Abstract
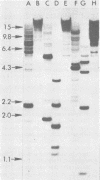
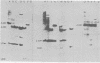
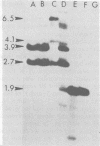
The structure of the endogenous murine leukemia virus (MuLV) sequences of NIH/Swiss mice was analyzed by restriction endonuclease digestion, gel electrophoresis, and hybridization to an MuLV nucleic acid probe. Digestion of mouse DNA with certain restriction endonucleases revealed two classes of fragments. A large number of fragments (about 30) were present at a relatively low concentration, indicating that each derived from a sequence present once in the mouse genome. A smaller number of fragments (one to five) were present at a much higher concentration and must have resulted from sequences present multiple times in the mouse genome. These results indicated that the endogenous MuLV sequences represent a family of dispersed repetitive sequences. Hybridization of these same digested mouse DNAs to nucleic acid probes representing different portions of the MuLV genome allowed construction of a map of the sites where restriction endonucleases cleave the endogenous MuLV sequences. Several independent recombinant DNA clones of endogenous MuLV sequences have been isolated from C3H mice (Roblin et al., J. Virol. 43:113-126, 1982). Analysis of these sequences shows that they have the structure of MuLV proviruses. The sites at which restriction endonucleases cleave within these proviruses appeared to be similar or identical to the sites at which these nucleases cleaved within the MuLV sequences of NIH/Swiss mice. This identity was confirmed by parallel electrophoresis. We conclude that the apparently complex pattern of endogenous MuLV sequences of NIH/Swiss mice consists largely of only two kinds of provirus, each repeated multiple times at dispersed sites in the mouse genome.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chattopadhyay S. K., Lander M. R., Gupta S., Rands E., Lowy D. R. Origin of mink cytopathic focus-forming (MCF) viruses:comparison with ecotropic and xenotropic murine leukemia virus genomes. Virology. 1981 Sep;113(2):465–483. doi: 10.1016/0042-6822(81)90175-6. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S. K., Lowy D. R., Teich N. M., Levine A. S., Rowe W. P. Qualitative and quantitative studies of AKR-type murine leukemia virus sequences in mouse DNA. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):1085–1101. doi: 10.1101/sqb.1974.039.01.124. [DOI] [PubMed] [Google Scholar]
- Chien Y. H., Verma I. M., Shih T. Y., Scolnick E. M., Davidson N. Heteroduplex analysis of the sequence relations between the RNAs of mink cell focus-inducing and murine leukemia viruses. J Virol. 1978 Oct;28(1):352–360. doi: 10.1128/jvi.28.1.352-360.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolberg D. S., Bacheler L. T., Fan H. Endogenous type C retroviral sequences of mice are organized in a small number of virus-like classes and have been acquired recently. J Virol. 1981 Oct;40(1):96–106. doi: 10.1128/jvi.40.1.96-106.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier F., Bünemann H., Grotjahn L. Analysis of calf-thymus satellite DNA: evidence for specific methylation of cytosine in C-G sequences. Eur J Biochem. 1977 Oct 17;80(1):175–183. doi: 10.1111/j.1432-1033.1977.tb11869.x. [DOI] [PubMed] [Google Scholar]
- Kozak C., Rowe W. P. Genetic mapping of xenotropic leukemia virus-inducing loci in two mouse strains. Science. 1978 Mar 31;199(4336):1448–1449. doi: 10.1126/science.204014. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Roblin R., Young J. M., Mural R. J., Bell T. E., Ihle J. N. Molecular cloning and characterization of murine leukemia virus-related DNA sequences from C3H/HeN mouse DNA. J Virol. 1982 Jul;43(1):113–126. doi: 10.1128/jvi.43.1.113-126.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy P. H., Weissbach A. DNA methylase from HeLa cell nuclei. Nucleic Acids Res. 1975 Oct;2(10):1669–1684. doi: 10.1093/nar/2.10.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Steffen D. L., Bird S., Weinberg R. A. Evidence for the Asiatic origin of endogenous AKR-type murine leukemia proviruses. J Virol. 1980 Sep;35(3):824–835. doi: 10.1128/jvi.35.3.824-835.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen D., Bird S., Rowe W. P., Weinberg R. A. Identification of DNA fragments carrying ecotropic proviruses of AKR mice. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4554–4558. doi: 10.1073/pnas.76.9.4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen D., Weinberg R. A. The integrated genome of murine leukemia virus. Cell. 1978 Nov;15(3):1003–1010. doi: 10.1016/0092-8674(78)90284-2. [DOI] [PubMed] [Google Scholar]
- Yoshimura F. K., Weinberg R. A. Restriction endonuclease cleavage of linear and closed circular murine leukemia viral DNAs: discovery of a smaller circular form. Cell. 1979 Feb;16(2):323–332. doi: 10.1016/0092-8674(79)90009-6. [DOI] [PubMed] [Google Scholar]