Abstract
Hepatitis C virus (HCV) infection is characterized by a strong propensity toward chronicity, autoimmune phenomena and lymphomagenesis, supporting a role for lymphocyte dysregulation during persistent viral infection. We have shown that HCV core protein inhibits T-cell functions through interaction with a complement receptor, gC1qR. Here, we further report that B cells also express gC1qR that can be bound by HCV core protein. Importantly, using flow cytometry, we demonstrated differential regulation of B and T lymphocytes by the HCV core–gC1qR interaction, with down-regulation of CD69 activation in T cells but up-regulation of CD69 activation and cell proliferation in B cells. HCV core treatment led to decreased interferon-γ production in CD8+ T cells but to increased immunoglobulin M and immunoglobulin G production as well as cell surface expression of costimulatory and chemokine receptors, including CD86 (B7-2), CD154 (CD40L) and CD195 (CCR5), in CD20+ B cells. Finally, we showed down-regulation of suppressor of cytokine signalling-1 (SOCS-1) using real-time reverse transcription–polymerase chain reaction, accompanied by up-regulation of signal transducer and activator of transcription-1 (STAT1) phosphorylation in B cells in response to HCV core protein, with the opposite pattern observed in HCV core-treated T cells. This study demonstrates differential regulation of B and T lymphocytes by HCV core and supports a mechanism by which lymphocyte dysregulation occurs in the course of persistent HCV infection.
Keywords: activation, B cells, hepatitis C, immunoglobulins, T cells
Introduction
Chronic hepatitis C virus (HCV) infects over 170 million people worldwide and is a major factor in the development of chronic hepatitis leading to cirrhosis and hepatocellular carcinoma. This virus exhibits a remarkable propensity toward chronic viral persistence. Chronic HCV infection is also associated with B-cell lymphoproliferative disorders, including most notably mixed cryoglobulinemia and B-cell non-Hodgkin lymphoma. Data suggest that B-cell clonal expansion or altered B-cell apoptosis may contribute to these lymphoproliferative disorders.1,2
Targeted studies have shown both B-cell and T-cell dysregulation in the setting of chronic HCV infection. Both proliferative capacity and effector function are impaired in HCV-specific CD4+/CD8+ T cells during chronic HCV infection, with an anergic/exhausted phenotype that lacks the ability to produce T helper type 1 cytokines.3,4 Dysfunction of T cells limits effective control of viral replication. Multiple mechanisms have been suggested by which T cells are dysregulated by HCV.5–7
Less well-described is the evidence for B-cell dysregulation that occurs during chronic HCV infection. This is manifest in several ways, including aberrant B-cell proliferation, decreased threshold for B-cell activation via the B-cell receptor complex, and overproduction of antibodies that are ineffective in controlling viral infection.8 A non-neoplastic expansion of B-cell clones expressing immunoglobulin M (IgM) has been noted in mixed cryoglobulinemia, a process that can be observed in more than 50% of individuals chronically infected with HCV. It appears that whereas T-cell function is suppressed during chronic HCV infection, B cells often exhibit activation and clonal expansion.
It is likely that a gene product(s) encoded by HCV directly affects B-cell and T-cell functions that are crucial for limiting virus replication. Several HCV-derived proteins, including the nucleocapsid core protein, may play a role in the impairment of host immunity either directly or through interaction with host molecules.9,10 It has been previously demonstrated that HCV core protein is necessary and sufficient to suppress host T-cell responses in a murine model.11 The molecular mechanism of HCV core-mediated immunomodulation was subsequently determined by identifying a host-binding partner, the C1q complement receptor gC1qR, on human T lymphocytes.6 C1q, the natural ligand for gC1qR, is the first molecule to be activated in the classical complement cascade and plays a critical role in modulating both innate and adaptive immunity.12
Binding of C1q to gC1qR on T lymphocytes leads to suppression of T-cell responsiveness;13 similarly, HCV core can inhibit T-cell responses through interaction with gC1qR.5,6,14 The engagement of circulating HCV core protein with gC1qR displayed on the surface of T lymphocytes may therefore provide the virus with a direct means of affecting host immunity.15 In light of the observations that free core particles circulate in the bloodstream of HCV-infected patients,16,17 our findings may be particularly salient to the pathogenesis of HCV. The role of gC1qR and HCV core in B-cell signalling, however, has not been elucidated.
Rampant T-cell or B-cell signalling can have disastrous biological consequences so lymphocyte signalling pathways are tightly controlled at multiple levels. Suppressor of cytokine signalling (SOCS) proteins represent a level of inhibitory machinery that is induced upon lymphocyte activation.18 This family consists of at least eight members and is characterized by an SH-2 domain and a unique C-terminal motif (the SOCS box). SOCS-1 and SOCS-3 in particular have been shown to bind to cytokine receptors or to receptor-associated Janus-associated kinases (JAKs) to inhibit activation of signal transducers and activators of transcription (STAT) members and ultimately interferon signalling.19,20 The SOCS proteins also regulate tumour necrosis factor-α-mediated cellular apoptosis by inhibiting phosphatidylinositol 3-kinase and p38 mitogen-activated protein kinase pathways.21 The role of SOCS proteins in regulating B-cell and T-cell responses in HCV infection has not been well studied.
In light of the immunomodulatory role of HCV core protein in lymphocyte signalling, we explored the possibility that HCV core might differentially regulate B-cell and T-cell signalling pathways. We found that B cells, like T cells and monocytes, express gC1qR and that HCV core can bind to this. There was differential regulation of B-cell and T-cell activation by HCV core, with up-regulation of activation markers in B cells but down-regulation in T cells. In terms of cellular function, HCV core treatment led to up-regulation of IgM and IgG production, cellular proliferation, as well as cell surface receptor expression including CD154, CD86 and CD195 in CD20+ B cells, but to decreased interferon-γ (IFN-γ) production in CD8+ T cells. Finally, we observed a differential regulation of SOCS-1 and STAT1 phosphorylation in B cells (reduced SOCS-1 expression/increased STAT1 phosphorylation) and T cells (increased SOCS-1 expression/reduced STAT1 phosphorylation).
Materials and methods
Cell isolation and culture
Human peripheral blood mononuclear cells (PBMC) were isolated from the peripheral blood of healthy donors by Ficoll density centrifugation with lympholyte-H (Cedarlane Labs, Hornby, ON, Canada). The B and T lymphocytes were further purified from isolated PBMC by incubation with a fluorescein isothiocyanate (FITC)-conjugated anti-CD3 or FITC-conjugated anti-CD20 antibody, followed by positive selection with anti-FITC magnetic beads (Miltenyi Biotec, Auburn, CA) following the manufacturer's instruction. Purified cells were washed twice and cultured with RPMI-1640 (Life Technologies, Gaithersburg, MD), containing 10% (volume/volume) fetal bovine serum (Life Technologies), penicillin–streptomycin (100 μg/ml for each drug; Life Technologies), l-glutamine (2 mm) and 2-mercaptoethanol (5·5 × 10−5 m; Life Technologies) at 37° with 5% CO2 in a humidified atmosphere.
