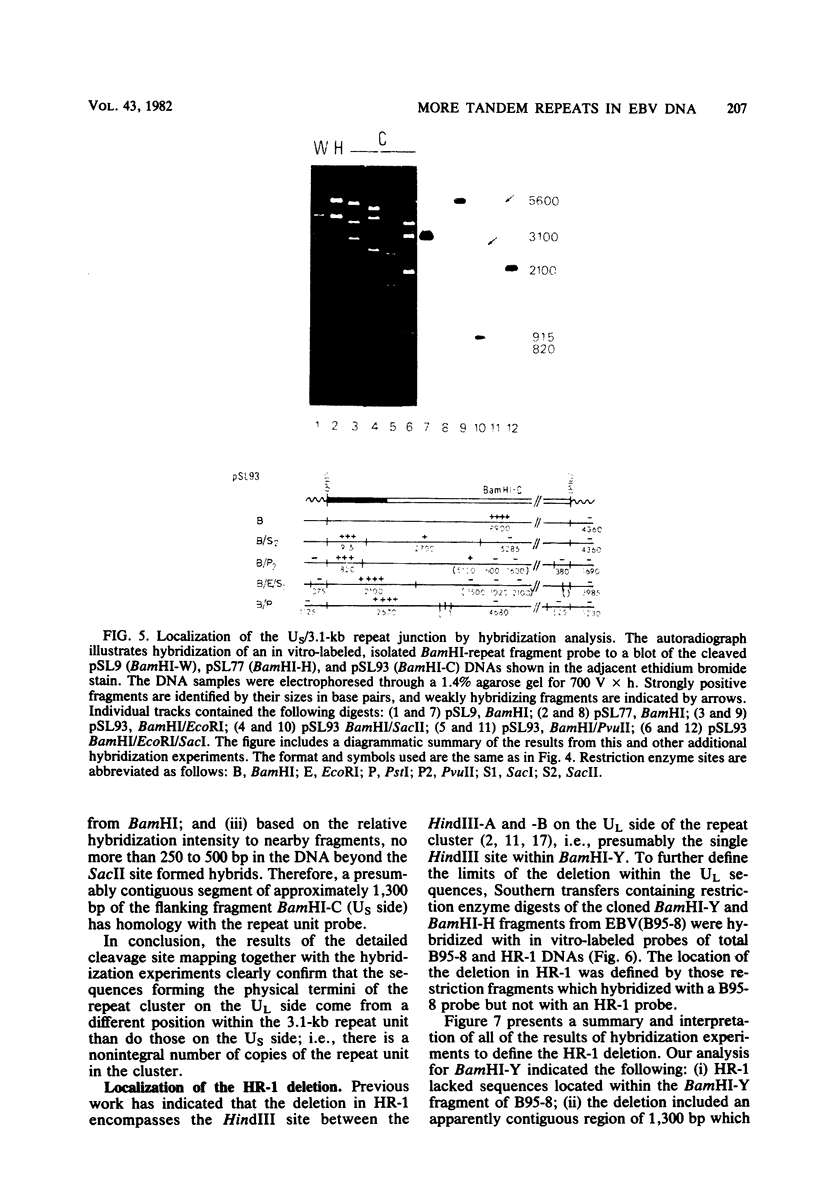
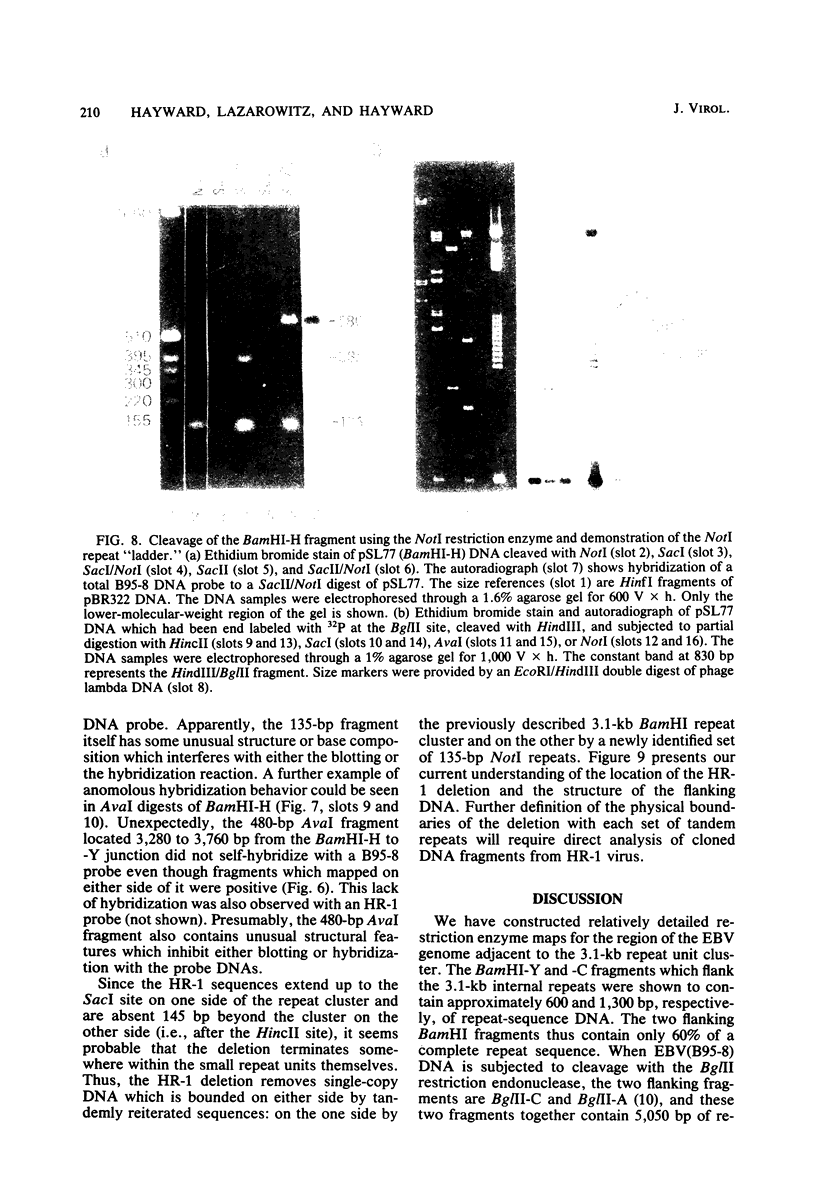
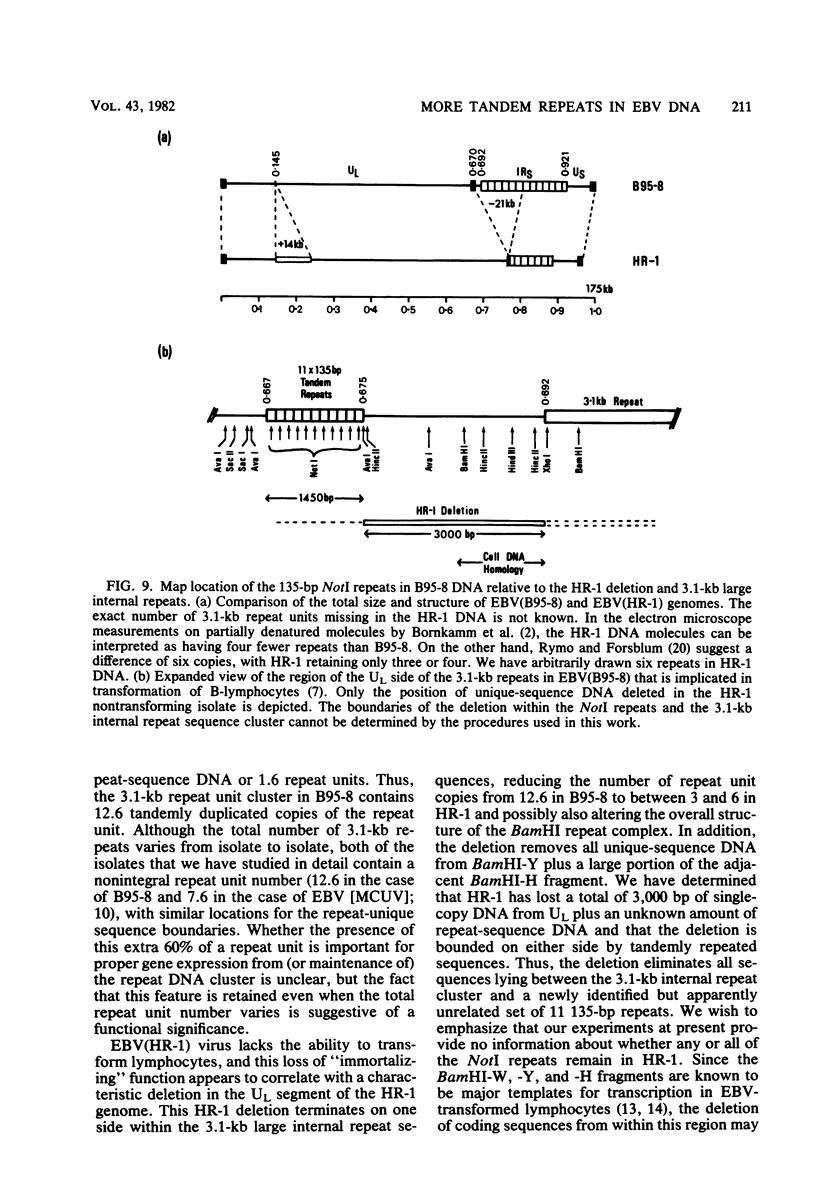
Abstract
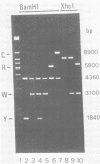
We used cloned BamHI fragments from Epstein-Barr virus strain B95-8 [EBV(B95-8)]DNA to obtain detailed restriction maps of the region of the genome adjacent to the large internal repeat cluster. These maps together with the results of hybridization experiments using a 3.1-kilobase repeat probe defined more precisely the location of the injection between the internal repeat cluster and the flanking unique-sequence DNA. On one side (UL), the repeat sequences extended 600 +/- 80 base pairs (bp) into BamHI-Y; on the other side (US), they extended 1,300 +/- 200 bp into BamHI-C. Therefore, EBV(B95-8) DNA contained a nonintegral number of 3.1-kilobase repeat units, namely, 12.6 copies. The mapping studies also revealed a second series of internal tandem repetitions in EBV(B95-8) DNA located within the BamHI-H fragment. This cluster comprised 11 copies of a 135-bp repeat unit which contained a single site for the NotI restriction endonuclease. Hybridization to these cloned EBV(B95-8) fragments using total EBV(HR-1) DNA as probe indicated that the deletion in EBV(HR-1) removed all 3,000 bp of unique-sequence DNA which lay between the large 3.1-kilobase and the small 135-bp repeat clusters. Thus, the deletion which destroyed the transforming ability in the EBV(HR-1) virus was bounded on either side by tandem repetitions.