Abstract
Barley, one of the major small grain crops, is especially important in climatically demanding agricultural areas of the world, with multiple uses within food, feed, and beverage. The barley endosperm is further of special scientific interest due to its three aleurone cell layers, with the potential of bringing forward the molecular understanding of seed development and cell specification from Arabidopsis and maize. Work done in Arabidopsis and maize indicate the presence of conserved seed developmental pathways where Crinkly4 (Cr4), Defective kernel1 (Dek1), and Supernumerary aleurone layer1 (Sal1) are key players. With the use of microscopy, a comprehensive phenotypic characterization of the barley defective seed5 (des5) mutant is presented here. The analysis further extends to molecular quantification of gene expression changes in the des5 mutant by qRT-PCR. Moreover, full-length genomic sequences of the barley orthologues were generated and these were annotated as HvDek1, HvCr4, and HvSal1. The most striking results in this study are the patchy reduction in number of aleurone cells, rudimentary anticlinal aleurone cell walls, and the specific change of HvCr4 expression compared to HvDek1 and HvSal1. The data presented support the involvement of Hvdes5 in establishing aleurone cells. Finally, how these results might affect the current model of aleurone and epidermal cell identity and development is discussed with a speculation regarding a possible role of Des5 in regulating cell division/ secondary cell wall building.
Keywords: Aleurone cell development, anticlinal cell wall mutation, barley endosperm, Crinkly4, Defective kernel1
Introduction
The cereal endosperm represents mankind's most important source of food, feed, and industrial raw materials. A thorough understanding of its development will strengthen the scientific basis for genetic improvement of agronomic performance, disease resistance, product quality, and diversity. Thus, given the economic importance, endosperm development mutants have been extensively studied, especially in maize. In addition, the endosperm is of special developmental interest given its simplicity with four cell types only; the surrounding aleurone cells, the main inner mass of starchy endosperm cells, the transfer cells, and the cells of the embryo surrounding region (Becraft et al., 2001a). Extensive maize mutant screens combined with the genomic sequence resources in Arabidopsis and rice, have shown that aleurone cell identity is controlled by the Dek1, Cr4, and Sal1 genes (Lid et al., 2002; Olsen, 2004). The triploid endosperm nucleus undergoes mitotic divisions without cytokinesis, resulting in a multinucleated syncytium. A cessation in nuclear divisions follows, and the syncytical cytoplasm becomes organized into nuclear cytoplasmic domains (NCDs) by radial microtubule systems (RMSs) emanating from the nuclei. This process is similar in all cereal species (Olsen, 2004). Anticlinal walls (perpendicular to the central cell wall) are initiated at multiple sites where RMSs interact along boundaries of the monolayer of NCDs. The outer edge of the first anticlinal cell wall joins the central cell wall, but the interior edges remain free in the cytoplasm facing the central vacuole. The ‘open-ended’ anticlinal cell walls are called alveoli. Nuclei in alveoli undergo periclinal mitosis followed by cytokinesis. The peripheral daughter cells, which will become aleurone, enter a different pathway with cortical hoop-like microtubules. Continuous periclinal divisions in alveoli lead to a complete cellularization of the seed. However, the rigid control of division planes is gradually lost and eventually occurs at random, leading to a complete cellularization with starchy endosperm cells in random planes. Anticlinal divisions occur in the periphery to accommodate enlargement of the developing grain. In barley, with three aleurone layers, divisions occur in both planes (Brown et al., 1994; Otegui and Staehelin, 2000). It has been demonstrated that alveoliation is the typical mechanism for cellularization of a syncytium peripheral to a large central vacuole, and in cereals this process dominates cellularization (Olsen et al., 1995; Brown et al., 1996; 1999; Nguyen et al., 2001).
Dek1 encodes a protein of 2159 amino acids, with 21 transmembrane regions in the N-terminus and a putative intracellular cysteine proteinase domain in the C-terminus (Lid et al., 2002). Due to the highly conserved proteinase domain DEK1 is considered to be a member of the calpain super family, and the gene is expressed in all mitotically active tissues (Lid et al., 2002; 2005). Calpaines are cysteine proteinases that are activated by a rise in calcium concentration and involved in mediating signal transduction leading to cell differentiation and proliferation (Sato and Kawashima, 2001). In vitro studies have further demonstrated that DEK1 has proteinase activity similar to the animal m-calpain, even in the absence of calcium (Wang et al., 2003).
Mutant dek1 grains completely lack aleurone cells, and homozygous mutants fail to germinate due to embryo arrest at the globular stage (Lid et al., 2005). Cr4 encodes a receptor-like kinase (RLK) protein similar to tumour necrosis factor receptors found in mammals (Becraft et al., 1996). The cr4 mutant has less severely affected kernels than dek1, with patchy aleurone layer development. In regions that lack aleurone, the peripheral part consists of starchy endosperm cells, reported to be caused by cell redifferentiation (Olsen et al., 1998; Becraft, 2001b). In addition, cr4 mutant plants have crinkly leaves and graft-like tissue fusions (Cao et al., 2005). The third gene that affects aleurone cell specification, Sal1, is a homologue of the human Charged vesicular body protein/Chromatin modulating protein1 gene (Chmp1), a member of the conserved family of E-class vacuolar protein-sorting genes implicated in membrane vesicle trafficking connected to the endocytotic machinery (Shen et al., 2003). Mutant sal1 seeds have up to seven layers of aleurone cells instead of the normal one layer present in wild-type (wt) maize. In addition, homozygous mutant kernels have highly defective embryos and do not germinate. This phenotype suggests a role for SAL1 in inhibiting aleurone differentiation in the interior of the seed. The expression and function of these genes in plant development is not limited to the endosperm. Homozygous cr4 mutants as well as weak alleles of dek1 and sal1 show pleiotropic effects, most severe in epidermal structures (Becraft et al., 2001b; Shen et al., 2003; Johnson et al., 2005; Lid et al., 2005). The Dek1 and Cr4 genes are necessary both to obtain and maintain aleurone cell identity in maize and Arabidopsis, while the ZmSal1 functions to restrict aleurone cell identity to the outer cell layer in maize. The dynamics of the interaction are not known, thus a model combining all available genetic, phenotypic, and molecular data has recently been published (Tian et al., 2007). Once aleurone cell fate specification has been initiated, the aleurone developmental pathway includes lateral movement of the activated signal, distributed through symplastic sub-domains mediated by CR4 in specialized plasmodesmata. The desired functional concentration of DEK1 and CR4 at the plasma membrane is maintained by internalization and degradation involving SAL1-positive endosomes (Tian et al., 2007). So far no uncoupled expression or phenotypically mutant differences between the Dek1 and Cr4 genes has been found, even though dek1 has the most dramatic phenotype, displaying a complete lack of aleurone cells in maize. Also, no possible order of action of these genes has been revealed.
The aleurone is one cell layer thick only in maize and wheat, varying between one and six cells thick in rice depending on developmental cell positioning, and finally three cells thick in barley. Further progress in understanding aleurone cell identity control is likely to come from barley genomics, given its intermediate physiology and difference concerning the number of aleurone cell layers. However, while awaiting full genome sequencing of barley, we depend on comparative genomics and sequencing of genes of specific interest to gain further insight into the genetic control of aleurone cell identity and specificity. The barley defective seed5 (des5) mutant was generated by applying Na-azide treatment to the barley cultivar Bomi (Bosnes et al., 1987; 1992), which resulted in a collection of 17 mutants.