Flow cytometry
To determine gC1qR expression on the B-cell surface, 1 × 106 isolated PBMC were first incubated with 1 μg α-gC1qR monoclonal antibody (a generous gift from Dr Young S. Hahn at University of Virginia) in 100 μl fluorescence-activated cell sorting (FACS) medium (RPMI-1640 supplemented with 10% fetal bovine serum and 1% NaN3) at 4° for 1 hr. The cells were washed twice in FACS medium at 200 g for 5 min at 4°, followed by incubation with 1 μg of phycoerythrin (PE)-labelled anti-mouse immunoglobulin secondary antibody (BD Pharmingen, San Diego, CA) in 100 μl FACS medium, and then double-stained with 20 μl FITC–anti-CD20 conjugate (BD Pharmingen). The cells were then washed three times and fixed with 1% paraformaldehyde in phosphate-buffered saline before flow cytometry (Becton Dickinson, San Jose, CA). The primary isotype controls were used to determine the level of background staining and 20 000 events were collected after gating on lymphocyte populations.
To determine core binding, various concentrations of β-gal–core protein (0·25, 0·5, 1, 2, 4 and 8 μg/ml; ViroGen, Watertown, MA; certified free of lipopolysaccharide) were incubated with 1 × 106 B cells purified by magnetic antibody cell sorting (MACS) at 37° for 2 hr. Core binding was determined using a procedure as described.22 First, cells were washed three times and resuspended with 1 μg anti-HCV core monoclonal antibody (ABR Inc., Golden, CO) in 100 μl FACS medium on ice for 1 hr. Second, the cells were washed three times and resuspended in 100 μl FACS medium containing 1 μg PE-conjugated anti-mouse immunoglobulin (BD Pharmingen) at 4° for 1 hr in the dark, then fixed and analysed by flow cytometry as described above.
To determine CD69 expression on activated B and T lymphocytes, 1 × 106 whole PBMC were stimulated with either 5 μg/ml phytohaemagglutinin (PHA; Sigma, St Louis, MO) or 1 μg/ml anti-CD3/CD28 antibodies (BD Pharmingen) for T cells or 1 μg/ml anti-CD40 antibody (BD Pharmingen) for B cells for 24 hr. The PBMC containing either activated B or T lymphocytes were simultaneously treated with 2 μg/ml HCV core or β-gal protein at 37° for 24 hr. The treated cells were then washed three times in FACS medium, resuspended in 100 μl FACS medium containing 1 μg PE–anti-CD69 conjugate, incubated at 4° for 1 hr in the dark, and double-stained with FITC–anti-CD4 or FITC–anti-CD20 conjugates as described above. To further characterize the specificity of the effect of core protein on CD69 expression on resting B cells, MACS-purified CD20+ B lymphocytes were treated with either HCV core (2 μg/ml, ViroGen), HCV NS3 (2 μg/ml, ViroGen), β-gal (2 μg/ml, Sigma), or C1q (2 μg/ml; Sigma) for 24 hr, and CD69 expression was measured as above.
To determine the effect of HCV core on intracellular IFN-γ production in T cells, 1 × 106 PBMC were stimulated with 1 μg/ml phorbol 12-myristate 13-acetate and 2 μg/ml calcium ionophore at 37° for 12 hr, followed by the addition of 2 μm monensin (BD GolgiStop™ protein transport inhibitor; BD Biosciences, San Jose, CA) for another 12 hr. The cells were washed with FACS medium and cell-surface-stained with FITC–anti-CD8 conjugate at 4° for 1 hr. After three washes with FACS medium, the cells were resuspended in 100 μl Fixation/Permeabilization solution (BD Cytofix/Cytoperm Kit; BD Pharmingen) at 4° for 20 min then washed twice in 1 × BD Perm/Wash buffer before being resuspended in the same buffer containing 0·4 μg/ml PE–anti-IFN-γ or isotype control antibody at 4° for 1 hr. This was followed by flow cytometric analysis.
To determine the effect of HCV core on the expression of the costimulatory molecules B7-2, CD40L, and chemokine receptor CCR5 on the surface of B cells, 1 × 106 isolated PBMC were activated with PHA and treated with β-gal–core or β-gal control. The PE–anti-CD86, PE–anti-CD154, PE–anti-CD195 and FITC–anti-CD20 double staining and flow cytometry analyses were carried out as above.
Proliferation assay
A human carboxy-fluorescein diacetate succinimidyl ester (CFSE) Flow Kit (Renovar Inc., Madison, WI) was used for the analysis of B-cell proliferation by flow cytometry. Briefly, 1 × 106 MACS-purified B cells were labelled with CFSE according to the manufacturer’s instructions, and then stimulated with PHA (5 μg/ml) and interleukin-2 (50 U/ml; eBioscience, San Diego, CA) in the presence of β-gal (2 μg/ml), C1q (50 μg/ml), or HCV core protein (2 μg/ml) for 5 days at 37° in a 5% CO2 atmosphere. The data were collected on a Becton Dickinson FACScan flow cytometer and analysed as histograms. The percentages of each distinct cell division peak, gated as M1, M2 and M3, were shown.
Reverse transcription–polymerase chain reaction (RT-PCR)
Anti-CD3/CD28 or PHA-activated PBMC or B and T lymphocytes (2 × 106 cells) were treated with or without HCV core protein (2 μg/ml; ViroGen) for various times (12, 24, 48 hr), and total RNA was isolated from these cells by the RNA isolation kit (Qiagen Sci. MD, Valencia, CA). A total of 1 μg RNA was treated with DNase to digest genomic DNA and 0·27 μg RNA was then reverse transcribed using murine leukaemia virus reverse transcriptase under the following conditions: 10 min at room temperature, 20 min at 42°, 5 min at 99°, and 5 min at 4°. After this, 1 μl of 1 : 10 series diluted cDNA, generated by the reverse transcription, was added to the PCR. The PCR was carried out using the following primer pairs: SOCS-1 sense 5′-ATG GTA GCA CAC AAC CAG GTG-3′, antisense 5′-TCA AAT CTG GAA GGG GAA GGA-3′; SOCS-3 sense 5′-CTC AAG ACC TTC AGC TCC AA-3′, antisense 5′-TTC TCA TAG GAG TCC AGG TG-3′; β-actin sense 5′-CGA GCG GGA AAT CGT GCG TGA CAT-3′, antisense 5′-CGT CAT ACT CCT GCT TGC TGA TCC ACA TCT-3′; for 35 cycles of 95° for 15 seconds, 58° for 15 seconds, 72° for 15 seconds, followed by a single 10-min extension at 72°. To control for genomic DNA contamination, equal amount of cDNA from each sample were amplified by PCR without RT. The resulting PCR products were separated on a 2% BioGel (Bio 101, Carsbad, CA) and viewed with a multimager. To examine whether gC1qR mediated the HCV core-induced induction of SOCS-1, a 1 : 10-diluted anti-gC1qR antibody or prebleeding control serum (kindly provided by Dr Y. S. Hahn) was coincubated with cells treated with core protein, and SOCS-1 messenger RNA expression was assessed as described above.
