Abstract
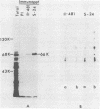
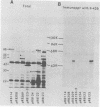
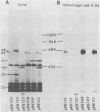
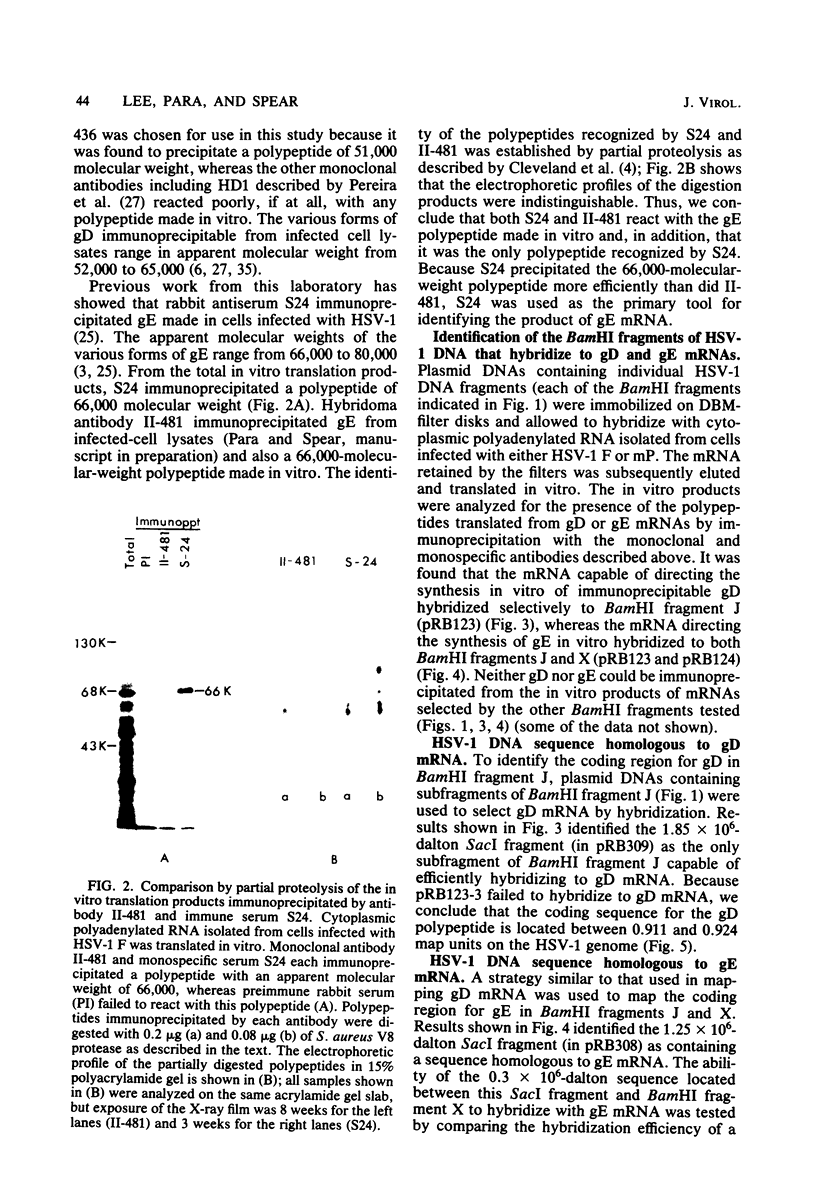
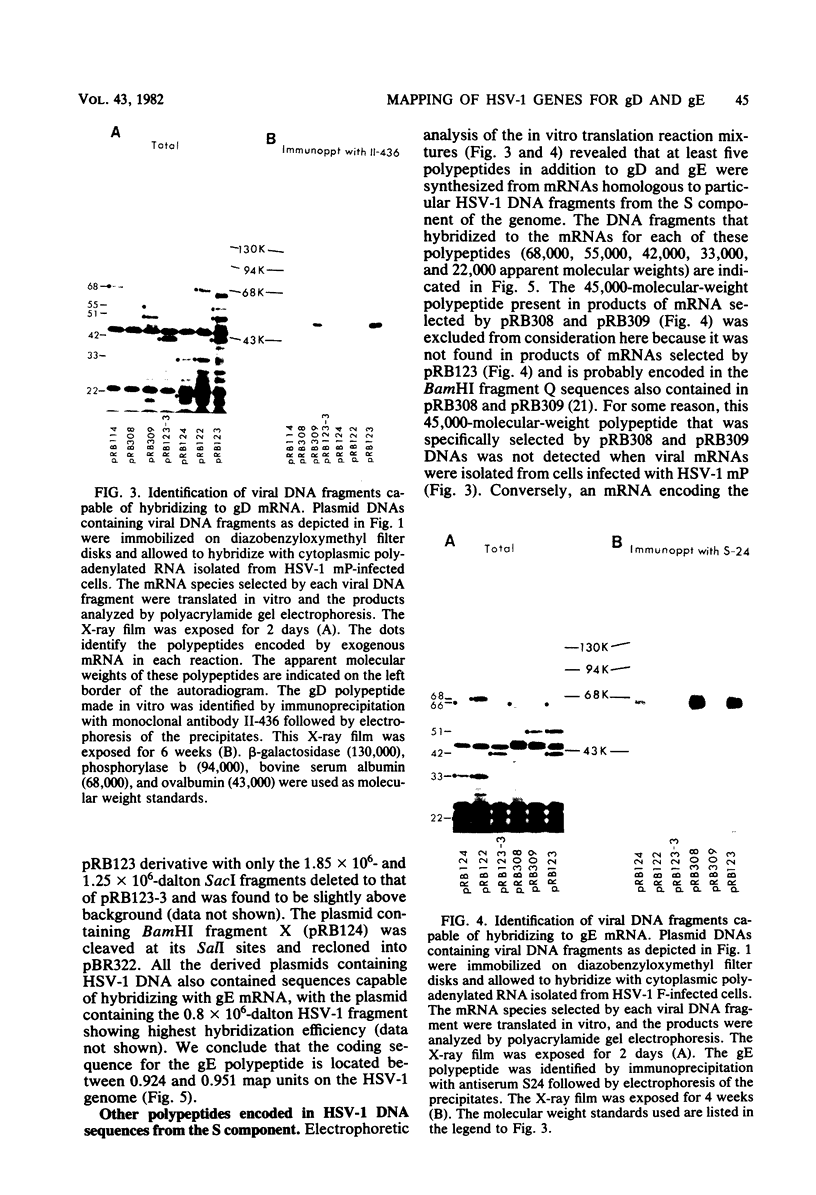
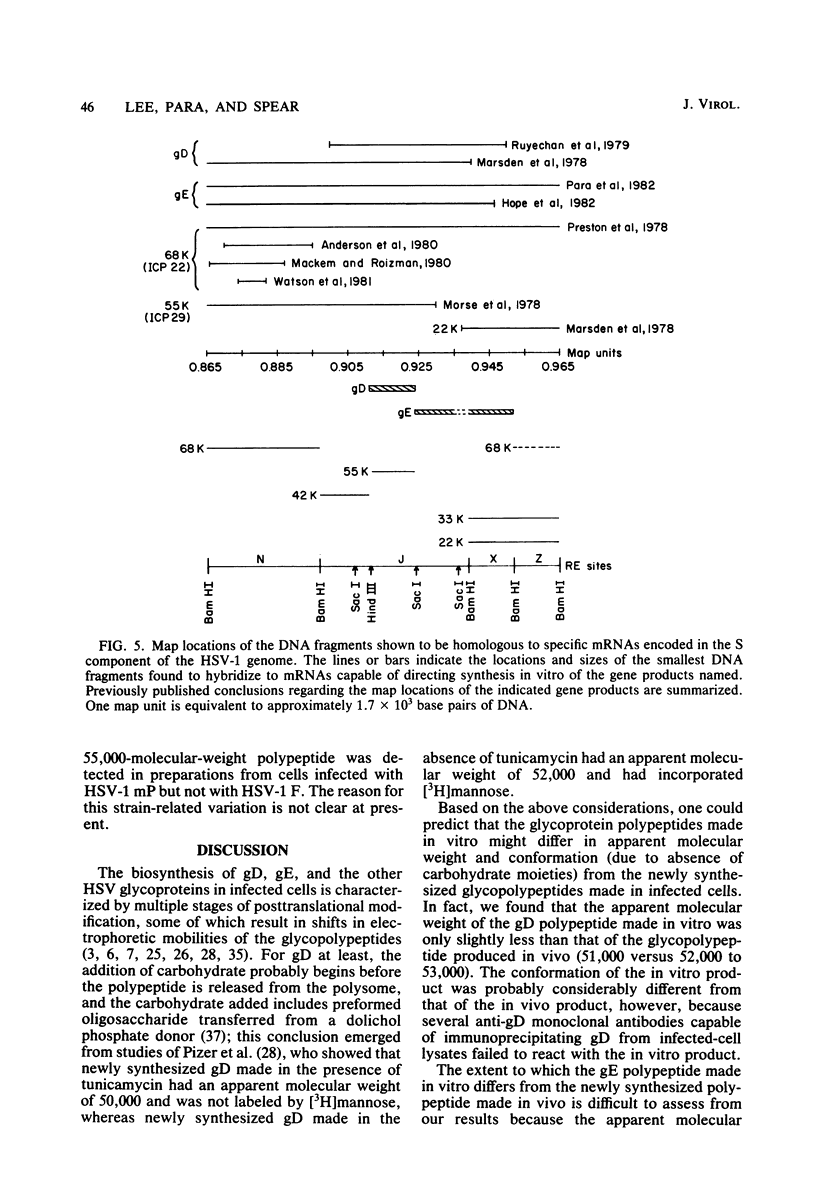
To map the structural genes for the gD and gE polypeptides and for other viral products encoded in the S component of herpes simplex virus type 1 DNA, we selected mRNAs capable of hybridizing to cloned viral DNA fragments and translated the mRNAs in vitro to determine which polypeptides were encoded therein. The gD and gE polypeptides were identified by immunoprecipitation with appropriate monoclonal and monospecific antibodies, whereas the other polypeptides were characterized only by their electrophoretic mobilities in polyacrylamide gels. We found that gD mRNA hybridized to a single SacI subfragment of BamHI fragment J, whereas gE mRNA hybridized to an adjacent SacI subfragment of BamHI fragment J and also to BamHI fragment X. These and other results permit the conclusion that the structural gene for gD is located between map coordinates 0.911 and 0.924, and the gene for gE is between map coordinates 0.924 and 0.951. We also found that mRNAs for polypeptides of 55,000, 42,000, 33,000, and 22,000 molecular weight hybridized to DNA fragments spanning the regions from map coordinates 0.911 to 0.924, 0.897 to 0.911, 0.939 to 0.965, and 0.939 to 0.965, respectively. Finally, in accord with the results of others, we found that mRNA for a 68,000-molecular-weight polypeptide hybridized to the two noncontiguous BamHI fragments N and Z, which share a reiterated DNA sequence.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson K. P., Costa R. H., Holland L. E., Wagner E. K. Characterization of herpes simplex virus type 1 RNA present in the absence of de novo protein synthesis. J Virol. 