Abstract
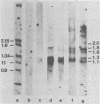
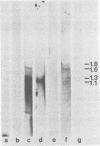
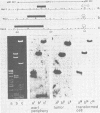
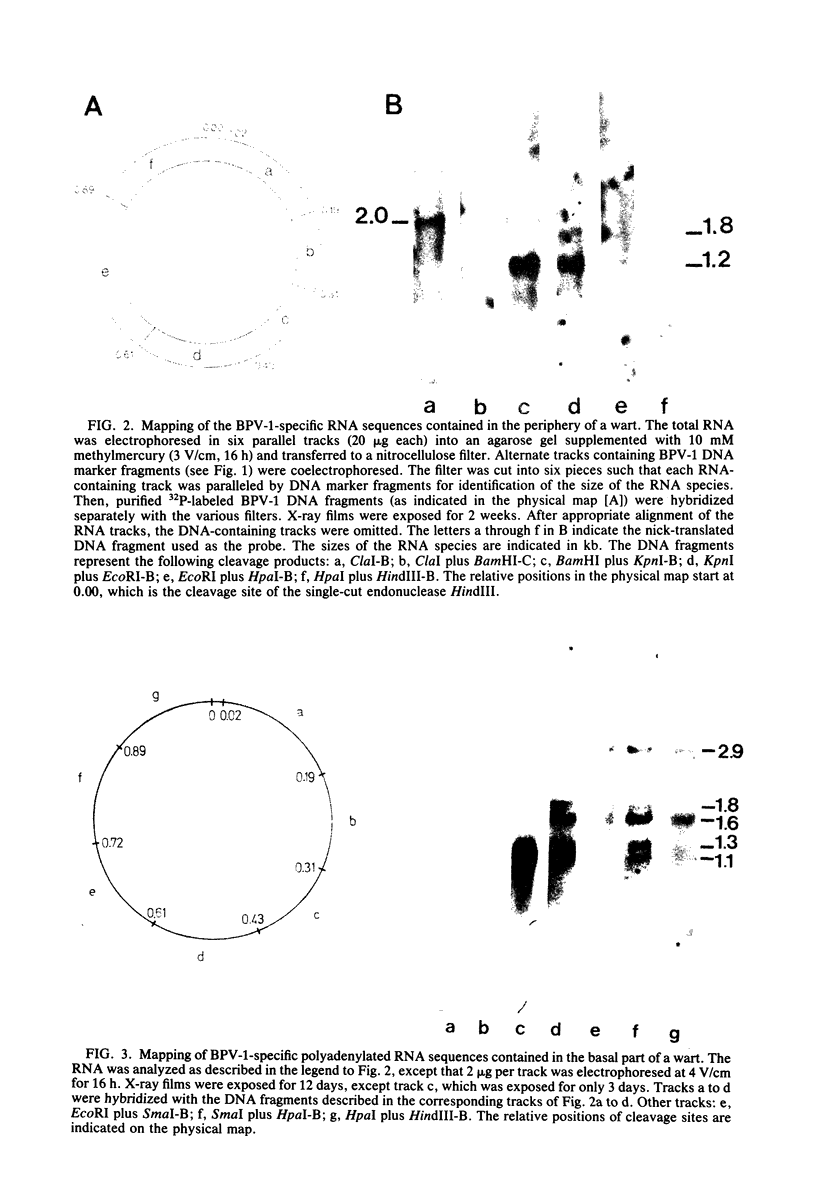
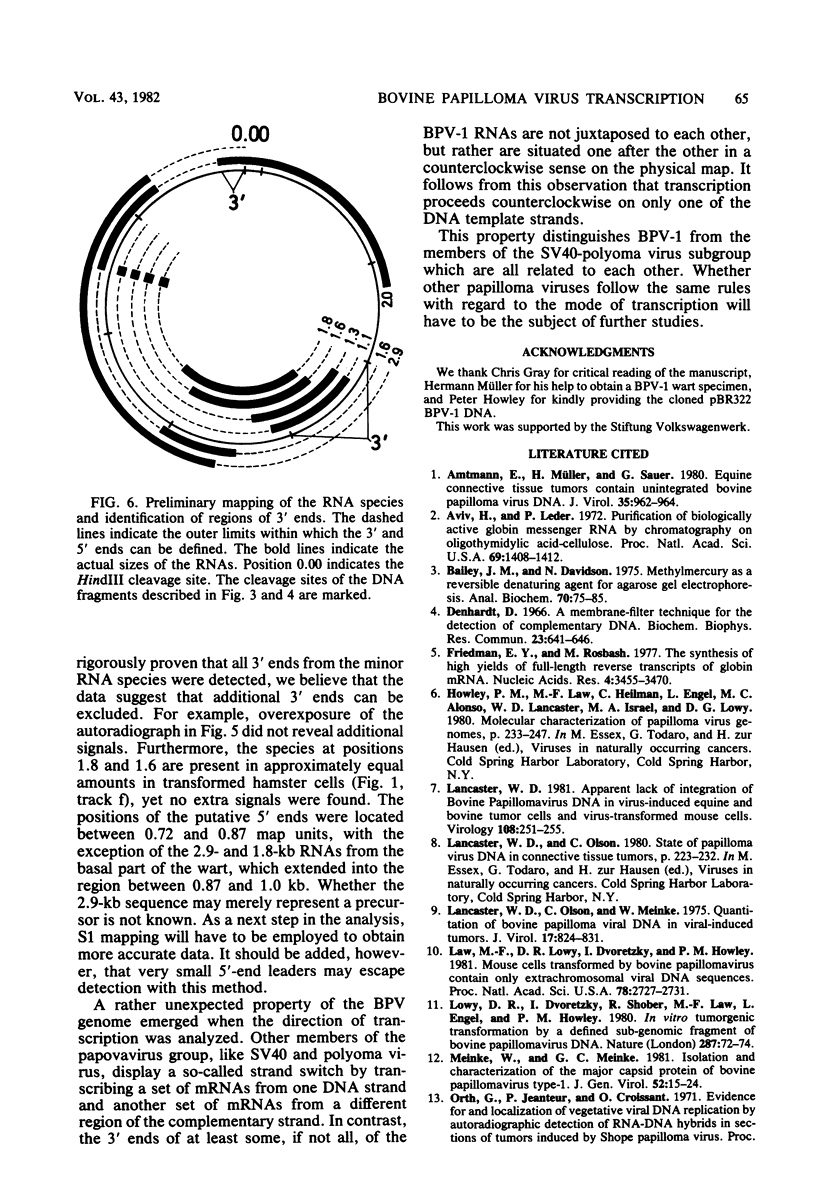
The bovine papilloma virus type 1 (BPV-1)-specific RNA species were identified in virus-induced bovine warts, hamster tumors, and transformed hamster and mouse cells. In each case two major species were present (1.1 and 1.3 kilobases [kb]). Also two species of 1.6 and 1.8 kb appearing in variable amounts were found. Only in the keratinized periphery of the warts, where virus replication takes place, was it possible to reveal an additional 2-kb RNA species. In this tissue, however, the 1.6-kb species was not detected. The basal part of a bovine wart contained an additional minor, 2.9-kb, BPV-1-specific RNA sequence. By hybridization with purified defined BPV-1 DNA fragments it was shown that most of the coding sequences of the 2-kb species were transcribed from a region between 0.02 and 0.19 map units. The majority of the coding sequences of the smaller species in transformed cells were located in the region between 0.31 and 0.61 map units. The putative 5′ ends mapped between 0.72 and 0.96 map units. Oligodeoxythymidylic acid-primed [32P]cDNA was synthesized from various RNA preparations to generate probes for the detection of 3′ termini of the polyadenylated BPV-1 RNAs. By hybridization across the BPV-1 genome only one signal between the map positions 0.30 and 0.40 was obtained when RNA from transformed cells and from a tumor was used as a template. In contrast, RNA from the periphery of a wart led to the detection of an additional signal which was confined to the region between 0.96 and 1.00 map units. From the arrangement of both the 3′ termini and the coding areas along the viral genome it appears that several RNA species are transcribed from one DNA strand.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amtmann E., Müller H., Sauer G. Equine connective tissue tumors contain unintegrated bovine papilloma virus DNA. J Virol. 