The quantitative real-time PCR was carried out using 2 μl of 1 : 10 diluted cDNA from the same RT reactions as above and using the Bio-Rad iCycler iQ Multicolor Real-Time PCR Detection System (Bio-Rad Life Science Research, Hercules, CA). The PCR was performed in a 50-μl volume using RT2 real-time™ SYBR Green Fluorescein PCR Master Mix (Super-Array Bioscience Corporation, Frederick, MD). All the PCR assays were performed in triplicate. The reaction conditions were 95° for 12 min; followed by 40 cycles at 95° for 15 seconds, 55° for 30 seconds and 72° for 30 seconds. β-Actin was amplified from all samples on each plate as a housekeeping gene to normalize the expression level of target between different samples, and to monitor the reproducibility of the assays. The reaction mixtures without template cDNA were used as negative controls. Threshold cycle numbers (CT) were determined using the Bio-Rad iCycle iQ Multicolor Real-Time PCR Detection System and transformed using the ΔCT comparative method. The amount of target, normalized to an endogenous reference and relative to a calibrator, was determined by the comparative CT method (ΔΔCT).
Immunoblotting
Both B and T lymphocytes were purified from human PBMC using magnetic beads (Miltenyi Biotec) in accordance with the manufacturer’s instructions. A total of 2 × 106 purified cells were activated with PHA (5 μg/ml; Sigma) in the presence or absence of 2 μg/ml β-gal–core at 37° in a 5% CO2 atmosphere for 24 hr. Cell lysate was prepared for 30 min at 4° with a lysis buffer (Life Technologies). Cell lysates were sonicated three times for 1 min each time. Cellular debris was pelleted by centrifugation at 16 000 g and supernatants were collected and frozen at −80°.
A total of 80 μg protein was denatured with sample loading buffer at 100° for 5 min and resolved by sodium dodecyl sulphate–polyacrylamide gel electrophoresis, followed by semidry transfer (GE Healthcare Biosciences, Piscataway, NJ) to a Hybond-P membrane (Amersham Biosciences, Arlington Heights, IL). After blocking in Blotto–Tween-20 (10 mm Tris–HCl, 0·9% NaCl, 0·1% Tween-20, 5% non-fat dried milk) at room temperature for 1 hr, the membrane was probed with polyclonal antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) to SOCS-1 (1 : 500), phosphor-STAT1 (1 : 1000), or β-actin (1 : 2000). After several 5-min washes with Tris-buffered saline–Tween and Tris-buffered saline, the membrane was incubated with horseradish peroxidase-conjugated rabbit anti-goat IgG secondary antibody (1 : 5000) and subsequently developed by enhanced chemiluminescence (ECL-plus; Amersham Biosciences) on X-OMAT-LS X-ray film (Kodak, Rochester, NY).
Statistical analysis
Data were shown as mean ± SD and the level of significance was determined using the analysis of variance program of Stata/SE 8.0 software. A P-value of < 0·05 was considered significant and P < 0·01 was considered very significant.
Results
HCV core directly binds to gC1qR expressed on B cells
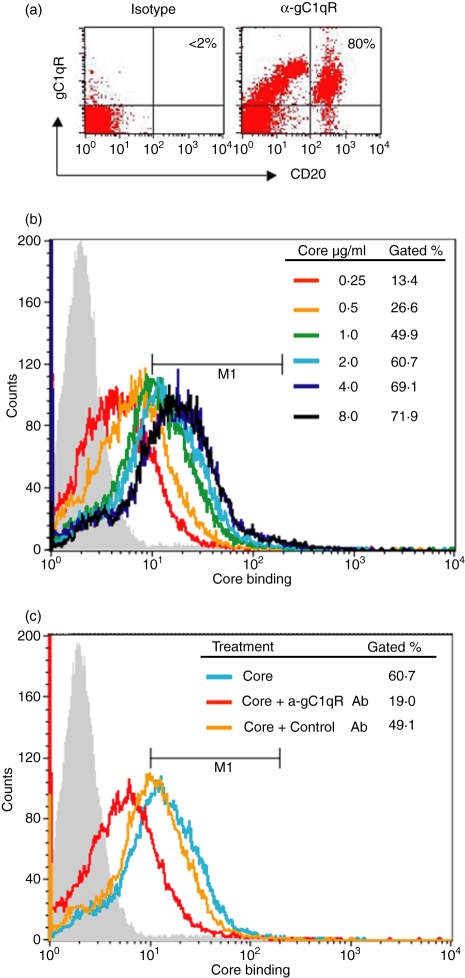
It was previously demonstrated that HCV core protein can bind directly to gC1qR on the surface of T lymphocytes and inhibit T-cell functions.22 To determine whether the HCV core–gC1qR interaction occurs in B cells, we examined the ability of HCV core to bind gC1qR on the surface of human B lymphocytes. We first analysed whether gC1qR is actually expressed on the surface of B lymphocytes using a monoclonal anti-gC1qR antibody by flow cytometry. As shown in Fig. 1(a), as was previously found on human T cells and monocytes, gC1qR was highly expressed on CD20+ B cells. We then examined core binding to purifed B cells by incubation with an escalating dose of recombinant β-gal–core followed by flow cytometry, as previously examined in T cells. As shown in Fig. 1(b), the core protein was found to bind to the surface of B lymphocytes in a dose-dependent manner. Notably, HCV core binding to the B-cell surface was saturated at a core concentration of 4 μg/ml, with a positive number close to the ratio of gC1qR+ B cells, suggesting that the binding sites for the core on B cells were fully occupied and that the binding of the core to the cell surface was mediated through a specific receptor. To determine whether gC1qR was involved in the binding of the HCV core to the surface of B lymphocytes, we further examined the ability of anti-gC1qR antibody to prevent core from associating with B cells. To this end, B cells were incubated with 2 μg/ml of HCV core and either a polyclonal anti-gC1qR antibody or a control antibody, and core binding was assessed as before. As shown in Fig. 1(c), core binding on B lymphocytes was blocked by anti-gC1qR antibody, whereas the binding was unaffected by the control antibody.
Figure 1.