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornkamm G. W., Delius H., Zimber U., Hudewentz J., Epstein M. A. Comparison of Epstein-Barr virus strains of different origin by analysis of the viral DNAs. J Virol. 1980 Sep;35(3):603–618. doi: 10.1128/jvi.35.3.603-618.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delius H., Bornkamm G. W. Heterogeneity of Epstein-Barr virus. III. Comparison of a transforming and a nontransforming virus by partial denaturation mapping of their DNAs. J Virol. 1978 Jul;27(1):81–89. doi: 10.1128/jvi.27.1.81-89.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer D. K., Miller G., Gradoville L., Heston L., Westrate M. W., Maris W., Wright J., Brandsma J., Summers W. C. Genome of a mononucleosis Epstein-Barr virus contains DNA fragments previously regarded to be unique to Burkitt's lymphoma isolates. Cell. 1981 May;24(2):543–553. doi: 10.1016/0092-8674(81)90345-7. [DOI] [PubMed] [Google Scholar]
- Given D., Kieff E. DNA of Epstein-Barr virus. VI. Mapping of the internal tandem reiteration. J Virol. 1979 Aug;31(2):315–324. doi: 10.1128/jvi.31.2.315-324.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Given D., Yee D., Griem K., Kieff E. DNA of Epstein-Barr virus. V. Direct repeats of the ends of Epstein-Barr virus DNA. J Virol. 1979 Jun;30(3):852–862. doi: 10.1128/jvi.30.3.852-862.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward G. S., Smith M. G. The chromosome of bacteriophage T5. I. Analysis of the single-stranded DNA fragments by agarose gel electrophoresis. J Mol Biol. 1972 Feb 14;63(3):383–395. doi: 10.1016/0022-2836(72)90435-4. [DOI] [PubMed] [Google Scholar]