Homozygote mutant seeds undergo seemingly normal development for 3 days (d) after fertilization, upon which the seeds start to appear shrunken. The des5 mutant is thus likely to provide a valuable link in the processes that control seed development. Here a thorough characterization of the mutant phenotype is presented which exhibits a reduced number of aleurone cells, a lack of anticlinal secondary cell walls, and morphologically defective aleurone and starchy endosperm cells. Furthermore, full-length genomic sequences of the HvDek1, HvCr4, and HvSal1 genes are presented and data, which for the first time indicate transcriptional regulation of Cr4, in a manner distinct from that of HvDek1 and HvSal1. The data point to an important role for HvDes5, in addition to HvDek1, HvCr4, and HvSal1, as a regulator of seed development.
Materials and methods
Mutation induction
Barley seeds were soaked in distilled water at 4 °C for 15–17 h. The distilled water was exchanged and oxygen was added for 6–17 h at 20 °C. The seeds were then treated with different concentrations of Na-azide in McIlvaine buffer and in the presence of oxygen for 2 h. After the mutation induction the seeds were washed in distilled water and stored in water at 4 °C (Bosnes et al., 1987).
Plant material
Plant tissues used in all experiments were isolated from barley (Hordeum vulgare cv. Bomi). Barley plants, both wt and des5 heterozygotes, were grown in pots containing peat-based compost in a greenhouse at 10–17 °C under a 16/8 h light/dark cycle. Seed development was calculated on the basis of days after pollination (DAP). When the anthers in the middle of the spike had opened and shed pollen, the spikes were tagged and dated (0 DAP). Heterozygous des5 plants were identified by the presence of defective and normal seeds in the same ear. For microscopy wild-type and des5 seeds were sampled at predetermined intervals after pollination. For molecular analysis seeds were harvested at days after pollination as indicated. The plant tissues were frozen in liquid nitrogen and stored at –80 °C until used.
Histology and microscopy analysis
For general anatomic examination, seeds harvested at 5, 10, 15, and 20 DAP were fixed overnight in 1.25% glutaraldehyde/2% paraformaldehyde in 50 mM PIPES-buffer pH 7.0 and rinsed in 50 mM PIPES-buffer pH 7.0. Specimens were dehydrated through a graded ethanol series, infiltrated, and embedded in LR White Hard Grade Acrylic Resin (Electron Microscopy Science, Fort Washington, PA) according to the manufacturer's instructions. One μm sections, prepared using an ultra microtome and diamond knife, were mounted on gelatin-coated slides and stained with Stevenel's Blue (del Cerro et al., 1980). For cryosectioning, grains were cut in half and frozen in Sakura TissueTek O.C.T. compound prior to sectioning. Cryosections (typical 12 μm) were prepared on a Microm HM560 cryostat, mounted on gelatin-coated slides and left unstained or stained with either Stevenel's Blue or Sudan IV. For Sudan IV staining, slides were washed with a 1:1 mixture of 87% glycerol in water, and subsequently a 1:1 mixture of 87% glycerol in ethanol. Sections were stained for 20 min in a 1:1 mixture of Sudan IV-saturated 96% ethanol and 99.5% glycerol, and washed in reverse order. Slides were examined with a Leitz Aristoplan microscope and images recorded using a Leica DC200 or DC300F digital camera. For scanning electron microscopy (SEM) material was fixed as above. Kernels were halved, and these, as well as root and leaf samples were dehydrated in a graded ethanol series, critical-point-dried using a CPD 030 (Bal-Tec), mounted on Al-stubs using double-faced carbon tabs (Agar Scientific), coated with approximately 500 Å Pt in a SC7640 sputter coater, and analysed using an EVO-50 Zeiss microscope. For transmission electron microscopy (TEM) analysis, seeds were harvested at 18 DAP and fixed overnight in 1.25% glutaraldehyde/2% paraformaldehyde in 50 mM HEPES buffer pH 7.2 and rinsed in 50 mM HEPES buffer pH 7.2. Specimens were dehydrated through a graded ethanol series and infiltrated and polymerized in Spurr's resin. Ultramicrotome sections were mounted on grids, observed in a Jeol JEM-1230 transmission electron microscope, and images were collected in a Gatan digital kamera MSC600CW.
Nucleic acid isolation
Genomic DNA was extracted from young leaves using the Nucleon Phytopure Plant DNA extraction kit (Amersham Biosciences, UK). Total RNA from young leaves was extracted using the RNeasy Plant Mini kit (Qiagen, Hilden, Germany). Due to the high starch content, total RNA from barley seeds was sequentially extracted with extraction buffer (50 mM TRIS pH 9, 200 mM NaCl, 1% sarcosyl, 20 mM EDTA, and 5 mM DTT) and phenol/chloroform/isoamylalcohol, and then with Trizol reagent (Invitrogen, Carlsbad, CA) and chloroform. Isolated RNA was purified and DNase-treated on columns using the RNeasy Plant Mini kit procedure (Qiagen, Hilden, Germany) and RQ1 RNase free DNase (Promega, Madison, WI). Concentration and integrity of total RNA were analysed with a Nanodrop ND-1000 spectrophotometer and on agarose gel. Quality of RNA for quantitative reverse transcription PCR (qRT-PCR) was also verified with standard RT-PCR of a control gene.
Identification and sequencing of HvDek1, HvCr4 and HvSal1
Cloning of the HvDek1 gene was done by screening a barley cDNA library with several primer pairs specific to the maize Dek1 cDNA (Lid et al., 2002). A PCR fragment obtained this way was cloned into a pCR4-TOPO vector following the manufacturer's recommendations and used as a probe screening an Hv_MBa BAC library from barley (Hordeum vulgare cv. Morex), obtained from CUGI, Clemson University (https://www.genome.clemson.edu/cgibin/orders?page=serviceHome&;service=barc).
Hybridization was performed according to protocols provided by CUGI. Positive hits were visualized by autoradiography, and clone identity was determined by using the filter layout and the duplication pattern code. Positive clones were obtained from CUGI and amplified in Luria Broth (LB) medium with chloramphenicol selection overnight. High quality BAC DNA was extracted using NucleoBond BAC100 kit (Macherey-Nagel, Duren, Germany). The first BAC DNA sequencing reaction was performed with an HvDek1 primer targeting the probed region, and full-length sequence was obtained by primer walking. The HvCr4 gene was cloned and sequenced from the same CUGI BAC library as HvDek1. Degenerate primers were used to obtain an initial cDNA sequence which served as a probe for the library, and primer walking was used to obtain the whole genomic sequence from the positive BAC clone. The genomic HvSal1 sequence was amplified using primers designed based on candidate barley EST sequences obtained from the databases. Amplified fragments were cloned into a pCR4-TOPO vector according to the manufacturer's recommendations, and were sequenced and aligned to make out the full coding region. All sequencing was done using the Big Dye 3.1 kit (Applied Biosystems, Foster City, CA) according to the manufacturer's recommendations, and automatic analysis was performed using ABI Prism 377 DNA Sequencher (Gene Codes Corporation, Ann Arbor, MI). Sequence editing, alignment, and interpretation were performed with the aid of Sequencher and Vector NTI 5 software (InforMax). Novel sequences were blasted against the public databases at ncbi.nih.gov, plantgdb.org (Altschul et al., 1997), and the HarvEST v1.59 barely EST database. The sequences were submitted with the following accession numbers: HvCr4, EU096088; HvDek1, EU096089; HvSal1, EU096087.