HCV core protein specifically binds to gC1qR displayed on the cell surface of human B lymphocytes. (a) gC1qR expression on human B lymphocytes. A total of 1 × 106 PBMC were incubated with 1 μg/ml of anti-gC1qR or an isotype control Ab followed by 1 μg/ml of PE–anti-mouse-Ig conjugate. The cells were then double-stained with fluorescein isothiocyanate (FITC)-anti-CD20 Ab. The percentage of gC1qR+ cells in the CD20+ cell population is shown in the right upper corner of the dot blot. (b) Dose-dependent core binding on B lymphocytes. Magnetic antibody cell sorting (MACS)-purified B cells were incubated with escalating concentrations of HCV core protein at 37º for 2 hr and core binding on the surface of B cells was analysed by flow cytometry as described in Materials and Methods. Data were displayed as overlying histograms and the percentages of cells positive for core binding as gated based on the isotype control (grey filled area, < 2%) were shown. The histogram shifted from left to right (increasing fluorescence intensity) as the core protein increased from lower to higher concentrations until saturation. (c) HCV core binds to B lymphocytes through gC1qR. MACS-purified B cells were incubated with 2 μg/ml of HCV core protein in the presence or absence of anti-gC1qR or control antibody at 37º for 2 hr and core binding on the surface of B cells was analysed as above. Data were displayed as overlying histograms and the percentages of cells positive for core binding as gated based on the isotype control (grey filled area, < 2%) were shown.
HCV core differentially regulates B- and T-lymphocyte functions
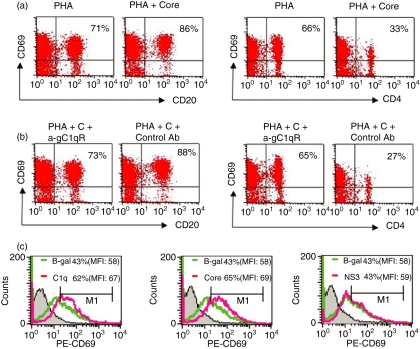
To determine the significance of HCV core–gC1qR interaction in B cells, we analysed the effect of HCV core on B-cell function and in parallel compared with the effect on T cells. CD69 has been used as an early marker for lymphocyte activation so we first analysed the effect of HCV core–gC1qR on CD69 expression on the surface of T and B lymphocytes. To this end, purified human PBMC were stimulated with PHA in the presence or absence of HCV core protein for 24 hr, and CD69 expression on the surface of CD4+ T cells, as well as on CD20+ B cells, was examined by flow cytometry. As shown in Fig. 2(a), HCV core protein inhibited CD69 expression on the CD4+ T cells, which was consistent with our previously reported results that core protein suppresses T-cell activation. Interestingly, in contrast to its effect on T cells, HCV core increased the expression of CD69 on the surface of CD20+ B cells. This differential regulation of B- and T-lymphocyte activation by HCV core was observed not only in core-treated PBMC stimulated by PHA-mitogenic activation but also in PBMC stimulated by anti-CD3/CD28 (T cells) or anti-CD40 (B cells) (data not shown). Importantly, this differential regulation by HCV core can be partially abrogated by simultaneously adding anti-gC1qR antibody to the culture system, but not by adding the control antibody (Fig. 2b, compare with Fig. 2a), suggesting that gC1qR is involved in this process.
Figure 2.
Differential regulation of B and T lymphocyte activation by HCV core protein. (a) CD69 expression on activated CD20+ B cells and CD4+ T cells in the presence of core. PBMC were activated with 5 μg/ml of phytohaemagglutinin (PHA) in the presence or absence of 2 μg/ml of core protein at 37º for 24 hr in a 5% CO2 atmosphere. CD69 expression on CD20+ B cells (upper panel) and CD4+ T cells (lower panel) was determined by fluorescence-activated cell sorting (FACS) with PE-conjugated anti-CD69 antibody and either fluorescein isothiocyanate (FITC)-conjugated anti-CD4 or FITC-conjugated anti-CD20 mcAb. The percentages of CD20+ and CD4+ lymphocytes positive for CD69 are shown. The results were reproducible in two independent experiments using different donors. (b) Abrogation of core-mediated differential regulation of B and T lymphocyte activation by α-gC1qR antibody. PHA-activated PBMC were treated with 2 μg/ml of HCV core in the presence of either 1:10 (volume/volume) α-gC1qR or control Ab at 37º for 24 h. CD69 expression on CD20+ B cells and CD4+ T cells was examined and shown as above. (c) Specificity of CD69 upregulation by HCV core. Magnetic antibody cell sorting (MACS)-purified CD20+ B lymphocytes were treated with either HCV core (2 μg/ml), HCV NS3 (2 μg/ml), β-gal (2 μg/ml), or C1q (2 μg/ml) for 24 hr, and CD69 expression was measured as above.
To further characterize the specificity of the effect of HCV core protein on resting B cells, purified CD20+ B lymphocytes were treated with either HCV core, HCV NS3, β-gal, or C1q protein for 24 hr, and CD69 expression was measured as described. As shown in Fig. 2(c), compared with the β-gal control in terms of percentage of CD69+ B cells or total mean fluorescence intensity, C1q, the natural ligand for gC1qR, up-regulated CD69 expression on the surface of B cells. Similarly, HCV core, but not HCV NS3, was found to increase CD69 expression as well as C1q-treated B cells.
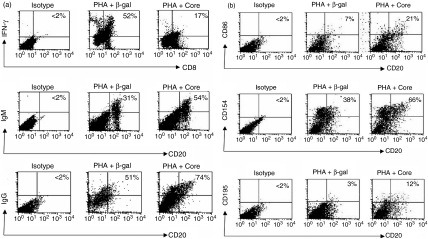
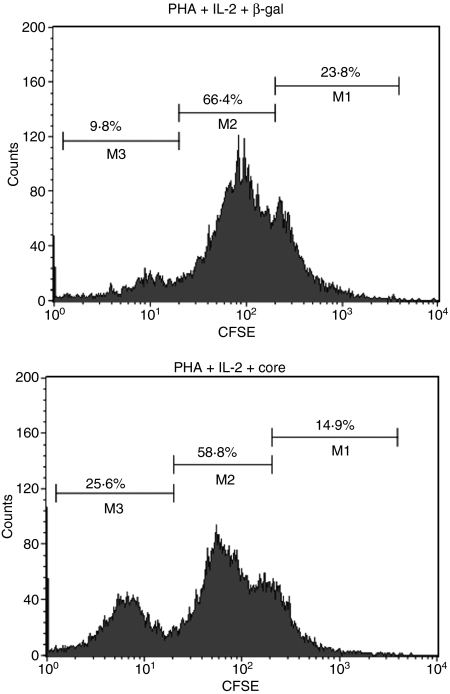
In addition to activation state, we also observed the effect of HCV core on the primary function of these cells. To this end, stimulated PBMC were treated with HCV core or β-gal control protein for 24 hr, and intracellular IFN-γ production by CD8+ T cells and IgM/IgG production by CD20+ B cells were analysed by flow cytometry. As shown in Fig. 3(a), and consistent with what we observed in terms of activation status, HCV core inhibited IFN-γ production by CD8+ T cells and enhanced IgM and IgG production by CD20+ B cells when compared to cells treated with β-gal control. These data are reproducible and hold true when we use PBMC isolated from at least two different donors in at least two separate experiments (data not shown). Additionally, we also examined the effect of HCV core on the expression of other cell surface markers, including costimulatory molecules such as CD86 (B7-2) and CD154 (CD40L), as well as chemokine receptors such as CD195 (CCR5), on CD20+ B cells. As shown in Fig. 3(b), HCV core increased the expression of CD86, CD154 and CD195 on the surface of CD20+ B cells compared with PHA-activated cells treated with β-gal control protein, suggesting a broad spectrum of activation of B lymphocytes by HCV core protein. We have previously reported by CFSE-Flow analysis, that T-cell proliferation was inhibited by HCV core protein. Here, we examined PHA-stimulated, CFSE-labelled, MACS-purified B-cell proliferation in response to β-gal, C1q and HCV core and found an increase in proliferation only in the B cells treated with HCV core proteon (Fig. 4), but not in C1q-treated B cells, which was consistent with the published data on C1q in B cells (data not shown).
