DISCUSSION
We initially isolated SKN7 in a genetic screen designed to identify new genes involved in late G1 transcription (Morgan et al., 1995b). Although Skn7 stimulated CLN2 expression through MCB and SCB promoter elements, the heat-shock-factor DNA-binding domain meant that it was unlikely to bind these elements directly. Skn7 overexpression was found to be lethal, and we have now exploited this finding to further investigate Skn7 interaction with the G1 transcription apparatus. Here we have isolated mutants resistant to overexpression of SKN7 and found that they mapped to the MBP1 gene. In turn, high overexpression of MBP1 is lethal and deletion of SKN7 relieves this lethality. These genetic data suggested that Skn7 and Mbp1 might physically interact. We have demonstrated this interaction: first, the two genes interact in the two-hybrid system; second, an in vitro synthesized Mbp1 protein is retained by a GST-Skn7 fusion protein. The failure to detect coimmunoprecipitation of Skn7 with Mbp1 is disappointing. However, there is also good evidence for a direct association of Skn7 with Yap1 (Morgan et al., 1997) and Rho1 (Alberts et al., 1998) and possibly also Sln1-Ypd1 (Li et al., 1998). In none of these cases has coimmunoprecipitation been demonstrated. Possibly only a small proportion of the total cellular Skn7 associates with any one of these proteins at one time, making coimmunoprecipitation difficult to detect.
We could not detect any interaction between Skn7 and Swi6 in GST-pulldown assays. This is in agreement with our genetic results, which showed that a SWI6 deletion does not suppress the Skn7-induced lethality. On the other hand, Swi4 did bind weakly to GST-Skn7. This may simply be the result of the high level of homology between Swi4 and Mbp1 (Koch et al., 1993), because we could detect no genetic evidence for a Swi4-Skn7 interaction in vivo. Thus, Skn7 interacts specifically with Mbp1 and is therefore directly associated with the G1 transcriptional machinery.
Gel-retardation experiments suggested that Skn7 is not a component of MBF. MBF is not supershifted by polyclonal antibodies directed against Skn7, and we could not detect Skn7, by Western blotting, in an affinity-purified MBF fraction (our unpublished results). However, Skn7 may still be an MBF-associated factor that is not necessary for DNA binding but only for transcription activation. Such a protein has already been described in fission yeast. In S. pombe, the Rep2 protein binds to Res2, an Mbp1 homologue, and can be immunoprecipitated with the Res2/Cdc10 complex, an MBF-like transcription factor (Nakashima et al., 1995). Rep2 is a transactivator that is absolutely required for Res2/Cdc10 activity, because the Res2/Cdc10 transcription factor is inactive in rep2− cells (Nakashima et al., 1995; Baum et al., 1997), although the Res2/Cdc10 complex can bind MCB elements on its own (Zhu et al., 1994, 1997; Baum et al., 1997). Just as Rep2 binds only to Res2 (Nakashima et al., 1995), Skn7 binds only to Mbp1. Moreover, Skn7 is a transcriptional activator (Brown et al., 1994; Morgan et al., 1995b, 1997) essential for the suppression by MBP1 of a mutant with crippled SBF activity (swi4ts swi6Δ). Thus, Skn7 could be a functional analogue of Rep2 in budding yeast. However, swi4 and skn7 mutations are not synthetically lethal (Morgan et al., 1995b), indicating that MBF is still active in skn7Δ cells. Moreover, only mbp1 mutations, but not swi6 mutations, are able to confer resistance to Skn7 overexpression, strongly suggesting that MBF activity is not affected by accumulation of the Skn7 protein in the cell. It is unlikely, therefore, that MBF activity is controlled by Skn7.
