Abstract
As the primary intracellular calcium sensor, calmodulin (CaM) regulates numerous and diverse proteins. Several mechanisms, including tissue specific expression, localization and sequestration, work in concert to limit the total number of available targets of calmodulin within a cell. While the free energies of binding of calmodulin-binding domains of regulated proteins by CaM have been shown to be highly similar, they result from vastly different enthalpic and entropic contributions. Here, we report the backbone and side-chain methyl dynamics of calcium-activated calmodulin in complex with a peptide corresponding to the CaM binding domain of calmodulin kinase kinase, along with the thermodynamic underpinnings of complex formation. The results show a considerable reduction in side-chain mobility throughout CaM upon binding the CaMKKα peptide which is consistent with the enthalpically driven nature of the binding. Site specific comparison to another kinase-derived peptide complex with similar thermodynamic values, reveals significant differences in dynamics largely localized to the hydrophobic binding sites.
Calmodulin is a highly conserved protein, capable of binding and regulating numerous proteins in response to increased levels of intracellular calcium (1, 2). Calcium-saturated calmodulin (CaM) is the dominant regulatory species though there are several cases where apocalmodulin is found to bind target proteins. Current estimates indicate well over 300 proteins contain a CaM binding motif (3). Critical among these are kinases which subsequently regulate gene transcription, muscle contraction, transport, and other cellular processes (4). Calcium/calmodulin-dependent protein kinase kinase (CaMKK) acts to enhance the activity of downstream CaM-dependent kinases (types I and IV) by phosphorylation of a single Thr in the so-called activation loop (5–7). CaMKI has recently been shown to function within the ERK signaling pathway (8) and CaMKI and CaMKIV differentially activate the transcription factors CREB, CREM, and ATF-1 (9–11). Two isoforms of CaMKK are known (α and β) each with abundant expression in neuronal, although CaMKKβ also has low level expression in the spleen, testis, and thymus (12, 13). Both CaMKK isoforms can phosphorylate CaMKI and CaMKIV in vitro; however, the remarkably similar neuronal expression patterns of CaMKKβ and CaMKIV suggest CaMKK isoforms may have, at least partially, non-overlapping target specificity and function (14, 15). This is exemplified by the requirement of CaMKKα for contextual long-term memory formation in the hippocampus while CaMKKβ is required for spatial long-term memory (16, 17).
The canonical CaM binding motif is approximately 20 amino acids and consists of two hydrophobic “anchors” bridged by an amphiphilic helix (Figure 1A). Typically, the amino-terminal anchor is adjacent to three or four basic residues with either 8 or 12 poorly conserved residues separating the two anchors in the primary sequence (18). Other than maintaining a hydrophobic character, the anchors are not well conserved. The NMR (PDB: 1CKK) and crystal (PDB: 1IQ5) structures of CaM in complex with peptides derived from rat CaMKKα and nematode CaMKK, respectively, revealed a novel variation of the canonical CaM binding motif (19, 20) (Figure 1B). The CaMKK isoforms are the only known CaM binding domains with 14 residues between the critical hydrophobic residues. Additionally, the basic residues are found near the carboxy-terminal anchor. In complex with CaM, the CaMKK peptides form a β-turn structure in order to accommodate the increased number of intervening residues. Finally, the orientation of the peptide relative to CaM is reversed from what is observed in most other CaM complexes, such that the amino-terminal hydrophobic anchor interacts with the amino-terminal domain of CaM (Figure 1B). Despite these differences, the global structure of CaM bound to CaMKK is highly similar to that of CaM bound to the smooth muscle myosin light chain kinase (smMLCKp) peptide (backbone 2.13Å rmsd) (21) and to the CaM kinase II peptide (backbone 1.43Å rmsd) (22), with the largest discrepancies occurring in the domain linker. The remarkable ability of CaM to accommodate diverse sequences appears to originate in the flexibility of the domain linker to independently position the globular domains and the abundance of long aliphatic side-chains, including four methionine residues per domain, which form the binding pockets for the anchor residues.