Figure 3.
Effect of HCV core on B and T lymphocyte functions. (a) HCV core on B and T lymphocyte function. Peripheral blood mononuclear cells (PBMC) were stimulated with PMA and calcium ionophore or phytohaemagglutinin (PHA) in the presence of β-gal-core or β-gal at 37º for 24 hr. The intracellular IFN-γ production in CD8+ T cells or IgM/IgG production on the surface of B cells was analysed by fluorescence-activated cell sorting (FACS) as described in Methods. (b) The effect of HCV core on the expression of co-stimulatory molecules and chemokine receptors on the surface of B lymphocytes. 1 × 106 PBMC were activated with PHA in the presence of β-gal-core or β-gal at 37º for 24 hr. PE–anti-CD86, PE–anti-CD154, or PE–anti-CD195 and fluorescein isothiocyanate (FITC)–anti-CD20 conjugate were used to examine the expression of these molecules on the surface of treated CD20+ B cells by FACS analysis.
Figure 4.
HCV core promotes B cell proliferation. 1 × 106 magnetic antibody cell sorting (MACS)-purified and carboxy-fluorescein diacetate succinimidyl ester (CFSE)-labeled CD20+ B cells were stimulated with PHA (5 μg/ml) and IL-2 (50 μg/ml) in the presence of β-gal (2 μg/ml) or HCV core (2 μg/ml) at 37º, 5% CO2 for 5 days. Data were collected by flow cytometer and represented as histograms. Percentages of distinct cell division peaks were gated as M1, M2, and M3. During each round of cell division, the relative intensity of the fluorescent dye is decreased by half.
HCV core regulates SOCS-1 expression differentially in T and B lymphocytes
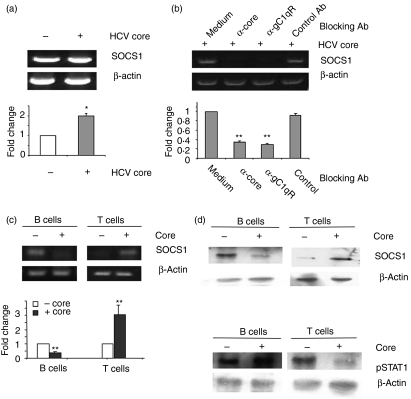
Since lymphocyte responses are negatively regulated by the SOCS proteins through inhibition of the JAK/STAT pathway,18–21 the differential regulation of B- and T-lymphocyte responses mediated by HCV core–gC1qR might be a result of the ability of the core protein to induce or suppress these negative regulators in the cells. To examine this possibility, we treated anti-CD3/CD28 or PHA-activated PBMC or purified T and B lymphocytes with HCV core and then detected SOCS-1 or SOCS-3 messenger RNA by RT-PCR. To determine the role of gC1qR in the core-induced induction of SOCS, anti-gC1qR or a control serum was added to the culture simultaneously with core, and SOCS gene expression was also detected. RT-PCR analysis of PBMC stimulated with anti-CD3/CD28 in the presence of HCV core for various times (12, 24 and 48 hr) indicated increased SOCS-1 gene expression (data not shown). As shown in Fig. 5(a), HCV core but not β-actin control induced SOCS-1 gene expression at 24 hr (left panel) and this core-mediated induction of SOCS-1 gene expression was diminished by the addition of anti-gC1qR (right panel) but not by the addition of control antibody. SOCS-3 gene expression was also up-regulated in T-cell receptor-activated PBMC treated with HCV core (data not shown). At least two repeated experiments using PBMC isolated from at least two different donors elicited the same effect on induction of SOCS-1 and SOCS-3 by core protein, whereas β-actin was not affected by the treatment. These results were confirmed by quantitative real-time PCR using the same RT reactions of activated T cells treated with HCV core.
Figure 5.
Differential regulation of suppressor of cytokine signalling-1 (SOCS-1) and signal transducer and activator of transcription-1 (STAT1) in B and T lymphocytes by HCV core. (a) gC1qR is involved in HCV core-induced induction of SOCS-1 gene expression in TCR-stimulated peripheral blood mononuclear cells (PBMCs). Left panel, up-regulation of SOCS-1 in anti-CD3/CD28-stimulated T cells by HCV core. PBMC were incubated with 1 μg/ml of anti-CD3/CD28 in the presence or absence of 2 μg/ml of β-gal-core for 24 hr, and SOCS-1 and β-actin genes were amplified as described in Materials and Methods. The fold changes of the amplified genes in cells treated with or without core were quantitatively analyzed by real-time reverse transcription–polymerase chain reaction (RT-PCR) and shown below, with the absence of core treatment deemed as one after normalization by β-actin reference control. *represents P < 0·05 between the means from triplicate experiments in the presence or absence of core treatment. Right panel, anti-CD3/CD28 stimulated PBMC were treated with HCV core in the presence or absence of α-core, α-gC1qR, or a control serum for 24 hr, and SOCS-1 gene amplification by RT-PCR or quantitative PCR was carried out as above. **represents P < 0·01 between the fold changes of SOCS-1 gene expression in the presence of blocking antibody compared with the absence of blocking antibody, which is deemed one after normalization by β-actin reference control. (b) Differential regulation of SOCS-1 gene expression. Magnetic antibody cell sorting (MACS)-purified B and T lymphocytes were stimulated with phytohaemagglutinin (PHA) or anti-CD3/CD28, respectively, in the presence or absence of core protein for 24 hr, and SOCS-1 gene amplification by RT-PCR or quantitative real-time PCR were performed as above. **represents P < 0·01 between the fold changes of the SOCS-1 gene in the presence of core compared with its absence, which is deemed one after normalization by the β-actin reference control. (c) Differential regulation of SOCS-1 protein expression in B and T lymphocytes by HCV core. MACS-purified B and T lymphocytes were stimulated with PHA or anti-CD3/CD28, respectively, in the presence or absence of core protein for 24 hr, and SOCS-1 protein expression in the treated cells was analyzed by immunoblotting. β-actin serves as loading control. (d) Differential regulation of STAT1 protein phosphorylation in B and T lymphocytes by HCV core. MACS-purified B and T lymphocytes were stimulated with PHA or anti-CD3/CD28, respectively, in the presence or absence of core protein for 24 hr, and STAT1 phosphorylation in the treated cells was analysed by immunoblotting. β-actin serves as loading control. Immunoblot results were reproducible in at least two independent experiments from two different donors.