Nonetheless, our genetic data clearly support the notion that Skn7 and Mbp1 form a functional transcription factor. High-copy-number MBP1 suppresses the temperature sensitivity of a swi4ts swi6Δ strain largely through activation of CLN1 and, to a lesser extent, CLN2, transcription. It is not surprising that there should be differences between CLN1 and CLN2 expression in these experiments, because CLN2 regulation is known to depend on SCB promoter elements and CLN1 regulation depends on MCB elements (Partridge et al., 1997). However, Mbp1 is known not to bind MCB elements on its own, MBF-binding activity being undetectable in swi6 mutants (Dirick et al., 1992; Lowndes et al., 1992). Importantly, the high-copy-number MBP1 suppression is dependent on SKN7, because a swi4ts swi6Δ skn7Δ strain transformed with MBP1 on a high-copy-number plasmid does not grow at 37°C. Moreover, the suppression is dependent on the presence of a functional DNA-binding domain in the Skn7 protein. In addition, whereas overexpression of Skn7 results in MBP1-mediated lethality, a wild-type strain carrying a pGAL-skn7ΔDBD plasmid grows normally on galactose (our unpublished results). The Skn7 DNA-binding domain, therefore, is clearly required for the functional interaction with Mbp1. So the two transcription factors probably cooperate to control CLN1 expression.
Note that the studies implicating MBP1 and SKN7 in the control of CLN gene expression were performed in strains with crippled SBF activity (Koch et al., 1993; Morgan et al., 1995b) and that deletions of skn7 or mbp1 in wild-type cells have no effect on G1 cyclin expression (Koch et al., 1993; Morgan et al., 1995b). The Skn7/Mbp1 transcription factor may be only a minor component of CLN1 regulation normally, e.g., binding the promoter only weakly and being readily displaced by SBF. Only under particular conditions in which SBF activity is reduced or not associated with CLN promoters might Skn7/Mbp1 control of CLN1 expression be physiologically significant. One such situation could be after oxidative stress (Morgan et al., 1997), but even under these conditions we could detect no novel species forming on the CLN1 or CLN2 promoters in band shifts (our unpublished results). Presumably, to detect the binding of Skn7/Mbp1 to DNA will require identification of more prominent physiological targets or determining more precisely the role of the Skn7-Mbp1 interaction in CLN1 expression.
Skn7 is a signaling protein involved in a wide range of processes that are apparently unrelated to one another, e.g., oxidative stress response, CLN expression, and maintenance of cell wall integrity (see INTRODUCTION). The nature of the signal involved remains obscure, but we note that two-component systems have only been found in cell wall–containing eukaryotes (reviewed by Morgan et al., 1995a). Moreover, Skn7 interacts with Rho1 (Alberts et al., 1998). Interestingly, Rho1 controls the PKC cascade (Nonaka et al., 1995), and its activity has recently been suggested to be responsive to cell wall integrity (Bickle et al., 1998). Finally, skn7 mutants are hypersensitive only to hydrogen peroxide and not to diamide (Krems et al., 1996; Morgan et al., 1997), and only the former is known to induce some form of cell wall damage. It is thus quite a strong possibility that Skn7 is part of a signal transduction pathway that is somehow regulated by cell wall integrity. According to the nature of the stress, Skn7 would then associate with a partner, e.g., Yap1 (Morgan et al., 1997) or Mbp1 (this work), to direct transcription of the appropriate targets. In this respect, it is noteworthy that the response regulator and the HR domain are both required for the interaction with Mbp1.
We could find no evidence for an effect of skn7Δ or mbp1Δ on cell wall structure per se. On the other hand, Skn7/Mbp1 may be involved in actin organization and/or bud emergence. Genetic interactions were detected between SKN7 and CDC42, which is directly involved in actin polarization (Pringle et al., 1995), and also with BEM1, another gene involved in actin polarization. In addition, overexpression of MBP1 or SKN7 prevented bud emergence, although in the case of SKN7 S phase was initiated in at least part of the population. Possibly, MBP1 overexpression interfered with normal expression of the MCB-regulated genes required for S phase as well as those required for bud emergence, whereas SKN7 overexpression only affected genes required for budding. Taken together, these data strongly suggest that Skn7/Mbp1 controls expression of at least one gene involved in bud emergence and/or actin organization.