Transcript analysis using real-time quantitative RT-PCR
Plant material for real-time transcript analysis of HvCr4, HvDek1, and HvSal1 was taken from wt and des5 mutant seeds (20–30 DAP) in two independent biological replicates. Quantitative RT-PCR was performed with three subsamples from each biological replication, resulting in a total of six experimental units. Each biological sample was extracted from 4–6 seeds. 966 ng total RNA from each sample was reverse transcribed using the Superscript III kit and RNase H treatment (Invitrogen, Carlsbad, CA) according to the manufacturer's recommendations. The reverse transcribed cDNA mix was then diluted 50 times and 5 μl were used as template for each qRT-PCR reaction. Transcript levels were analysed using a 7500 Real-time PCR machine (Applied Biosystems, Foster City, CA). PCR reactions were performed using the Platinum SYBR Green qPCR SuperMix UDG with ROX kit (Invitrogen, Carlsbad, CA), in 25 μl reaction volumes, but otherwise following the manufacturer's recommendations. Cycling conditions were standard conditions according to the 7500 System software: 50 °C 2 min, 95 °C 10 s, and 45 cycles of 95 °C 15 s, and 60 °C 1 min. Primers (Invitrogen, Carlsbad, CA) were designed using Primer Express 1.5 (Applied Biosystems) or Primer3 (http://www-genome.wi.mit.edu/genome_software/other/primer3.html). All primers were used at a concentration of 200 nM, at which efficiency ranged from 83% to 94%. Target specificity of each primer was ensured using dissociation curve analysis after the real-time PCR program. Individual threshold cycles (Ct values) were located from 22 to 33 cycles. Relative transcript levels of HvCr4, HvDek1, and HvSal1 were determined using the Standard curve method according to the User Bulletin 2 (ABI PRISM sequence detection system; PE-Applied Biosystems). This method was chosen in order to account for the varying primer efficiencies. The relative values of steady-state gene expression in Fig. 6 were normalized to ubiquitin, Mub1 (Acc. No. M60175), as the endogenous reference. Real-time primers were as follows: HvUb-F, CAAGTGCGGACTCACCTATG; HvUb-R, TCGCGATAGGTAAAAGAGCAG; HvDek1-F, TTATCCACCTGAAATGCGCTATT; HvDek1-R, TGGCAACCACCTGCACTGTA; HvSal1-F, GCAGATCTTCGACCTCAAGTTCA; HvSal1-R, CCTTGAGCTTCTGGTCCTTCTC; HvCr4-F, GACCAGCGGATCGAGTACAG; HvCr4-R, ACCGCAGACCATCTCCTATG.
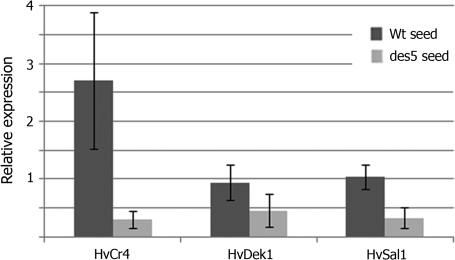
Fig. 6.
Real-time qRT-PCR analysis of HvCr4, HvDek1, and HvSal1 transcript levels in wt and des5 seeds. Expression is shown relative to ubiquitin as the internal control. RNA was extracted from two biological samples, and real-time qRT-PCR was performed on three subsamples from each biological sample. Data represent means ±SD for six repetitions. ANOVA t test confirmed with *P <0.05 significant down-regulation of HvCr4 in des5 seeds (n=6) as opposed to wt (n=6).
Phylogenetic analysis
Multiple sequence alignments of full-length protein sequences and phylogenetic analysis according to the Neighbor–Joining method were constructed using ClustalX (v.2.0.3; Thompson et al., 1997). An unrooted radial phylogenetic tree was displayed using TreeView (v.1.6.6; Page, 1996). Statistical confidence was calculated by bootstrap analysis based on 1000 reiterations, of which none of the values were below 630 (values not shown). GenBank accession numbers for proteins are HvDEK1; EU096089, ZmDEK1; AY061804, OsDEK1; AP004161, OJ1311_H06.4, AtDEK1; AC027034, BAC F7A10.23, HvSAL1; EU096087, AtSAL1 (At1g73030), AtSAL2 (At1g17730), ZmSAL1; NP_001105218, OsSAL1(NP_001058177), HvCR4; EU096088, ACR4; NP_191501, AtCRR1; NM_111813.3, AtCRR2; NM_129475.3, AtCRR3; NM_115453.2, AtCRK1; NM_124159.2, OsCR4; AB057787, OsCRR1; AL606452.2, OsCRR2; AP004584.3, OsCRR3; AC129720.2, OsCRR4; AC123524.2, ZmCR4; NP_001105395.
Results
Identification and genetic characterization showing des5 to be a single gene mutation
The control of aleurone identity in maize is set up by the ZmDek1, ZmCr4, and ZmSal1 genes, after the first periclinal division of the peripheral cells, when initiating cellularization. Barley, with its three periclinal divisions and three aleurone cell layers surrounding the seed starchy endosperm and embryo, is more complex and therefore of particular interest scientifically. The des5 mutant was previously identified in our laboratory as a defective xenia (dex) mutant, with wt and mutant seeds on the same ear (Bosnes et al., 1987, 1992). Xenia is Greek for hospitality and is used to name plants that have seeds of different genotypes at the same ear. Historically this mutant was for convenience named B5 (barley5), mutant number 5 in a collection of 17. At the start of this study the mutant was renamed des5 (defective seed5), a name that actually resembles the phenotype observed and described in this paper. Mutant seeds are easily detected at mature stages as being smaller than wt seeds with a crinkly or defective appearance. At maturity (30 DAP) homozygous mutant seeds appear shrunken, with wrinkled and approximately half-sized seeds compared to wt. The reduced size is due to developmental arrest of the endosperm and a prematurely aborted embryo. Upon self-pollination of heterozygote plants a close to 3:1 segregation ratio was observed. The Pearson's chi square test generated a chi value of the mutation being 12.04 of the total 16.02 (see Supplementary data at JXB online). Such tests resulting in chi values above 3.84 are commonly interpreted as justification for rejecting the null hypothesis. Based on this we claim that this chi square test supports a 3:1 segregation ratio, since when using such a large data set this problem is not unexpected. Even a seemingly small deviation from the expected ratio would significantly distort the 3:1 segregation. Unfortunately, the homozygote and heterozygote classes were not kept separate, thereby losing a degree of freedom and failing to obtain the strongest P-value. However, in summary, the results strongly indicate segregation consistent with a Mendelian inheritance pattern of a recessive single-gene mutation.