To determine whether differential regulation of B- and T-lymphocyte responses by the HCV core–gC1qR interaction was associated with the core protein's ability to induce SOCS in these cells, B and T lymphocytes from PBMC were further purified using MACS separation beads, and the highly purified B and T cells were activated and treated with HCV core as above. As shown in Fig. 5(b), SOCS-1 expression was inhibited in B cells and enhanced in T cells, whereas β-actin was not affected in these cells upon treatment with core protein. We found the same differential regulation of SOCS-3 expression in purified B and T lymphocytes by HCV core protein (data not shown). Again, these results were confirmed by the quantitative real-time RT-PCR. Notably, purified B and T cells treated with C1q showed no differences in SOCS-1 production by semi-quantitative RT-PCR despite multiple attempts (data not shown), suggesting that whereas both C1q and HCV core bind gC1qR, the downstream events may differ.
We also detected the SOCS-1 protein expression by immunoblotting in purified B and T lymphocytes treated with HCV core as above. As shown in Fig. 5(c), similar to what was observed with messenger RNA expression, SOCS-1 protein was inhibited in B cells and induced in T cells treated with HCV core, while β-actin was not affected by the treatment. Since SOCS-1 protein regulates lymphocyte responses through inhibition of the JAK/STAT pathway, we further examined STAT1 phosphorylation in MACS-purified/PHA-activated B cells and T-cell receptor-activated T cells exposed to core protein by immunoblotting. As shown in Fig. 5(d), STAT1 phosphorylation was up-regulated in B cells and down-regulated in T cells, but β-actin was not affected in these cells, by treatment with HCV core. This suggested that the differential regulation of B- and T-lymphocyte responses by HCV core was associated with its ability to induce or suppress SOCS expression, which in turn, alters JAK/STAT signalling.
Discussion
Recent reports suggest that HCV infection leads to immunodysregulation that involves multiple aspects of the host immune response.8,23 In this study, we found that the nucleocapsid core protein of HCV differentially regulates the functions of both B- and T-lymphocytes. This involved regulation of lymphocyte activation markers, effects on measures of cellular activation and alteration in adapter and kinase pathways fundamental to lymphocyte function. Our previous studies and current data would suggest that HCV core protein may be a major viral antigen involved in immunomodulating host responses.
Over 90% of individuals with mixed cryoglobulinemia are found to have HCV infection.24,25 Mixed cryoglobulinemia is the major extrahepatic manifestation of HCV infection and is found in 30–50% of HCV patients, who also have a high prevalence of non-Hodgkin B-cell lymphoma.26–29 Investigators have suggested that persistent HCV antigen may stimulate crucial cell signalling pathways, leading to T-cell-dependent B-cell expansion and predisposing the patient to autoimmune and lymphoproliferative disorders. For example, monoclonal expansion of B cells in the periphery as well as the livers of patients with HCV-associated mixed cryoglobulinemia has been observed.30,31 In addition, successful treatment of HCV infection with IFN/ribavirin, or elimination of B cells with novel anti-CD20 chimeric antibodies such as rituximab, leads to resolution of mixed cryoglobulinemia, regression of IgM overproduction and clearance of B-cell clones.32–36
Our data clearly support a role for up-regulation of B-cell activation and immunoglobulin production in response to HCV core protein. We found that HCV core could bind gC1qR, a known receptor for HCV core that is expressed on CD20+ B cells. Blocking the HCV core–gC1qR interaction led to inhibition of core binding. This appeared to be partial inhibition that may have resulted from incomplete blockade by a partially effective anti-gC1qR antibody. Our experimental design involved simultaneous addition of HCV core and anti-gC1qR antibody, and it is possible that preincubation with antibody might have improved the inhibition. The HCV core is also quite well known for its ability to bind other receptors, particularly in the tumour necrosis factor family, which we were not targeting but which may provide sites for core binding.
Treatment with HCV core protein increased cell surface expression of IgM, IgG and key B-cell activation markers including CD69, CD40L, B7-2, CCR5 and, ultimately, B-cell proliferation. This augmented effect on proliferation was observed following fairly potent B-cell stimulation in two independent experiments and is consistent with the observed up-regulation of markers of activation. Conversely, T cells exhibited a suppressive pattern, with decreased CD69 expression and diminished IFN production. It has been shown that HCV core can indeed alter T-cell functions via gC1qR.6,7 Loss of effective T-cell responses, and in particular the development of an anergic phenotype, may contribute to the poor control of viral replication that is observed during chronic HCV infection.37 This resulting persistent viral replication may in turn drive antigen-mediated B-cell expansion. Intriguingly, HCV core is secreted from infected cells and circulates in the bloodstream of infected individuals at levels consistent with those used in our experiments.16,17 In addition, the amount of free core protein or core protein expressed on the surface of infected cells is greater in the microenvironment of the liver, where virus replication occurs quite vigorously in early infection. The HCV core protein therefore appears to be present in the setting of clinical infection and could play a role in antigen-mediated B-cell expansion.
The SOCS proteins are considered to be tumor suppressors and represent a powerful mechanism for regulating the JAK/STAT pathways and, ultimately, cytokine production and cell proliferation.18 Our results suggest that B-cell activation by HCV core is associated with up-regulated phosphorylation of STAT1 and suppression of SOCS-1 expression; T-cell suppression by HCV core exhibits the opposite pattern. SOCS-1 and SOCS-3 are known to be potent negative regulators of T-cell proliferation and IFN-γ production via the JAK/STAT pathway, and so our findings of SOCS up-regulation and STAT1 down-regulation in T cells are perhaps not surprising and are in line with several studies focusing on IFN resistance.19,20
While the effects of HCV core appear to be mediated via gC1qR, we do not as yet know if gC1qR expression levels differ in T or B cells in HCV-infected individuals or if such a difference alters core-mediated signalling. In addition, the blood level of C1q in HCV-infected patients remains unknown, and so the interesting question of how signalling by C1q versus HCV core differs, and if there is a synergistic or antagonistic effect between C1q and HCV core that might disrupt T-cell and B-cell responses in vivo, remains. Given the fact that C1q-deficient subjects, are prone to develop autoimmune disorders, there is support for C1q functioning as an immunomodulator in vivo. We suspect, however, that the binding of C1q to the N-terminal region of gC1qR (rather than the C-terminal region, where core binds) may affect its signalling. We have previously discussed this in studies focusing on T cells and have consistently found that the equal molar ratios of HCV core and C1q led to more intense responses by HCV core, perhaps supporting a distinct pathway or mechanism of signalling.22 The fact that we found no difference in SOCS-1 expression in response to C1q in B or T cells despite clear differences in response to HCV core may also support different downstream signalling mechanisms.