1980 Apr;34(1):9–27. doi: 10.1128/jvi.34.1.9-27.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson K. P., Stringer J. R., Holland L. E., Wagner E. K. Isolation and localization of herpes simplex virus type 1 mRNA. J Virol. 1979 Jun;30(3):805–820. doi: 10.1128/jvi.30.3.805-820.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucke R. B., Spear P. G. Membrane proteins specified by herpes simplex viruses. V. Identification of an Fc-binding glycoprotein. J Virol. 1979 Dec;32(3):779–789. doi: 10.1128/jvi.32.3.779-789.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland D. W., Fischer S. G., Kirschner M. W., Laemmli U. K. Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J Biol Chem. 1977 Feb 10;252(3):1102–1106. [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Properties of a supercoiled deoxyribonucleic acid-protein relaxation complex and strand specificity of the relaxation event. Biochemistry. 1970 Oct 27;9(22):4428–4440. doi: 10.1021/bi00824a026. [DOI] [PubMed] [Google Scholar]
- Cohen G. H., Long D., Eisenberg R. J. Synthesis and processing of glycoproteins gD and gC of herpes simplex virus type 1. J Virol. 1980 Nov;36(2):429–439. doi: 10.1128/jvi.36.2.429-439.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle R., Courtney R. J. gA and gB glycoproteins of herpes simplex virus type 1: two forms of a single polypeptide. J Virol. 1980 Dec;36(3):665–675. doi: 10.1128/jvi.36.3.665-675.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg R. J., Ponce de Leon M., Cohen G. H. Comparative structural analysis of glycoprotein gD of herpes simplex virus types 1 and 2. J Virol. 1980 Aug;35(2):428–435. doi: 10.1128/jvi.35.2.428-435.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejercito P. M., Kieff E. D., Roizman B. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol. 1968 May;2(3):357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- Goldberg M. L., Lifton R. P., Stark G. R., Williams J. G. Isolation of specific RNA's using DNA covalently linked to diazobenzyloxymethyl cellulose or paper. Methods Enzymol. 1979;68:206–220. doi: 10.1016/0076-6879(79)68016-3. [DOI] [PubMed] [Google Scholar]