ACKNOWLEDGMENTS
We thank all of our colleagues in the Division of Yeast Genetics, particularly J.-C. Igual and J.B.A. Millar for helpful advice and discussion. We are especially indebted to G. Banks and A. Spanos, who made the first observation that overexpression of Mbp1 is lethal. We are grateful to A. Alberts and R. Treisman for communicating results before publication. We thank R. Treisman, N. Lowndes, K. Nasmyth, J. Pringle, and D. Johnson for sending plasmids and strains. N.B. was supported by an International Travelling Research Fellowship Award from the Wellcome Trust.
REFERENCES
- Alberts A, Bouquin N, Johnston LH, Treisman R. Analysis of RhoA-binding proteins reveals an interaction domain conserved in heterotrimeric G protein beta subunits and the yeast response-regulator protein Skn7. J Biol Chem. 1998;273:8616–8622. doi: 10.1074/jbc.273.15.8616. [DOI] [PubMed] [Google Scholar]
- Baum B, Wuarin J, Nurse P. Control of S-phase periodic transcription in the fission yeast mitotic cycle. EMBO J. 1997;16:4676–4688. doi: 10.1093/emboj/16.15.4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton BK, Tinkelenberg AH, Jean D, Plump SD, Cross FR. Genetic analysis of Cln/Cdc28 regulation of cell morphogenesis in budding yeast. EMBO J. 1993;12:5267–5275. doi: 10.1002/j.1460-2075.1993.tb06222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickle M, Delley P-A, Schmidt A, Hall MN. Cell wall integrity modulates RHO1 activity via the exchange factor ROM2. EMBO J. 1998;17:2235–2245. doi: 10.1093/emboj/17.8.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourret RB, Borkovich KA, Simon MI. Signal transduction pathways involving protein phosphorylation in prokaryotes. Annu Rev Biochem. 1991;60:401–441. doi: 10.1146/annurev.bi.60.070191.002153. [DOI] [PubMed] [Google Scholar]
- Brown JL, Bussey H, Stewart RC. Yeast Skn7p functions in a eukaryotic two-component regulatory pathway. EMBO J. 1994;13:5186–5194. doi: 10.1002/j.1460-2075.1994.tb06849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JL, North S, Bussey H. SKN7, a yeast multicopy suppressor of a mutation affecting cell wall β-glucan assembly, encodes a product with domains homologous to prokaryotic two-component regulators and to heat-shock transcription factors. J Bacteriol. 1993;175:6908–6915. doi: 10.1128/jb.175.21.6908-6915.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho RJ, et al. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol Cell. 1998;2:65–73. doi: 10.1016/s1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- Cvrckova F, Nasmyth K. Yeast G1 cyclins CLN1 and CLN2 and a GAP-like protein have a role in bud formation. EMBO J. 1993;12:5277–5286. doi: 10.1002/j.1460-2075.1993.tb06223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirick L, Böhm T, Nasmyth K. Roles and regulation of Cln-Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. EMBO J. 1995;14:4803–4813. doi: 10.1002/j.1460-2075.1995.tb00162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirick L, Moll T, Auer H, Nasmyth K. A central role for SWI6 in modulated cell cycle Start-specific transcription in yeast. Nature. 1992;357:508–513. doi: 10.1038/357508a0. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia coli vectors constructed with in vitro mutagenized yeast genes lacking six-bp restriction sites. Gene, 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters and LacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- Igual J-C, Johnson AL, Johnston LH. Coordinated regulation of gene expression by the cell cycle transcription factor SWI4 and the protein kinase C MAP kinase pathway for yeast cell integrity. EMBO J. 1996;15:5001–5013. [PMC free article] [PubMed] [Google Scholar]