The role of these regulatory pathways in B cells, however, has not been studied, and our findings represent the first report of how a hepatitis C virus gene product affects SOCS proteins and JAK/STAT signalling in B cells. It is possible that chronic antigen exposure to circulating HCV core, while dysregulating T-cell responses and promoting an anergic state via up-regulation of SOCS proteins, also leads to down-regulated SOCS-1 and/or SOCS-3 expression in B cells. This might drive the B-cell clonal expansion that is currently thought to be the impetus behind the development of mixed cryoglobulinemia and non-Hodgkin lymphoma. Interestingly, several investigators have found either inactivation or suppression of SOCS-1 in the setting of uncontrolled growth and malignancies, including notably multiple myeloma but also breast, pancreatic, and ovarian cancer.38–40 How these proteins might be involved in the development of B-cell clonal expansion, mixed cryoglobulinemia, and non-Hodgkin lymphoma in patients who are chronically infected with HCV is unknown but currently under investigation.
Acknowledgments
We thank our colleagues for constructive criticism and comments. Particularly, we appreciate Dr Young S. Hahn (of the University of Virginia) for generously providing anti-gC1qR antibody and control serum. This work was supported in part by East Tennessee State University major research grants to Z.Q.Y. (RDC #07-002M) and to J.P.M. (RDC #06-002M) and by a National Institutes of Health National Institute of Allergy and Infectious Disease grant to J.P.M./Z.Q.Y. (R15AI072750).
References
- 1.Sansonno D, Dammacco F. Hepatitis C virus, cryoglobulinaemia, and vasculitis: immune complex relations. Lancet Infect Dis. 2005;5:227–36. doi: 10.1016/S1473-3099(05)70053-0. [DOI] [PubMed] [Google Scholar]
- 2.Racanelli V, Sansonno D, Piccoli C, D'Amore FP, Tucci FA, Dammacco F. Molecular characterization of B cell clonal expansions in the liver of chronically hepatitis C virus-infected patients. J Immunol. 2001;167:21–9. doi: 10.4049/jimmunol.167.1.21. [DOI] [PubMed] [Google Scholar]
- 3.Gerlach JT, Diepolder HM, Jung MC, et al. Recurrence of hepatitis C virus after loss of virus-specific CD4(+) T-cell response in acute hepatitis C. Gastroenterology. 1999;117:933–41. doi: 10.1016/s0016-5085(99)70353-7. [DOI] [PubMed] [Google Scholar]
- 4.Shoukry NH, Grakoui A, Houghton M, Chien DY, Ghrayeb J, Reimann KA, Walker CM. Memory CD8+ T cells are required for protection from persistent hepatitis C virus infection. J Exp Med. 2003;197:1645–55. doi: 10.1084/jem.20030239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yao ZQ, King E, Prayther D, Yin D, Moorman JP. PD-1/PDL-1 pathway involvement in hepatitis C-mediated T cell dysfunction. Viral Immunol. 2007;20:276–87. doi: 10.1089/vim.2006.0096. [DOI] [PubMed] [Google Scholar]
- 6.Kittlesen DJ, Chianese-Bullock KA, Yao ZQ, Braciale TJ, Hahn YS. Interaction between complement receptor gC1qR and hepatitis C virus core protein inhibits T-lymphocyte proliferation. J Clin Invest. 2000;106:1239–49. doi: 10.1172/JCI10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yao ZQ, Eisen-Vandervelde A, Ray S, Hahn YS. HCV core/gC1qR interaction arrests T cell cycle progression through stabilization of the cell cycle inhibitor p27Kip1. Virology. 2003;314:271–82. doi: 10.1016/s0042-6822(03)00419-7. [DOI] [PubMed] [Google Scholar]
- 8.King E, Trabue C, Yin D, Yao ZQ, Moorman JP. Hepatitis C. The complications of immune dysfunction. Exp Rev Clin Immunol. 2007;3:145–57. doi: 10.1586/1744666X.3.2.145. [DOI] [PubMed] [Google Scholar]
- 9.Eisen-Vandervelde AL, Waggoner SN, Yao ZQ, Cale EM, Hahn CS, Hahn YS. Hepatitis C virus core selectively suppresses interleukin-12 synthesis in human macrophages by interfering with AP-1 activation. J Biol Chem. 2004;279:43479–86. doi: 10.1074/jbc.M407640200. [DOI] [PubMed] [Google Scholar]
- 10.Ray RB, Ray R. Hepatitis C virus core protein: intriguing properties and functional relevance. FEMS Microbiol Lett. 2001;202:149–56. doi: 10.1111/j.1574-6968.2001.tb10796.x. [DOI] [PubMed] [Google Scholar]
- 11.Large MK, Kittlesen DJ, Hahn YS. Suppression of host immune response by the core protein of hepatitis C virus: possible implications for hepatitis C virus persistence. J Immunol. 1999;162:931–8. [PubMed] [Google Scholar]
- 12.Ghebrehiwet B, Lim BL, Kumar R, Feng X, Peerschke EI. gC1q-R/p33, a member of a new class of multifunctional and multicompartmental cellular proteins, is involved in inflammation and infection. Immunol Rev. 2001;180:65–77. doi: 10.1034/j.1600-065x.2001.1800106.x. [DOI] [PubMed] [Google Scholar]
- 13.Chen A, Gaddipati S, Hong Y, Volkman DJ, Peerschke EI, Ghebrehiwet B. Human T cells express specific binding sites for C1q. Role in T cell activation and proliferation. J Immunol. 1994;153:1430–40. [PubMed] [Google Scholar]
- 14.Yao ZQ, Nguyen DT, Hiotellis AI, Hahn YS. Hepatitis C virus core protein inhibits human T lymphocyte responses by a complement-dependent regulatory pathway. J Immunol. 2001;167:5264–72. doi: 10.4049/jimmunol.167.9.5264. [DOI] [PubMed] [Google Scholar]
- 15.Yao ZQ, Ray S, Eisen-Vandervelde A, Waggoner S, Hahn YS. Hepatitis C virus: immunosuppression by complement regulatory pathway. Viral Immunol. 2001;14:277–95. doi: 10.1089/08828240152716547. [DOI] [PubMed] [Google Scholar]
- 16.Maillard P, Krawczynski K, Nitkiewicz J, et al. Nonenveloped nucleocapsids of hepatitis C virus in the serum of infected patients. J Virol. 2001;75:8240–50. doi: 10.1128/JVI.75.17.8240-8250.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masalova OV, Atanadze SN, Samokhvalov EI, et al. Detection of hepatitis C virus core protein circulating within different virus particle populations. J Med Virol. 1998;55:1–6. doi: 10.1002/(sici)1096-9071(199805)55:1<1::aid-jmv1>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 18.Alexander WS. Suppressors of cytokine signalling (SOCS) in the immune system. Nat Rev. 2002;2:410–6. doi: 10.1038/nri818. [DOI] [PubMed] [Google Scholar]