- Hayward S. D., Nogee L., Hayward G. S. Organization of repeated regions within the Epstein-Barr virus DNA molecule. J Virol. 1980 Jan;33(1):507–521. doi: 10.1128/jvi.33.1.507-521.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller M., Dambaugh T., Kieff E. Epstein-Barr virus DNA. IX. Variation among viral DNAs from producer and nonproducer infected cells. J Virol. 1981 May;38(2):632–648. doi: 10.1128/jvi.38.2.632-648.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. C., Hayward G. S., Roizman B. Anatomy of herpes simplex virus DNA VII. alpha-RNA is homologous to noncontiguous sites in both the L and S components of viral DNA. J Virol. 1977 Jan;21(1):268–276. doi: 10.1128/jvi.21.1.268-276.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King W., Thomas-Powell A. L., Raab-Traub N., Hawke M., Kieff E. Epstein-Barr virus RNA. V. Viral RNA in a restringently infected, growth-transformed cell line. J Virol. 1980 Nov;36(2):506–518. doi: 10.1128/jvi.36.2.506-518.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King W., Van Santen V., Kieff E. Epstein-Barr virus RNA. VI. Viral RNA in restringently and abortively infected Raji cells. J Virol. 1981 May;38(2):649–660. doi: 10.1128/jvi.38.2.649-660.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kintner C. R., Sugden B. The structure of the termini of the DNA of Epstein-Barr virus. Cell. 1979 Jul;17(3):661–671. doi: 10.1016/0092-8674(79)90273-3. [DOI] [PubMed] [Google Scholar]
- Raab-Traub N., Dambaugh T., Kieff E. DNA of Epstein-Barr virus VIII: B95-8, the previous prototype, is an unusual deletion derivative. Cell. 1980 Nov;22(1 Pt 1):257–267. doi: 10.1016/0092-8674(80)90173-7. [DOI] [PubMed] [Google Scholar]