- Johnston LH. Cell cycle control of gene expression in yeast. Trends Cell Biol. 1992;2:353–357. doi: 10.1016/0962-8924(92)90041-k. [DOI] [PubMed] [Google Scholar]
- Johnston LH, Lowndes NF. Cell cycle control of DNA synthesis in budding yeast. Nucleic Acids Res. 1992;20:2403–2410. doi: 10.1093/nar/20.10.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketala T, Brown JL, Stewart RC, Bussey H. Yeast Skn7p activity is modulated by the Sln1p-Ypd1p osmosensor and contributes to regulation of the HOG pathway. Mol Gen Genet. 1998;259:372–378. doi: 10.1007/s004380050824. [DOI] [PubMed] [Google Scholar]
- Koch C, Moll T, Neuberg M, Nasmyth K. A role for the transcription factors Mbp1 and Swi4 in progression from G1 to S phase. Science. 1993;261:1551–1557. doi: 10.1126/science.8372350. [DOI] [PubMed] [Google Scholar]
- Krems B, Charizanis C, Entian K-D. The response regulator-like protein Pos9/Skn7 of Saccharomyces cerevisiae is involved in oxidative stress resistance. Curr Genet. 1996;29:327–334. doi: 10.1007/BF02208613. [DOI] [PubMed] [Google Scholar]
- Lew DJ, Reed SI. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Ault A, Malone CL, Raitt D, Dean S, Johnston LH, Deschenes RJ, Fassler JS. The yeast histidine protein kinase, Sln1p, mediates phosphotransfer to two response regulators, Ssk1p and Skn7p. EMBO J. 1998;17:6952–6962. doi: 10.1093/emboj/17.23.6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowndes NF, Johnson AL, Breeden L, Johnston LH. SWI6 protein is required for transcription of the periodically expressed DNA synthesis genes in budding yeast. Nature. 1992;357:505–508. doi: 10.1038/357505a0. [DOI] [PubMed] [Google Scholar]
- Lowndes NF, Johnson AL, Johnston LH. Coordination of expression of DNA synthesis genes in budding yeast by a cell-cycle regulated trans factor. Nature. 1991;350:247–248. doi: 10.1038/350247a0. [DOI] [PubMed] [Google Scholar]
- Madden K, Sheu YJ, Baetz K, Andrews B, Snyder M. SBF cell cycle regulator as a target of the yeast PKC-MAP kinase pathway. Science. 1997;275:1781–1784. doi: 10.1126/science.275.5307.1781. [DOI] [PubMed] [Google Scholar]
- McInerny CJ, Patridge JF, Mikesell GE, Breeden L. A novel Mcm1-dependent element in the SWI4, CLN3, CDC6 and CDC47 promoters activates M/G1-specific transcription. Genes Dev. 1997;11:1277–1288. doi: 10.1101/gad.11.10.1277. [DOI] [PubMed] [Google Scholar]
- Mendenhall MD, Hodge AE. Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1998;62:1191–1243. doi: 10.1128/mmbr.62.4.1191-1243.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan BA, Banks GR, Toone WM, Raitt D, Kuge S, Johnston LH. The Skn7 response regulator controls gene expression in the oxidative stress response of the budding yeast Saccharomyces cerevisiae. EMBO J. 1997;16:1035–1044. doi: 10.1093/emboj/16.5.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan BA, Bouquin N, Johnston LH. Two-component signal transduction systems in budding yeast MAP a different pathway? Trends Cell Biol. 1995a;5:453–457. doi: 10.1016/s0962-8924(00)89114-x. [DOI] [PubMed] [Google Scholar]
- Morgan BA, Bouquin N, Merrill GF, Johnston LH. A yeast transcription factor bypassing the requirement for SBF and DSC1/MBF in budding yeast has homology to bacterial signal transduction proteins. EMBO J. 1995b;14:5679–5689. doi: 10.1002/j.1460-2075.1995.tb00255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JAH. Molecular Analysis of the Yeast Two-Micron Plasmid. Ph.D. Thesis. Heidelberg, Germany: European Molecular Biology Laboratory; 1987. [Google Scholar]