Figure 1.
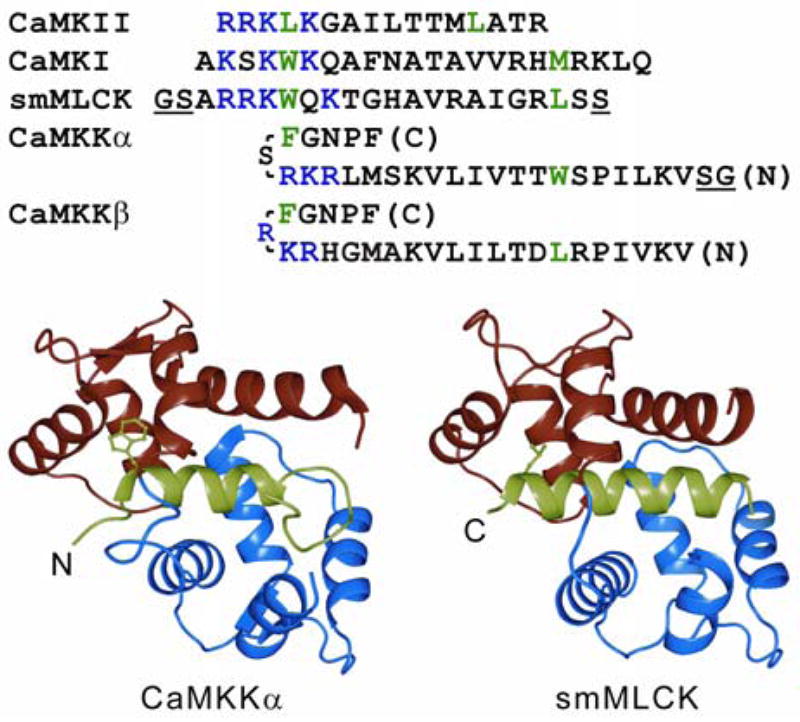
CaM binding domain sequence and structure alignment. (A) The primary sequences of CaM binding domains are shown with hydrophobic anchors in green and the cluster of positively charged residues in blue. Note the inverted direction of CaMKK isoforms. The underlined GS pairs are non-native residues present in the peptides used for our analysis; additionally, the C-terminal serine of smMLCK was omitted. (B) The NMR-based structure of CaM:CaMKKα is shown on the left and the crystal structure of CaM:smMLCK on the right. The amino and carboxy CaM EF-hand domains are colored red and blue, respectively, with the peptides in green. Key hydrophobic anchors are displayed in ball and stick representation. Ribbon diagrams were created with PyMol (http://www.pymol.org).
The adaptability of CaM to bind diverse sequences is juxtaposed by highly similar binding affinities for such targets. Numerous competitive binding and calorimetric studies have shown the affinity of many CaM binding peptides approach −50 kJ/mol. Furthermore, calorimetry has clearly established that binding of these high affinity target peptides can be enthalpically or entropically driven (23). This impressive variability of system thermodynamic parameters presents the possibility that changes induced by binding a target peptide are manifested in both calmodulin structure (enthalpy) and dynamics (entropy). We have previously shown that binding of a peptide corresponding to the calmodulin binding domain of smMLCK causes a striking change in CaM methyl bearing side-chain dynamics with relatively little change in backbone amide N-H dynamics (24). More importantly, a quantitative interpretation of the change in dynamics suggests that the change in the residual entropy of calmodulin comprises a significant fraction of the total change in entropy due to binding of the domain (24). This raises the important question of whether the conformational entropy of calmodulin influences target selection and/or duration of CaM mediated signaling.