- Raab-Traub N., Pritchett R., Kieff E. DNA of Epstein-Barr virus. III. Identification of restriction enzyme fragments that contain DNA sequences which differ among strains of Epstein-Barr virus. J Virol. 1978 Aug;27(2):388–398. doi: 10.1128/jvi.27.2.388-398.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rymo L., Forsblom S. Cleavage of Epstein-Barr virus DNA by restriction endonucleases EcoRI, HindIII and BamI. Nucleic Acids Res. 1978 Apr;5(4):1387–1402. doi: 10.1093/nar/5.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymo L., Lindahl T., Adams A. Sites of sequence variability in Epstein-Barr virus DNA from different sources. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2794–2798. doi: 10.1073/pnas.76.6.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skare J., Strominger J. L. Cloning and mapping of BamHi endonuclease fragments of DNA from the transforming B95-8 strain of Epstein-Barr virus. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3860–3864. doi: 10.1073/pnas.77.7.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O., Birnstiel M. L. A simple method for DNA restriction site mapping. Nucleic Acids Res. 1976 Sep;3(9):2387–2398. doi: 10.1093/nar/3.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sugden B. Comparison of Epstein-Barr viral DNAs in Burkitt lymphoma biopsy cells and in cells clonally transformed in vitro. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4651–4655. doi: 10.1073/pnas.74.10.4651. [DOI] [PMC free article] [PubMed] [Google Scholar]