The des5 mutation has adverse effects on seed morphology during development
In wt barley seeds, at 5 DAP, the peripheral part is composed of a two cells thick aleurone layer, one with isodiametric, cytoplasmically dense aleurone cells, and a sub-aleurone layer of larger, less dense, isodiametric cells and thinner cell walls. In most des5 kernels the aleurone layer at this early stage consists of one layer of isodiametric aleurone-like cells, and a several cells thick sub-aleurone layer. The resulting phenotype is a variation in number of aleurone cells, typically varying from no aleurone cells to a one cell thick aleurone layer only, within the same seed. At 12 DAP the reduced size of the des5 compared with wt seeds is noticeable (data not shown). Wt seeds exhibit, in general, a three cells thick aleurone layer. The defective des5 seeds are less plump and the peripheral part is composed of one layer of aleurone-like cells, broader and less dense than observed for wt seeds, and a sub-aleurone layer composed of cells with an appearance intermediate between that of aleurone and starchy endosperm cells.
At 20 DAP, wt seeds exhibit completely cellularized seeds with a three cell layers thick aleurone. The des5 seeds exhibit a reduced number of cells at this stage, with peripheral cells lacking the typical aleurone cell appearance (Fig. 1a, b). The peripheral cells appear broader and larger (data not shown), less dense and phenotypically more similar to starchy endosperm cells (Fig. 1b). Although the overall morphological determination of the peripheral cells into aleurone cells seems to be weaker or delayed compared with wt, occasionally as many as four layers of aleurone-like cells may be found in the des5 mutant. However, the overall mutant phenotype is a reduction in the number of aleurone cells. Finally, the number of cell divisions is reduced, resulting in a reduced number of starchy endosperm cells. The reduced number of aleurone cells is due to the overall reduction of the wt three layers thick aleurone, and a reduction of starchy endosperm cells is due to reduced filling from prematurely arrested cellularization. Given the incomplete cellularization, the remaining unfilled centre vacuole cavity makes the seeds appear shrunken.
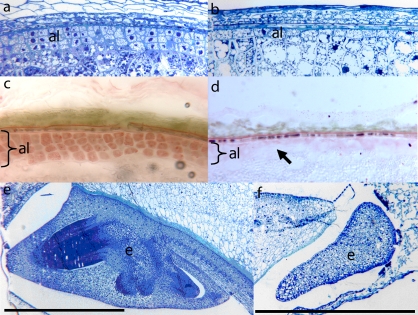
Fig. 1.
Microsections showing the peripheral cell layers of wt (a) and des5 kernels (b). (c, d) Sudan Red stained cryosections of wt (c) and des5 kernels (d). Sudan red is a specific stain for lipid accumulation, and often used as a marker for the lipid-rich aleurone cells. The arrow indicates missing aleurone cell layers in the mutant. (e, f) Microsections showing the embryo of wt (e) and des5 kernels (f). The des5 embryo arrests at an early stage. Al, aleurone; e, embryo. Black scale bars are 1 mm long.
A similar phenotype was also observed for mature des5 seeds (data not shown). Staining of cryosections with Sudan IV show lipid accumulation in all three aleurone layers in 20 DAP wt seeds (Fig. 1c), whereas des5 seeds show an overall reduction in the number of aleurone cells (Fig. 1d).
Consistent with previously described aleurone mutants, embryo development is also affected in des5 seeds (Fig. 1f). At 20 DAP the defective des5 embryo appears amorphous and arrested at an early stage (Fig. 1f). Similar to previously published morphological data from barley (Brown et al., 1994) the arrest has taken place prior to true cell differentiation. Taken together, the results show that the des5 mutation affects cell differentiation and division of the aleurone cells and cell division of starchy endosperm cells, as well as causing an early arrest of the developing embryo.
The des5 mutation affects anticlinal cell wall thickening of aleurone cells
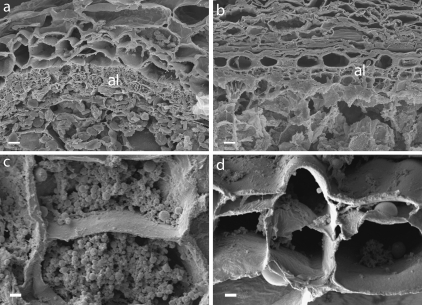
There is a striking morphological difference between cell walls of aleurone and starchy endosperm cells. To investigate the effects of the des5 mutation on aleurone cell development closer, sections of wt and des5 seeds were analysed using scanning electron microscopy (SEM). In addition to the variation in the number of aleurone cells (layers), the size, density, and number of aleurone granules in the cells differed in the des5 mutant compared with wt (Fig. 2). The des5 aleurone cells seemed collapsed, appearing as a pile of multiple periclinal cell walls stacked together (Fig. 2d). The loss of cell material and the appearance of empty aleurone cells might be due to the chemical fixation, but this still suggests a biochemical difference between wt and des5 cells leading to the observed phenotype. High resolution micrographs give the impression of empty cells in the des5 mutant (Fig. 2d) as compared to wt (Fig. 2c). This has also been observed in tissue with mosaic-induced reverting aleurone cells in maize, caused by chromosome breakage mutations of dek1 (Becraft and Asuncion-Crabb, 2000).
Fig. 2.
Effects of the des5 mutation on aleurone cell morphology. (a, b) wt (a) and des5 (b) aleurone cells at 12 DAP. Mutant aleurone cells are unorganized and lack the cubical shape and rigidness compared to wt. (c, d) High-resolution micrograph close-ups show aleurone cells lacking cell content (d), in contrast to the wt cell (c). Al, aleurone. Scale bars indicate 10 μm (a, b) and 2 μm (c, d).
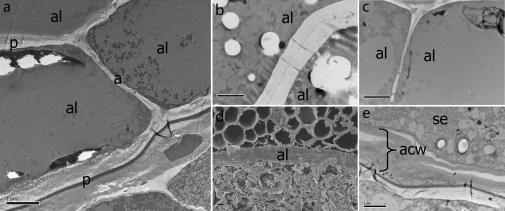
The apparently collapsed aleurone cells, possibly resulting from changes in aleurone cell wall composition or strength, were investigated by transmission electron microscopy (TEM). Interestingly, TEM micrographs clearly show that the anticlinal cell walls between connecting aleurone cells are significantly thinner in the des5 mutant compared to the wt (Fig. 3a, b, c). Judging from the micrographs they seem to consist of the middle lamella and primary cell wall only. This may be one of the physical factors involved in the collapse of new aleurone cells. Consistent with this, stacks of cell walls at the junction between the innermost aleurone cells and the starchy endosperm cells were also observed in des5 mutant seeds in TEM micrographs, which were not present in wt seeds (Fig. 3d, e). When comparing the cell wall thickness of connecting starchy endosperm cells of des5 and wt no such difference was found (data not shown). This is the first reported mutation affecting the development of anticlinal cell walls in aleurone to our knowledge, making it especially interesting for further progress in understanding seed development.
Fig. 3.
Transmission electron micrographs. (a) Anticline and periclinal cell walls between connecting aleurone cells in the des5 mutant. Anticlinal walls consist of middle lamella and primary wall only. (b, c) Anticlinal cell wall between aleurone cells in wt (b) and des5 seeds (c), respectively. (d, e) Stacks of collapsed cell walls between the innermost aleurone cell and starchy endosperm are present in the des5 mutant. Al, aleurone; se, starchy endosperm; acw, aleurone cell wall. Scale bars indicate 2 μm (a, b, d, e) and 5 μm (c, f).