Materials and Methods
Expression and purification of CaMKKαp
De novo synthesis of the CaMKKαp template was carried out by extending two partially overlapping oligonucleotides, corresponding to the CaMKKα amino acid sequence: VKLIPSWTTVILVKSMLRKRSFGNPF, under standard PCR conditions. This template was PCR amplified with primers containing NdeI and BamHI restriction sites which were subsequently used to insert the CaMKKαp DNA into a modified pET32 vector. Transformed BL21(DE3) E. coli grown in rich media were induced to over-express the thioredoxin fusion protein with 1mM IPTG when the cultures reached an OD600 of ~1. The protein was found exclusively in the insoluble fraction of chemically lysed cells, necessitating extraction with 8M urea. Significant enrichment of the fusion protein was achieved with His·Bind (Novagen) resin under denaturing conditions. In the absence of denaturant, we found the fusion protein was highly soluble below pH 4.5, but precipitated at near neutral pH. Therefore, the eluted protein was acidified with glacial acetic acid and dialyzed extensively against acidified water (pH 3.75) at 4 °C. In order to retain fusion protein solubility for enzymatic digest, it was slowly titrated into excess CaM (6mM CaCl2, 20mM Mops, pH 7.2) at room temperature. After equilibration, the peptide was cleaved with thrombin (Sigma) over a period of 16–24 hrs at 22 °C. The digest was quenched by addition of 20% acetonitrile/0.1%TFA and the peptide was isolated on a C18 reverse phase HPLC column (Vydac) using a linear 20–60% acetonitrile gradient. The mass and purity were confirmed by MALDI-TOF mass spectrometry. The freeze-dried peptide was stored at −20 °C until use. Purification of wild type chick CaM expressed in BL21(DE3) E. coli was accomplished using phenyl sepharose as described previously (25).
Isothermal titration calorimetry
The thermodynamic parameters governing the association of CaMKKαp with CaM were determined using isothermal titration calorimetry. Measurements were performed with a VP-ITC (Microcal) thermostatically regulated at 35 °C. Data was analyzed with Origin(v.5) software. Binary complex formation was achieved by titrating 200 μM CaM into the sample cell containing 40 μM CaMKKαp. In order to match the conditions of the NMR samples, CaM was dialyzed overnight against 1L of NMR buffer (100mM KCl, 6mM CaCl2, 10mM imidazole, pH 6.5, and 0.02% azide). This buffer was then used to re-constitute CaMKKαp. In order to reduce aggregation, the buffer was first acidified to pH 4.5, then adjusted back to 6.5 after addition of the peptide. All solutions were filtered and degassed prior to use and the pH of each solution was verified before each run. Protein concentrations were determined spectrophotometrically.
NMR sample preparation
For resonance assignment experiments, CaM was expressed in M9 media containing 2 g/L 13C-glucose and 0.1% 15N ammonium chloride. Backbone amide relaxation experiments were acquired on CaM isolated from M9 media containing 2 g/L 13C-glucose and 1 g/L 15N ammonium chloride. Side-chain methyl relaxation experiments were collected on CaM expressed in M9 media containing 55% D2O, 2 g/L 13C-glucose and 1 g/L 15N ammonium chloride. All NMR experiments were conducted on samples titrated with CaMKKαp that were allowed to equilibrate with mixing for at least 16 hours upon reaching saturation. At each titration point, as the peptide solution mixed with the buffered CaM, it formed visible aggregates that diminished within seconds as CaM bound CaMKKαp. The time required for the aggregates to disappear increased along the titration, eventually resulting in small amounts of insoluble CaMKKαp as the titration approached 1:1. Complex formation was monitored by changes in the 1H-15N HSQC spectrum. Excess CaMKKαp was not soluble under these conditions and after overnight incubation a minor population (~4%) of unbound CaM persisted. Aggregated peptide was removed by centrifugation prior to the NMR experiments. Samples were stable for several months; however, most data was acquired on samples made within one month.