- Nakashima N, Tanaka K, Sturm S, Okayama H. Fission yeast Rep2 is a putative transcriptional activator subunit for the cell cycle ‘start’ function of Res2-Cdc10. EMBO J. 1995;14:4794–4802. doi: 10.1002/j.1460-2075.1995.tb00161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. Control of the yeast cell cycle by the Cdc28 protein kinase. Curr Opin Cell Biol. 1993;5:166–179. doi: 10.1016/0955-0674(93)90099-c. [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Dirick L. The role of SWI4 and SWI6 in the activity of G1 cyclins in yeast. Cell. 1991;66:995–1013. doi: 10.1016/0092-8674(91)90444-4. [DOI] [PubMed] [Google Scholar]
- Nonaka H, Tanaka K, Hirano H, Fujiwara T, Kohno H, Umikawa M, Mino A, Takai Y. A downstream target of RHO1 small GTP-binding protein is PKC1, a homolog of protein kinase C, which leads to activation of the MAP kinase cascade in Saccharomyces cerevisiae. EMBO J. 1995;14:5931–5938. doi: 10.1002/j.1460-2075.1995.tb00281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paravicini G, Cooper M, Friedli L, Smith DJ, Carpentier JL, Klig LS, Payton MA. The osmotic integrity of the yeast cell requires a functional PKC1 gene product. Mol Cell Biol. 1992;12:4896–4905. doi: 10.1128/mcb.12.11.4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson JS. Signal transduction schemes in bacteria. Cell. 1993;73:857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- Partridge JF, Mikesell GE, Breedan L. Cell cycle-dependent transcription of CLN1 involves Swi4 binding to MCB-like elements. J Biol Chem. 1997;272:9071–9077. doi: 10.1074/jbc.272.14.9071. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Bi E, Harkins HA, Zahner JE, De Virgilio C, Chant J, Corrado K, Fares H. Establishment of cell polarity in yeast. Cold Spring Harbor Symp Quant Biol. 1995;60:729–744. doi: 10.1101/sqb.1995.060.01.079. [DOI] [PubMed] [Google Scholar]
- Quon KC, Marczynski GT, Shapiro L. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- Siegmund RF, Nasmyth KA. The Saccharomyces cerevisiae Start-specific transcription factor Swi4 interacts through the ankyrin repeats with the mitotic Clb2/Cdc28 kinase and through its conserved carboxy terminus with Swi6. Mol Cell Biol. 1996;16:2647–2655. doi: 10.1128/mcb.16.6.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell-cycle regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Cell Biol. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart D, Wittenberg C. CLN3, not positive feedback, determines the timing of CLN2 transcription in cycling cells. Genes Dev. 1995;9:2780–2794. doi: 10.1101/gad.9.22.2780. [DOI] [PubMed] [Google Scholar]
- Yu G, Deschenes RJ, Fassler JS. The essential transcription factor, Mcm1, is a downstream target of Sln1, a yeast ‘two-component’ regulator. J Biol Chem. 1995;270:8739–8743. doi: 10.1074/jbc.270.15.8739. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Tadeka T, Nasmyth K, Jones N. pct1+, which encodes a new DNA-binding partner of p85cdc10, is required in meiosis in the fission yeast Schizosaccharomyces pombe. Genes Dev. 1994;8:885–898. doi: 10.1101/gad.8.8.885. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Tadeka T, Whitehall S, Peat N, Jones N. Functional characterization of the fission yeast Start-specific transcription factor Res2. EMBO J. 1997;16:1023–1034. doi: 10.1093/emboj/16.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziman M, O’Brien JM, Ouellette LA, Church WR, Johnson DI. Mutational analysis of CDC42Sc, a Saccharomyces cerevisiae gene that encodes a putative GTP-binding protein involved in the control of cell polarity. Mol Cell Biol, 1991;11:3537–3544. doi: 10.1128/mcb.11.7.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]