The des5 mutation alters composition in starchy endosperm cells
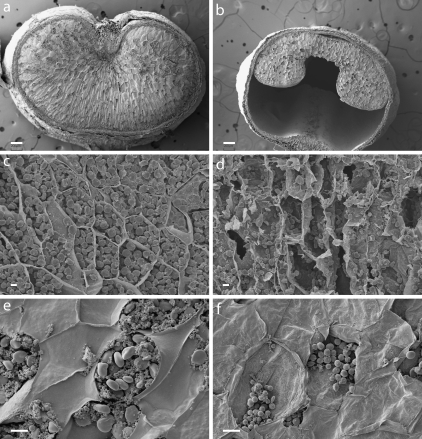
Quantification of starchy endosperm cells in the barley seed is hampered by the random division planes during cellularization, which complicates direct comparisons between seeds. des5 mutant seeds have a shrunken appearance and whole seed micrographs support that this is due to arrested cellularization leading to reduced cell numbers, and hence incomplete seed filling, with a considerable central vacuole remaining (Fig. 4a, b). To investigate the causes for this further, individual starchy endosperm cells were analysed using SEM. While the size of starchy endosperm cells appears normal in the des5 mutant, sections through the cells reveal fundamental structural differences between the des5 mutant and wt. The des5 mutant contains a considerably reduced amount of starch granules and protein substance (Fig. 4c, d). While wt cells are completely packed with starch granules, giving rise to a considerable tension in the cell walls, the des5 mutant starchy endosperm cells have fewer and smaller starch granules, leading to a more shrunken appearance of the cells. To avoid cell content loss during the fixation process, new sections were made on previously dried and coated material, and the tissue was studied again with SEM. High resolution micrographs clearly show the two types of granules in wt starchy endosperm cells (Fig. 4e), the larger A-type granules and the smaller B-type granules. In addition to protein, these starch granules are the major content of starchy endosperm cells in barley. In the des5 starchy endosperm cells however, one type of starch granules only was seen, and they are not similar to either A- or B-type wt granules (Fig. 4f). They might be either an intermediate granule type, or simply smaller A-type granules, in which case des5 lack B-type starch granules.
Fig. 4.
Effects of the des5 mutation on starchy endosperm cell morphology. (a, b) Whole seed micrographs of wt (a) and des5 (b) seeds. des5 seeds show an arrest in cellularization. The large central vacuole, in addition to the reduced cellularization, leads to the shrunken apperance. (c, d) Starchy endosperm cells of wt and des5. (d) des5 cells show an altered composition. (e, f) Starch granules of wt and des5 mutant. (f) des5 starchy endosperm cells have a reduced amount and altered composition of A- and B-type starch granules (f) compared to wt (e). Scale bars indicate 200 μm (a, b), 30 mm (d, e), and 10 mm (c, f).
Identification and annotation of HvCr4, HvDek1 and HvSal1
Given the lethal nature of the des5 mutation, sequencing of the HvDek1, HvCr4, and HvSal1 genes is impossible in the mutant. Cloning of the point mutated gene responsible for des5 is also a demanding and long-term goal. In order to enable an investigation of the relationship between the des5 mutation and previously described seed developmental genes, the putative barley orthologues of Cr4, Dek1, and Sal1, were cloned and named HvCr4, HvDek1, and HvSal1, respectively.
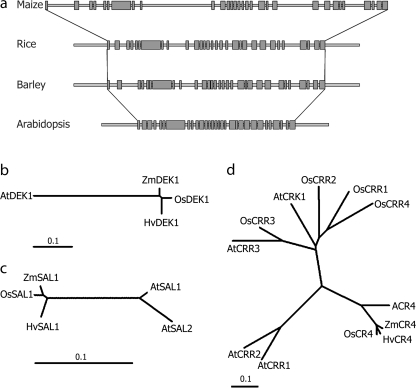
The initial barley cDNA fragment amplified using ZmDek1 primers was 91.3% identical to the ZmDek1 cDNA. Annotation of the full-length HvDek1 (Acc. No. EU096089) was assisted by comparison with Dek1 sequences from maize, Arabidopsis, and rice, respectively, with which it shares a high degree of sequence conservation (Fig. 5a). BLAST against the HarvEST v1.59 EST database also supported the presence of a single Dek1 gene in the barley genome. This was confirmed with Southern blot analysis (data not shown) and is in agreement with all plant genomes analysed to date, with the exception of Populus having two copies (LT Olsen, unpublished results). The HvDek1 gene is 14 009 bp long, consisting of 30 exons within the coding region, giving rise to a predicted protein of 2160 amino acids. Structurally it shares 93% sequence identity with OsDEK1 at the protein level. Comparison of HvDek1 with Dek1 of maize, rice, and Arabidopsis is summarized in Table 1, and visualized phylogenetically in Fig. 5b. The number of coding exons is conserved among Arabidopsis and the cereals.
Fig. 5.
Analysis of barley Dek1, Cr4, and Sal1. (a) HvDek1 gene structure. Overall comparison of exon–intron structure of HvDek1 coding region with Dek1 from maize, rice, and Arabidopsis. The number of exons (grey boxes; 30) is conserved in Arabidopsis and the cereals. (b, c, d) Phylogenetic analysis showing the evolutionary relationship within the families of DEK1, SAL1, and CR4, respectively. Multiple sequence alignments of full-length protein sequences and phylogenetic analysis according to the Neighbor–Joining method were constructed using ClustalX. Displayed are unrooted radial phylogenetic trees. Statistical confidence was ensured by bootstrap analysis based on 1000 reiterations, of which none of the values were below 630 (values not shown). Scale bars indicate number of substitutions per site.
Table 1.
Comparison of Dek1 from Arabidopsis and the cereals
The HvCr4 gene (Acc. No. EU096088) was cloned and sequenced from the same BAC library as HvDek1. A cDNA sequence obtained with degenerate primers was used as the initial probe. Using the FGENESH software, unique translation start and stop codons were predicted, indicating HvCr4 to consist of a single-exon coding region of 2694 bp, coding for an 897 amino acids long protein. tBLASTn search in the NCBI database uncovered one full-length published Cr4 cDNA clone only, from H. vulgare cv. Haruna Nijo (Acc. No. AK251439). BLAST search against the HarvEST v1.59 database also indicates one copy only of HvCr4 in the barley genome, however Southern blot analysis (LT Olsen, data not shown) suggests the presence of two, or possibly three, copies. Alignment with AK251439 and barley ESTs indicated a 5′ UTR of approximately 470 bp, containing a putative intron of 138 bp. Although the Crinkly family of RLK genes in rice and Arabidopsis are known to consist of five members (Cao et al., 2005), phylogenetic analysis showed that the predicted HvCR4 protein sequence shares significantly higher similarity to the CRINKLY 4 orthologues, OsCR4 and ACR4, as well as to ZmCR4, and is therefore likely to represent the true Cr4 orthologue in barley (Fig. 5d). HvCR4 shares 88, 84, and 59% sequence identity to CR4 from rice, maize, and Arabidopsis, respectively.