- HOGGAN M. D., ROIZMAN B. The isolation and properties of a variant of Herpes simplex producing multinucleated giant cells in monolayer cultures in the presence of antibody. Am J Hyg. 1959 Sep;70:208–219. doi: 10.1093/oxfordjournals.aje.a120071. [DOI] [PubMed] [Google Scholar]
- Heine J. W., Honess R. W., Cassai E., Roizman B. Proteins specified by herpes simplex virus. XII. The virion polypeptides of type 1 strains. J Virol. 1974 Sep;14(3):640–651. doi: 10.1128/jvi.14.3.640-651.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland L. E., Anderson K. P., Stringer J. R., Wagner E. K. Isolation and localization of herpes simplex virus type 1 mRNA abundant before viral DNA synthesis. J Virol. 1979 Aug;31(2):447–462. doi: 10.1128/jvi.31.2.447-462.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honess R. W., Roizman B. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J Virol. 1974 Jul;14(1):8–19. doi: 10.1128/jvi.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope R. G., Palfreyman J., Suh M., Marsden H. S. Sulphated glycoproteins induced by herpes simplex virus. J Gen Virol. 1982 Feb;58(Pt 2):399–415. doi: 10.1099/0022-1317-58-2-399. [DOI] [PubMed] [Google Scholar]
- Kessler S. W. Rapid isolation of antigens from cells with a staphylococcal protein A-antibody adsorbent: parameters of the interaction of antibody-antigen complexes with protein A. J Immunol. 1975 Dec;115(6):1617–1624. [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D. Quantitative film detection of 3H and 14C in polyacrylamide gels by fluorography. Eur J Biochem. 1975 Aug 15;56(2):335–341. doi: 10.1111/j.1432-1033.1975.tb02238.x. [DOI] [PubMed] [Google Scholar]