1980 Sep;35(3):962–964. doi: 10.1128/jvi.35.3.962-964.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J. M., Davidson N. Methylmercury as a reversible denaturing agent for agarose gel electrophoresis. Anal Biochem. 1976 Jan;70(1):75–85. doi: 10.1016/s0003-2697(76)80049-8. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Friedman E. Y., Rosbash M. The syntheiss of high yields of full-length reverse transcripts of globin mRNA. Nucleic Acids Res. 1977 Oct;4(10):3455–3471. doi: 10.1093/nar/4.10.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster W. D. Apparent lack of integration of bovine papillomavirus DNA in virus-induced equine and bovine tumor cells and virus-transformed mouse cells. Virology. 1981 Jan 30;108(2):251–255. doi: 10.1016/0042-6822(81)90433-5. [DOI] [PubMed] [Google Scholar]
- Lancaster W. D., Olson C., Meinke W. Quantitation of bovine papilloma viral DNA in viral-induced tumors. J Virol. 1976 Mar;17(3):824–831. doi: 10.1128/jvi.17.3.824-831.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law M. F., Lowy D. R., Dvoretzky I., Howley P. M. Mouse cells transformed by bovine papillomavirus contain only extrachromosomal viral DNA sequences. Proc Natl Acad Sci U S A. 1981 May;78(5):2727–2731. doi: 10.1073/pnas.78.5.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowy D. R., Dvoretzky I., Shober R., Law M. F., Engel L., Howley P. M. In vitro tumorigenic transformation by a defined sub-genomic fragment of bovine papilloma virus DNA. Nature. 1980 Sep 4;287(5777):72–74. doi: 10.1038/287072a0. [DOI] [PubMed] [Google Scholar]
- Meinke W., Meinke G. C. Isolation and characterization of the major capsid protein of bovine papilloma virus type 1. J Gen Virol. 1981 Jan;52(Pt 1):15–24. doi: 10.1099/0022-1317-52-1-15. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- SMITHIES L. K., OLSON C. CORRELATION OF IMMUNOFLUORESCENCE AND INFECTIVITY IN THE DEVELOPING BOVINE CUTANEOUS PAPILLOMA. Cancer Res. 1964 May;24:674–681. [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]