The genomic HvSal1 sequence (Acc. No. EU096087) was amplified using primers designed based on barley EST sequences. Using the FGENESH software, unique translational start and stop codons were predicted. This resulted in an open reading frame with no introns, coding for a putative protein of 204 amino acids. tBLASTn search in the NCBI database uncovered one full-length published Sal1 cDNA clone only, from H. vulgare cv. Haruna Nijo (Acc. No. AK252514). BLAST search in the HarvEST v1.59 database indicates the presence of one copy of Sal1 in the barley genome, possibly with additional members of considerably lower similarity. Alignment with AK252514 indicates the presence of a large intron of 2181 bp in the 3’ UTR region, followed by a small second exon. At the protein level HvSAL1 shares 96% and 87% sequence identity with SAL1 from maize and Arabidopsis, respectively. Phylogenetically, HvSAL1 clusters closely together with SAL1 from maize and rice, and is more closely related to AtSAL1 than AtSAL2 (Fig. 5c). Based on the high degree of sequence conservation of all three genes, and supported by BLAST searches and phylogenetic analysis, it is concluded that these are true orthologues of Dek1, Cr4, and Sal1, identified in maize, rice, and Arabidopsis.
Molecular characterization by transcription profiling
HvDek1, HvCr4, and HvSal1 transcripts exhibit low expression in all cell types including the embryo of wt seeds observed by in situ hybridization (data not shown). This confirms previous studies using Lynx MPSS (Brenner et al., 2000), and in situ hybridization showing the ubiquitous expression of all three genes indicating a functional role in all cell types (Lid et al., 2002; 2005). In situ hybridization was not feasible on des5 mutant seeds, due to the nature and loose consistency of these seeds it is not technically feasible to prepare sections of sufficient quality for in situ hybridization in des5 seeds.
The des5 mutation causes a significant reduction of HvCr4 transcript expression in the seed
Performing a preliminary microarray analysis (data not shown), putative interesting effects on the HvCr4 and HvSal1 genes were identified in the des5 mutant. In order to monitor the expression levels of HvCr4, HvDek1, and HvSal1 in wt and mutant backgrounds, real-time quantitative RT-PCR (qRT-PCR) was performed. Relative mRNA levels of the genes were analysed in wt and mutant seeds (20–30 DAP), and were normalized to ubiquitin as the internal control. From the expression analysis, the main finding was a significant and specific down-regulation of HvCr4 in des5 seeds compared with wt seeds (Fig. 6; P <0.05). The difference in HvCr4 expression in wt and mutant seeds was also repeatedly noticed in regular semi-quantitative RT-PCR upon gel fractionation of the PCR products (data not shown). This result suggests a connection between des5 and HvCr4 regulatory pathways. The altered regulation of HvCr4 could explain some of the des5 mutant phenotype.
Discussion
The barley B5 mutant, here renamed des5, has a particularly interesting phenotype, expected to bring the genetic understanding of aleurone control known from Arabidopsis and maize, forward. Here a phenotypic and transcriptional characterization of the des5 mutant is presented.
The des5 seed mutant display a lack of secondary anticlinal alerone cell walls
Microscopy analyses of des5 developmentally arrested mutant seeds showed that the major phenotypic effects were variations in the number of aleurone cells, the absence of anticlinal secondary cell walls between aleurone cells, and mutated starchy endosperm starch granules. In wt barley, the aleurone is three cells thick, and the aleurone cells have a characteristic regular cuboid shape set up by rigid cell walls. In the des5 mutant, however, the number of cells in the layer, and hence the total number of cells, is mostly reduced, although occasionally as many as a later four cell thick layer may be found. Furthermore, the aleurone cells appear empty, and microscopy revealed the absence of wild-type strengthened anticlinal cell walls between connecting aleurone cells. The cell walls only consist of the middle lamella and the primary cell wall. In contrast to primary cell walls, which are synthesized in basically all plant cells, lignified secondary walls develop in cells that have ceased to expand (Turner et al., 2001). After the cessation of expansion and division, a secondary wall is synthesized within the bounds of the primary wall (Mitsuda et al., 2005). The des5 aleurone cells have a wider appearance, consistent with possible absence of final cell expansion.
The des5 collapsed aleurone cells contradict reverted cell identity as previously reported
Interestingly, an additional phenotypic feature was observed in the des5 mutants that appeared to be piled periclinal cell walls stacked next to each other at the junction between the innermost aleurone and the starchy endosperm cells (Fig. 3d, e). This observation suggests that the initial aleurone cells in the des5 mutant do not revert to starchy endosperm cells due to a switch in cell fate, as previously reported for aleurone mutants. The aleurone cells that loose their cell identity seem rather to collapse, leaving the remaining cell walls stacked next to the remaining space that is filled by the neighbouring starchy endosperm cells (Becraft and Ansuncion-Crabb, 2000; Becraft et al., 2002; Johnson et al., 2005). This finding is consistent with observations made in our laboratory when studying microscopy sections of dek1 mutated aleurone cells in maize (data not shown).
Previously described aleurone mutants, all of which des5 shares some similarity with, include dek1 completely lacking aleurone cells (Lid et al., 2002), cr4 displaying patchy aleurone cells (Becraft et al., 1996), disorganized aleurone 1 and 2 (dil1 and dil2) having defective aleurone cell morphology (Lid et al., 2004), and sal1 with up to seven aleurone layers (Shen et al., 2003). Other known maize endosperm mutants with similarities to the barley des5 mutant are bareback and naked that display variable defects in the starchy endosperm (Becraft and Asuncion-Crabb, 2000). In both mutants, the endosperm is usually opaque and often white, with a floury texture. These features are also found in the des5 mutant seeds, where the endosperm appears loose and shrunken. It is therefore possible that, in these mutants, the penetration of the positional signal varies around the perimeter of the endosperm due to its variable physical properties. However, if defects in the physical properties of the endosperm should be the primary cause of variable penetration of the positional cue, a phenotypic effect on aleurone organization would be expected in all mutants with floury endosperm. From extensive screening for aleurone mutants in both maize and barley, only occasional floury endosperm mutants have defects in aleurone organization as well. The loose endosperm in the des5 mutant is therefore not likely to be the primary cause of the aleurone phenotype.
The des5 mutation affects the starch granules of the endosperm
Although cell walls between starchy endosperm cells are unaffected in the des5 mutant, and the cells have wt size, they appear shrunken, partially due to an aberrant type of misshapen starch granules; irregular in shape and intermediate in size. Since the A-granules have much higher amylase content than B-granules, there is reason to believe that the biochemical property of des5 starchy endosperm is also altered. The onset of large and small granule differentiation is initiated at different stages during endosperm development. In wt barley endosperm, A-type granules initiate at approximately 5–10 d after anthesis followed approximately 10 d later by a second wave of granule initiation that gives rise to the B-type granules (Burton et al., 2002). The genetic mechanisms controlling the structure and function of the different starch granules are, however, poorly understood (Williams and Duffus, 1977; Shewry, 1992). Recent results suggest that as well as suppressing phytoglycogen synthesis, isoamylase in the wt endosperm plays a role in determining number, and also to some degree the morphology, of starch granules (Burton et al., 2002). The altered granule composition in des5 is believed to be an indirect effect, due to a disruption in starch metabolism in early endosperm development, possibly as a result of dysfunctional aleurone cells or cell divisions.
The des5 mutation also affects embryo development similarly to other aleurone mutations
The des5 mutation is lethal and whether this is due to lethal defects within the barley embryo and/or failure of the endosperm tissue to sustain further development is not known. Monocotyledonous species such as barley develop embryos with a complex architecture, with the embryonic axis displayed laterally relative to the scutellum (Roth, 1955). The early developmental arrest of the des5 mutant embryo is a common effect in all mutations affecting aleurone cell development. The complex development of barley embryos can be divided into three stages: (i) a differentiation stage in which various tissues are formed and the basic body plan of the seedling is established (0–19 d after flowering); (ii) a maturation stage which is primarily determined by the formation of protein and lipid bodies (20–39 DAF); and (iii) a desiccation stage which eventually yields the dormant grain (Nielsen et al., 2006). By visual inspection of embryos using SEM it is suggested that the des5 mutant embryos arrest at an early differentiation stage (0–10 DAF).