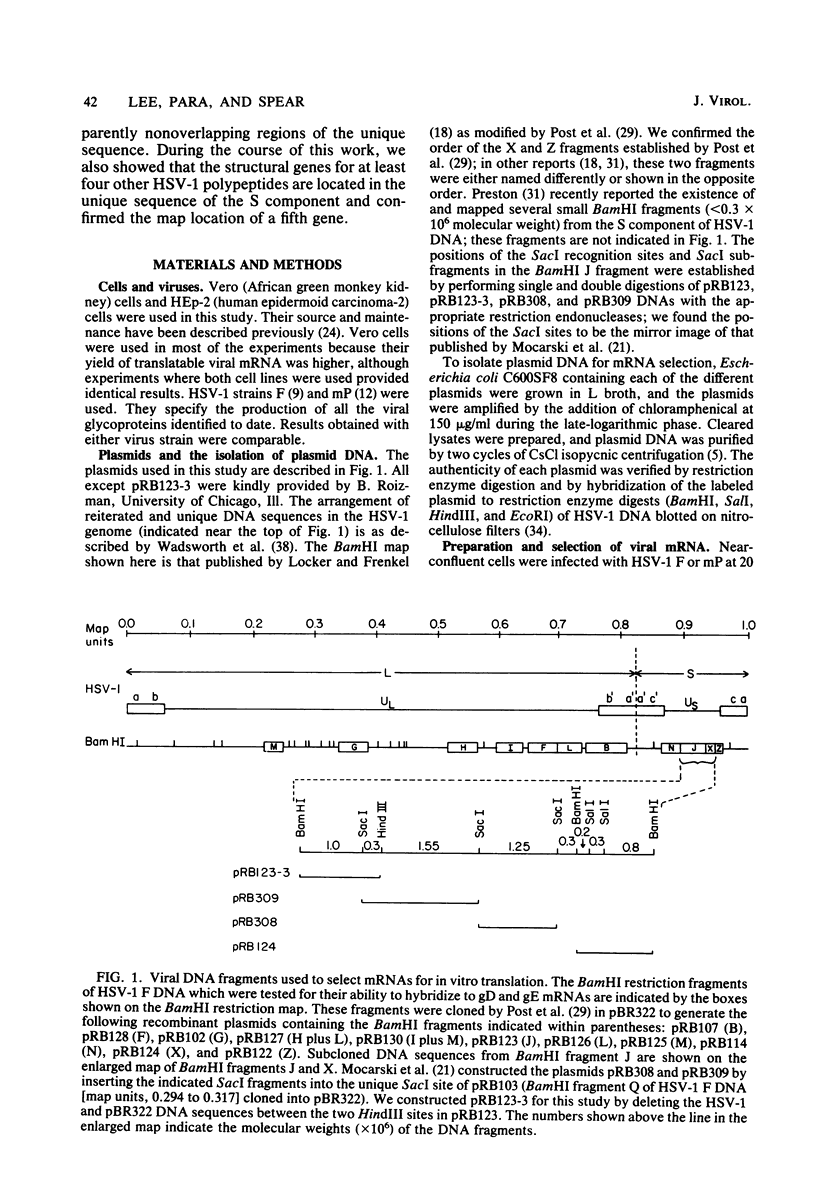
- Locker H., Frenkel N. BamI, KpnI, and SalI restriction enzyme maps of the DNAs of herpes simplex virus strains Justin and F: occurrence of heterogeneities in defined regions of the viral DNA. J Virol. 1979 Nov;32(2):429–441. doi: 10.1128/jvi.32.2.429-441.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackem S., Roizman B. Regulation of herpesvirus macromolecular synthesis: transcription-initiation sites and domains of alpha genes. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7122–7126. doi: 10.1073/pnas.77.12.7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden H. S., Stow N. D., Preston V. G., Timbury M. C., Wilkie N. M. Physical mapping of herpes simplex virus-induced polypeptides. J Virol. 1978 Nov;28(2):624–642. doi: 10.1128/jvi.28.2.624-642.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski E. S., Post L. E., Roizman B. Molecular engineering of the herpes simplex virus genome: insertion of a second L-S junction into the genome causes additional genome inversions. Cell. 1980 Nov;22(1 Pt 1):243–255. doi: 10.1016/0092-8674(80)90172-5. [DOI] [PubMed] [Google Scholar]
- Morse L. S., Pereira L., Roizman B., Schaffer P. A. Anatomy of herpes simplex virus (HSV) DNA. X. Mapping of viral genes by analysis of polypeptides and functions specified by HSV-1 X HSV-2 recombinants. J Virol. 1978 May;26(2):389–410. doi: 10.1128/jvi.26.2.389-410.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrild B., Ludwig H., Rott R. Identification of a common antigen of herpes simplex virus bovine herpes mammillitis virus, and B virus. J Virol. 1978 Jun;26(3):712–717. doi: 10.1128/jvi.26.3.712-717.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Para M. F., Baucke R. B., Spear P. G. Glycoprotein gE of herpes simplex virus type 1: effects of anti-gE on virion infectivity and on virus-induced fc-binding receptors. J Virol. 