- 19.Song MM, Shuai K. The suppressor of cytokine signaling (SOCS) 1 and SOCS3 but not SOCS2 proteins inhibits interferon-mediated antiviral and antiproliferative activities. J Biol Chem. 1998;273:35056–62. doi: 10.1074/jbc.273.52.35056. [DOI] [PubMed] [Google Scholar]
- 20.Bode JG, Ludwig S, Ehrhardt C, Albrecht U, Erhardt A, Schaper F, Heinrich PC, Haussinger D. IFN-alpha antagonistic activity of HCV core protein involves induction of suppressor of cytokine signaling-3. FASEB J. 2003;17:488–90. doi: 10.1096/fj.02-0664fje. [DOI] [PubMed] [Google Scholar]
- 21.Morita Y, Naka T, Kawazoe Y, et al. Signals transducers and activators of transcription (STAT)-induced STAT inhibitor-1 (SSI-1)/suppressor of cytokine signaling-1 (SOCS-1) suppresses tumor necrosis factor alpha-induced cell death in fibroblasts. Proc Natl Acad Sci USA. 2000;97:5405–10. doi: 10.1073/pnas.090084797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yao ZQ, Eisen-Vandervelde A, Waggoner SN, Cale EM, Hahn YS. Direct binding of hepatitis C virus core to gC1qR on CD4+ and CD8+ T cells leads to impaired activation of Lck and Akt. J Virol. 2004;78:6409–19. doi: 10.1128/JVI.78.12.6409-6419.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eisen-Vandervelde AL, Yao ZQ, Hahn YS. The molecular basis of HCV-mediated immune dysregulation. Clin Immunol. 2004;111:16–21. doi: 10.1016/j.clim.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 24.Agnello V. Hepatitis C virus infection and type II cryoglobulinemia: an immunological perspective. Hepatology. 1997;26:1375–9. doi: 10.1053/jhep.1997.v26.ajhep0261375. [DOI] [PubMed] [Google Scholar]
- 25.Ferri C, Greco F, Longombardo G, et al. Antibodies to hepatitis C virus in patients with mixed cryoglobulinemia. Arthritis Rheum. 1991;34:1606–10. doi: 10.1002/art.1780341221. [DOI] [PubMed] [Google Scholar]
- 26.Duberg AS, Nordstrom M, Torner A, Reichard O, Strauss R, Janzon R, Back E, Ekdahl K. Non-Hodgkin's lymphoma and other nonhepatic malignancies in Swedish patients with hepatitis C virus infection. Hepatology. 2005;41:652–9. doi: 10.1002/hep.20608. [DOI] [PubMed] [Google Scholar]
- 27.Germanidis G, Haioun C, Dhumeaux D, Reyes F, Pawlotsky JM. Hepatitis C virus infection, mixed cryoglobulinemia, and B-cell non-Hodgkin's lymphoma. Hepatology. 1999;30:822–3. doi: 10.1002/hep.510300323. [DOI] [PubMed] [Google Scholar]
- 28.Kashyap A, Nademanee A, Molina A. Hepatitis C and B-cell lymphoma. Ann Intern Med. 1998;128:695. doi: 10.7326/0003-4819-128-8-199804150-00022. [DOI] [PubMed] [Google Scholar]
- 29.Ohsawa M, Shingu N, Miwa H, et al. Risk of non-Hodgkin's lymphoma in patients with hepatitis C virus infection. Int J Cancer. 1999;80:237–9. doi: 10.1002/(sici)1097-0215(19990118)80:2<237::aid-ijc12>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 30.Franzin F, Efremov DG, Pozzato G, Tulissi P, Batista F, Burrone OR. Clonal B-cell expansions in peripheral blood of HCV-infected patients. Br J Haematol. 1995;90:548–52. doi: 10.1111/j.1365-2141.1995.tb05582.x. [DOI] [PubMed] [Google Scholar]
- 31.Vallat L, Benhamou Y, Gutierrez M, et al. Clonal B cell populations in the blood and liver of patients with chronic hepatitis C virus infection. Arthritis Rheum. 2004;50:3668–78. doi: 10.1002/art.20594. [DOI] [PubMed] [Google Scholar]
- 32.D'Amico E, Chincoli C, Cacciatore P, et al. Effects of combined antiviral therapy on asymptomatic mixed cryoglobulinemia in naive patients with chronic hepatitis C virus infection: a preliminary study. Dig Dis Sci. 2005;50:2344–7. doi: 10.1007/s10620-005-3059-x. [DOI] [PubMed] [Google Scholar]
- 33.Mazzaro C, Zorat F, Caizzi M, et al. Treatment with peg-interferon alfa-2b and ribavirin of hepatitis C virus-associated mixed cryoglobulinemia: a pilot study. J Hepatol. 2005;42:632–8. doi: 10.1016/j.jhep.2004.10.031. [DOI] [PubMed] [Google Scholar]
- 34.Mazzaro C, Zorat F, Comar C, et al. Interferon plus ribavirin in patients with hepatitis C virus positive mixed cryoglobulinemia resistant to interferon. J Rheumatol. 2003;30:1775–81. [PubMed] [Google Scholar]
- 35.Zaja F, De Vita S, Russo D, Michelutti A, Fanin R, Ferraccioli G, Baccarani M. Rituximab for the treatment of type II mixed cryoglobulinemia. Arthritis Rheum. 2002;46:2252–4. doi: 10.1002/art.10345. (author reply 4–5) [DOI] [PubMed] [Google Scholar]
- 36.Scheinfeld N. A review of rituximab in cutaneous medicine. Dermatol Online J. 2006;12:3. [PubMed] [Google Scholar]
- 37.Wedemeyer H, He XS, Nascimbeni M, et al. Impaired effector function of hepatitis C virus-specific CD8+ T cells in chronic hepatitis C virus infection. J Immunol. 2002;169:3447–58. doi: 10.4049/jimmunol.169.6.3447. [DOI] [PubMed] [Google Scholar]
- 38.Yamamoto M, Nishimoto N, Davydova J, Kishimoto T, Curiel DT. Suppressor of cytokine signaling-1 expression by infectivity-enhanced adenoviral vector inhibits IL-6-dependent proliferation of multiple myeloma cells. Cancer Gene Ther. 2006;13:194–202. doi: 10.1038/sj.cgt.7700873. [DOI] [PubMed] [Google Scholar]
- 39.Sutherland KD, Lindeman GJ, Choong DY, Wittlin S, Brentzell L, Phillips W, Campbell IG, Visvader JE. Differential hypermethylation of SOCS genes in ovarian and breast carcinomas. Oncogene. 2004;23:7726–33. doi: 10.1038/sj.onc.1207787. [DOI] [PubMed] [Google Scholar]
- 40.Komazaki T, Nagai H, Emi M, et al. Hypermethylation-associated inactivation of the SOCS-1 gene, a JAK/STAT inhibitor, in human pancreatic cancers. Jpn J Clin Oncol. 2004;34:191–4. doi: 10.1093/jjco/hyh035. [DOI] [PubMed] [Google Scholar]