The des5 mutant supports our recent model explaining the control of aleurone cell identity
In summary, it is believed the main phenotypic effects of the des5 mutation is related to the aleurone. Aleurone cell identity needs positional cues and signalling, and absent starch cell identity seems to be the default pathway. It has also been shown that most genes preferentially expressed in the aleurone are expressed in the embryo, most probably due to their mutual interaction (Aalen et al., 1994; Becraft et al., 2002; Opsahl-Sorteberg et al., 2004; Sreenivasulu et al., 2008). How barley obtains and controls a three cells thick aleurone layer while most other plants exhibit just a one cell thick layer is so far unknown. ZmSal1 restricts development of an aleurone layer that is multiple cells thick in maize, and HvSal1 might be involved in the same process in barley. If so, one should expect des5 to have an increased expression of HvSal1; however, this is not the case. des5 could represent a mutation in an unknown gene affecting aleurone cell identity, either interacting with or acting independently of the Dek1, Cr4, and/or Sal1 genes, thus affecting the distribution of positional cues believed to be mediated through CR4 during aleurone signalling (Olsen et al., 1998; Becraft and Asuncion-Crabb, 2000; Lid et al., 2002).
In order to relate des5 to the already known mechanisms for regulation of aleurone differentiation, and possibly further elucidate the nature of the des5 mutation, the expression of HvCr4, HvDek1, and HvSal1 was investigated in barley wt and the des5 mutant. The analysis showed a significant down-regulation of HvCr4 in des5 mutant seeds as compared to wt. Since in situ hybridization showed constitutive expression of Cr4 transcript throughout the seed in wt barley (data not shown), this reflects a true down-regulation of HvCr4 rather than an overall reduction in the number of aleurone cells in the mutant. We have recently shown CR4, the receptor-like kinase with similarity to the ligand binding domain of TNFR (Tumor Necrosis Factor Receptor), to be enriched in expanded plasmodesmata (PD) in anticlinal aleurone cell walls, possibly securing aleurone cell identity by signalling through these symplastic sub-domains (Tian et al., 2007). It is therefore reasonable to believe that, as a cause or an effect of des5 mutation, reduced cell walls might reduce Cr4 expression by hampering the deposition of CR4 protein to PD, consequently disturbing regulatory feedback loops and HvCr4 expression.
The current model for aleurone cell specification involves DEK1 acting as a sensor for positional information, signalling to the cells where in the organism they are located; surface or internal. In this model DEK1 responds to a positional signal triggering aleurone fate, CR4 acts to counteract lateral inhibition of aleurone cell fate between aleurone cells, and proper concentration of DEK1 and CR4 in plasma membranes is maintained by internalization and degradation via trafficking through SAL1-positive endosomes (Tian et al., 2007). Hence, in the context of our data and current literature, we are tempted to speculate on whether DEK1, being the exclusive calpain in plants as well as highly conserved through evolution, might activate NAC proteins resulting in multiple effects on plant development (Kim et al., 2006). The des5 mutation could represent a defective NAC transcription factor, or another component involved in this signalling, affecting aleurone secondary cell wall formation (supported by our unpublished microarraydata showing reduced expression of a number of NAC like genes). Aberrant cell walls would, in turn, affect CR4 localization (and possibly indirectly Cr4 transcript level), resulting in loss of aleurone cell fate. Fate-less aleurone cells would be expected to collapse due to the lack of proper anticlinal cell walls. Final cloning of the des5 mutated gene is expected to improve our understanding of aleurone cell control and overall seed development.
Supplementary data
We have included a Supplementary Table 1, available at JXB online, showing the chi-square calculation for segragation of des5 mutant seeds.
Supplementary Material
Acknowledgments
We thank Professor Odd-Arne Olsen for providing des5 seeds. We thank Peter Sekkelsten for conscientious plant care, and Elin Ørmen and Trygve Krekling for excellent technical microscopy assistance. The transmission electron microscopy was performed at the Molecular Imaging Center (FUGE, Norwegian Research Council), University of Bergen. The qRT-PCR analysis was done at the Norwegian Forest and Landscape Institute under the guidance of Dr Carl Gunnar Fossdal, and statistical analysis was carried out by Professor Odd-Arne Rognli at the Norwegian University of Life Sciences. We thank CUGI for providing the BAC filters and useful help in clone identification. This study was supported by grant 159031/I20 from the Norwegian Research Council.
Glossary
Abbreviations
- Cr4
Crinkly4
- Dek1
Defective kernel1
- des5
defective seed5
- NAC
NAM/ATAF/CUC
- qRT-PCR
real-time quantitative RT-PCR
- Sal1
Supernumerary aleurone layer1
References
- Aalen RB, Opsahl-Ferstad H-G, Linnestad C, Olsen O-A. Transcripts encoding an oleosin and a dormancy-related protein are present in both the aleurone layer and the embryo of developing barley (Hordeum vulgare L.) seeds. The Plant Journal. 1994;5:385–396. doi: 10.1111/j.1365-313x.1994.00385.x. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becraft PW, Asuncion-Crabb Y. Positional cues specify and maintain aleurone cell fate in maize endosperm development. Development. 2000;127:4039–4048. doi: 10.1242/dev.127.18.4039. [DOI] [PubMed] [Google Scholar]
- Becraft PW, Brown RC, Lemmon BE, Olsen O-A, Opsahl-Ferstad H-G. Endosperm development. In: Bhojwani SS, editor. Current trends in the embryology of angiosperms. Dordrecht, The Nederlands: Kluwer Academic Publishers; 2001a. pp. 353–374. [Google Scholar]
- Becraft PW, Kang SH, Suh SG. The maize CRINKLY4 receptor kinase controls a cell-autonomous differentiation response. Plant Physiology. 2001b;127:486–496. [PMC free article] [PubMed] [Google Scholar]
- Becraft PW, Li K, Dey N, Asuncion-Crabb Y. The maize Dek1 gene functions in embryonic pattern formation and cell fate specification. Development. 2002;129:5217–5225. doi: 10.1242/dev.129.22.5217. [DOI] [PubMed] [Google Scholar]
- Becraft PW, Stinard PS, McCarty DR. CRINKLY 4, A TNFR- like receptor kinase involved in maize epidermal differentiation. Science. 1996;273:1406–1409. doi: 10.1126/science.273.5280.1406. [DOI] [PubMed] [Google Scholar]
- Bosnes M, Harris EL, Aigeltinger L, Olsen O-A. Morphology and ultrastructure of 11 barley shrunken endosperm mutants. Theoretical and Applied Genetics. 1987;74:177–187. doi: 10.1007/BF00289966. [DOI] [PubMed] [Google Scholar]