1982 Jan;41(1):129–136. doi: 10.1128/jvi.41.1.129-136.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Para M. F., Baucke R. B., Spear P. G. Immunoglobulin G(Fc)-binding receptors on virions of herpes simplex virus type 1 and transfer of these receptors to the cell surface by infection. J Virol. 1980 May;34(2):512–520. doi: 10.1128/jvi.34.2.512-520.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Para M. F., Goldstein L., Spear P. G. Similarities and differences in the Fc-binding glycoprotein (gE) of herpes simplex virus types 1 and 2 and tentative mapping of the viral gene for this glycoprotein. J Virol. 1982 Jan;41(1):137–144. doi: 10.1128/jvi.41.1.137-144.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L., Klassen T., Baringer J. R. Type-common and type-specific monoclonal antibody to herpes simplex virus type 1. Infect Immun. 1980 Aug;29(2):724–732. doi: 10.1128/iai.29.2.724-732.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizer L. I., Cohen G. H., Eisenberg R. J. Effect of tunicamycin on herpes simplex virus glycoproteins and infectious virus production. J Virol. 1980 Apr;34(1):142–153. doi: 10.1128/jvi.34.1.142-153.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post L. E., Conley A. J., Mocarski E. S., Roizman B. Cloning of reiterated and nonreiterated herpes simplex virus 1 sequences as BamHI fragments. Proc Natl Acad Sci U S A. 1980 Jul;77(7):4201–4205. doi: 10.1073/pnas.77.7.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post L. E., Roizman B. A generalized technique for deletion of specific genes in large genomes: alpha gene 22 of herpes simplex virus 1 is not essential for growth. Cell. 1981 Jul;25(1):227–232. doi: 10.1016/0092-8674(81)90247-6. [DOI] [PubMed] [Google Scholar]
- Preston V. G., Davison A. J., Marsden H. S., Timbury M. C., Subak-Sharpe J. H., Wilkie N. M. Recombinants between herpes simplex virus types 1 and 2: analyses of genome structures and expression of immediate early polypeptides. J Virol. 1978 Nov;28(2):499–517. doi: 10.1128/jvi.28.2.499-517.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston V. G. Fine-structure mapping of herpes simplex virus type 1 temperature-sensitive mutations within the short repeat region of the genome. J Virol. 1981 Jul;39(1):150–161. doi: 10.1128/jvi.39.1.150-161.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruyechan W. T., Morse L. S., Knipe D. M., Roizman B. Molecular genetics of herpes simplex virus. II. Mapping of the major viral glycoproteins and of the genetic loci specifying the social behavior of infected cells. J Virol. 1979 Feb;29(2):677–697. doi: 10.1128/jvi.29.2.677-697.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spear P. G. Membrane proteins specified by herpes simplex viruses. I. Identification of four glycoprotein precursors and their products in type 1-infected cells. J Virol. 1976 Mar;17(3):991–1008. doi: 10.1128/jvi.17.3.991-1008.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struck D. K., Lennarz W. J. Evidence for the participation of saccharide-lipids in the synthesis of the oligosaccharide chain of ovalbumin. J Biol Chem. 1977 Feb 10;252(3):1007–1013. [PubMed] [Google Scholar]
- Watson R. J., Sullivan M., Vande Woude G. F. Structures of two spliced herpes simplex virus type 1 immediate-early mRNA's which map at the junctions of the unique and reiterated regions of the virus DNA S component. J Virol. 1981 Jan;37(1):431–444. doi: 10.1128/jvi.37.1.431-444.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westmoreland D., Watkins J. F. The IgG receptor induced by herpes simplex virus: studies using radioiodinated IgG. J Gen Virol. 1974 Jul;24(1):167–178. doi: 10.1099/0022-1317-24-1-167. [DOI] [PubMed] [Google Scholar]