- Bosnes M, Weideman F, Olsen OA. Endosperm differentiation in barley wild-type and sex mutants. The Plant Journal. 1992;2:661–674. [Google Scholar]
- Brenner S, Johnson M, Bridgham J, et al. Gene expression analysis by massive parallel signature sequencing (MPPS) on microbeads arrays. Nature Biotechnology. 2000;18:630–634. doi: 10.1038/76469. [DOI] [PubMed] [Google Scholar]
- Brown RC, Lemmon BE, Nguyen H, Olsen O-A. Development of endosperm in Arabidopsis thaliana. Sexual Plant Reproduction. 1999;12:32–42. [Google Scholar]
- Brown RC, Lemmon BE, Olsen O-A. Endosperm development in barley: microtubule involvement in the morphogenetic pathway. The Plant Cell. 1994;6:1241–1252. doi: 10.1105/tpc.6.9.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RC, Lemmon BE, Olsen O-A. Development of the endosperm in rice (Oryza sativa L.) cellularization. Journal of Plant Research. 1996;109:301–313. [Google Scholar]
- Burton RA, Jenner H, Carrnagis L, et al. Starch granule initiation and growth are altered in barley mutants that lack isoamylase activity. The Plant Journal. 2002;31:97–112. doi: 10.1046/j.1365-313x.2002.01339.x. [DOI] [PubMed] [Google Scholar]
- Cao X, Li K, Suh SG, Guo T, Becraft PW. Molecular analysis of the CRINKLY4 gene family in Arabidopsis thaliana. Planta. 2005;220:645–657. doi: 10.1007/s00425-004-1378-3. [DOI] [PubMed] [Google Scholar]
- del Cerro M, Cogen J, del Cerro C. Stevenel's Blue, an excellent stain for optical microscopical study of plastic embedded tissues. Microscopica Acta. 1980;83:117–121. [PubMed] [Google Scholar]
- Johnson KL, Degnan KA, Ross Walker J, Ingram GC. AtDEK1 is essential for specification of embryonic epidermal cell fate. The Plant Journal. 2005;44:114–127. doi: 10.1111/j.1365-313X.2005.02514.x. [DOI] [PubMed] [Google Scholar]
- Kim YS, Kim SG, Park JE, Park HY, Lim MH, Chua NH, Park CM. A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. The Plant Cell. 2006;18:3132–3144. doi: 10.1105/tpc.106.043018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lid SE, Al RH, Krekling T, Meeley RB, Ranch J, Opsahl-Ferstad H-G, Olsen O-A. The maize disorganized aleurone layer 1 and 2 (dil1, dil2) mutants lack control of the mitotic division plane in the aleurone layer of developing endosperm. Planta. 2004;218:370–378. doi: 10.1007/s00425-003-1116-2. [DOI] [PubMed] [Google Scholar]
- Lid SE, Gruis D, Jung R, Lorentzen JA, Ananiev E, Chamberlin M, Niu X, Meeley R, Nichols S, Olsen O-A. The defective kernel1 (dek1) gene required for aleurone cell development in the endosperm of maize grains encodes a membrane protein of the calpain gene superfamily. Proceedings of the National Academy of Sciences, USA. 2002;99:5460–5465. doi: 10.1073/pnas.042098799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lid SE, Olsen L, Nestestog R, Aukerman M, Brown RC, Lemmon B, Mucha M, Opsahl-Sorteberg H-G, Olsen O-A. Mutation in the Arabidopsis thaliana DEK1 calpain gene perturbs endosperm and embryo development while over-expression affects organ development globally. Planta. 2005;221:339–351. doi: 10.1007/s00425-004-1448-6. [DOI] [PubMed] [Google Scholar]
- Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. The NAC Transcription Factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. The Plant Cell. 2005;17:2993–3006. doi: 10.1105/tpc.105.036004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H, Brown RC, Lemmon BE. Pattern of cytoskeletal organization reflects distinct developmental domains in endosperm of Coronopus didymus (Brassicsceae) International Journal of Plant Science. 2001;162:1–14. [Google Scholar]
- Nielsen ME, Lok F, Nielsen HB. Distinct developmental defence activations in barley embryos identified by transcriptome profiling. Plant Molecur Biology. 2006;61:589–601. doi: 10.1007/s11103-006-0034-7. [DOI] [PubMed] [Google Scholar]
- Olsen O-A. Nuclear endosperm development in cereals and Arabidopsis thaliana. The Plant Cell. 2004;16:214–227. doi: 10.1105/tpc.017111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen O-A, Brown RC, Lemmon BE. Pattern and process of wall formation of developing endosperm. BioEssays. 1995;17:803–812. [Google Scholar]
- Olsen O-A, Brown RC, Lemmon BE. A model for aleurone development. Trends in Plant Science. 1998;3:168–169. [Google Scholar]
- Opsahl-Sorteberg H-G, Divon HH, Nielsen PS, Kalla R, Hammon-Kosach M, Shimamoto K, Kohli A. Identification of a 49-bp fragment of the HvLTP2 promoter directing aleurone cell specific expression. Gene. 2004;341:49–58. doi: 10.1016/j.gene.2004.06.059. [DOI] [PubMed] [Google Scholar]
- Otegui M, Staehelin LA. Cytokinesis in flowering plants, more than one way to divide a cell. Current Opinion in Plant Biology. 2000;3:493–502. doi: 10.1016/s1369-5266(00)00119-9. [DOI] [PubMed] [Google Scholar]
- Page RDM. TREEVIEW: an application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Roth I. Zur morphologishen Deutung des Grasembryos und verwandter Embryotypen. Flora. 1955;144:163–212. [Google Scholar]
- Sato K, Kawashima S. Calpain function in the modulation of signal transduction molecules. Biological Chemistry. 2001;382:743–751. doi: 10.1515/BC.2001.090. [DOI] [PubMed] [Google Scholar]
- Shen B, Li C, Min Z, Meeley RB, Tarczynski MC, Olsen O-A. sal1 determines the number of aleurone cell layers in maize endosperm and encodes a class E vacuolar sorting protein. Proceedings of the National Academy of Sciences, USA. 2003;100:6552–6557. doi: 10.1073/pnas.0732023100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewry PR. Barley: genetics, biochemistry, molecular biology and biotechnology. Biotechnology in agriculture No. 5. Wallingford, UK: CAB International; 1992. [Google Scholar]
- Sreenivasulu N, Usadel B, Winter A, et al. Barley grain maturation and germination: metabolic pathway and regulatory network commonalities and differences highlighted by new MapMan/PageMan profiling tools. Plant Physiology. 2008;146:1738–1758. doi: 10.1104/pp.107.111781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q, Olsen L, Sun B, et al. Subcellular localization and functional domain studies of DEFECTIVE KERNEL 1 in maize and Arabidopsis thaliana suggests a model for aleurone cell fate specification involving CRINKLY 4 and SUPERNUMERARY ALEURONE LAYER 1. The Plant Cell. 2007;19:1–19. doi: 10.1105/tpc.106.048868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SR, Taylor N, Jones L. Mutations of the secondary cell wall. Plant Molecular Biology. 2001;47:209–219. [PubMed] [Google Scholar]
- Wang C, Barry JK, Min Z, Tordsen G, Rao AG, Olsen O-A. The calpain domian of the maize DEK1 protein contains the conserved catalytic triad and functions as a cystein protease. Journal of Biological Chemistry. 2003;278:34467–34474. doi: 10.1074/jbc.M300745200. [DOI] [PubMed] [Google Scholar]
- Williams JM, Duffus CM. The development of endosperm amyloplasts during grain maturation in barley. Journal of the Institute of Brewing. 1977;84:47–50. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.