NMR spectroscopy
All NMR experiments were performed at 35 °C on Varian Inova spectrometers. Sequential backbone and methyl bearing side-chain assignment experiments were performed at 750 MHz. Gradient-enhanced HNCACB and CBCA(CO)NH spectra were sufficient to assign the backbone amides (26–28). The methyl optimized (H)CCH3-TOCSY was used to correlate 1H and 13C resonances of methyl groups with the carbon resonances obtained in the backbone experiments (29). Methionine epsilon methyl resonances were assigned using a two dimensional HMBC experiment (30) with ambiguous assignments confirmed with information from a 13C-edited 1H-1H NOESY. Chemical shift data from the BMRB (code:4270) was used to guide the assignment process. NMR data were processed with nmrPipe (31); Sparky was used for visualization and assignment bookkeeping (32).
The 15N relaxation parameters of backbone amide bond vectors were measured with two dimensional HSQC based experiments (33). Nine time points spanning 23 to 984ms for T1 and 7.8 to 124ms for T2 were acquired with an additional three duplicate time points in order to assess uncertainty in measured peak intensities. For NOE uncertainties, the RMS noise of a spectral region without a peak was used. All relaxation rates were measured at 500 and 750 MHz on the same sample. A total of 512 (1H) by 160 (15N) complex data points were acquired for each relaxation time point.
Side-chain methyl dynamics were obtained from longitudinal (IZCZIZ and IZCZ) and transverse (IZCZDY) relaxation rates of 13CH2D isotopomers (34). Each rate was determined by nine time points between 2.6 and 70ms (IZCZIZ), 0.7 and 20ms (IZCZDY), and 2.6 and 80ms (IZCZ). Deuterium relaxation experiments were performed at 500 and 750 MHz on a single sample which had a CaM concentration matched to the sample used to record 15N relaxation data. Each two dimensional 1H-13C HSQC acquired with 512 by 84 complex points.
Relaxation data calculations
Relaxation rates were obtained from maximum peak intensities fitted to single exponential decays using the Levenberg-Maquardt routine. The {1H}-15N NOE was calculated as the peak intensity ratio from saturated and non-saturated experiments. Determination of dynamic parameters was accomplished by fitting the relaxation rates to the simple model free spectral density function (35) with an in-house exhaustive grid search algorithm (36). The global rotational correlation time, τm, was first determined using 15N relaxation data of well-ordered residues assuming an isotropic rotational diffusion. Residues were excluded if the NOE value was less than 0.65 or the T1T2 product exceeded one standard deviation (37). Subsequently, squared generalized order parameters and internal correlation times, τe, were fitted for all spectrally resolved residues. Methyl generalized order parameters, , are reported after accounting for motion about the methyl symmetry axis, O2/0.111. The fitting errors were determined with 150 Monte Carlo simulations.
Results
NMR relaxation analysis offers a unique experimental approach to obtain site specific information describing the dynamic behavior of macromolecules in solution. In addition, owing to the implicit “counting of states” inherent in a dynamical description one has, in principle, access to the residual conformational entropy of proteins (38, 39). Using calmodulin as a model system, we have set out to assess changes in dynamics (entropy) among different CaM complexes and examine the relationship to changes in total system entropy. We have previously reported the subnanosecond backbone and side-chain dynamics of CaM and the CaM:smMLCKp complex (24). We now build upon this information using the CaM binding domain of CaMKKα.
Isothermal Titration Calorimetry
The thermodynamics of the binding of CaMKKαp by CaM has been examined by isothermal titration calorimetry (Figure 2). CaMKKαp binds with high affinity to CaM; the KD was determined to be approximately 4nM (ΔG= −49.8 kJ/mol) at 35 °C. The heat of binding was found to be −140 kJ/mol and the corresponding change in system entropy was +90 kJ/mol at 35 °C. Thus, the binding is enthalphically driven. Slight deviations from perfect sigmoidal curvature are noted just ahead of and after the transition. This feature was reproduced in several experiments and it is most likely an effect of reversible, microscopic CaMKKαp aggregation. At near neutral pH, even under the dilute conditions for ITC, CaMKKαp is in equilibrium between monomeric and aggregated forms. Conversion of aggregated CaMKKαp to the soluble peptide is apparently an endothermic process.
Figure 2.
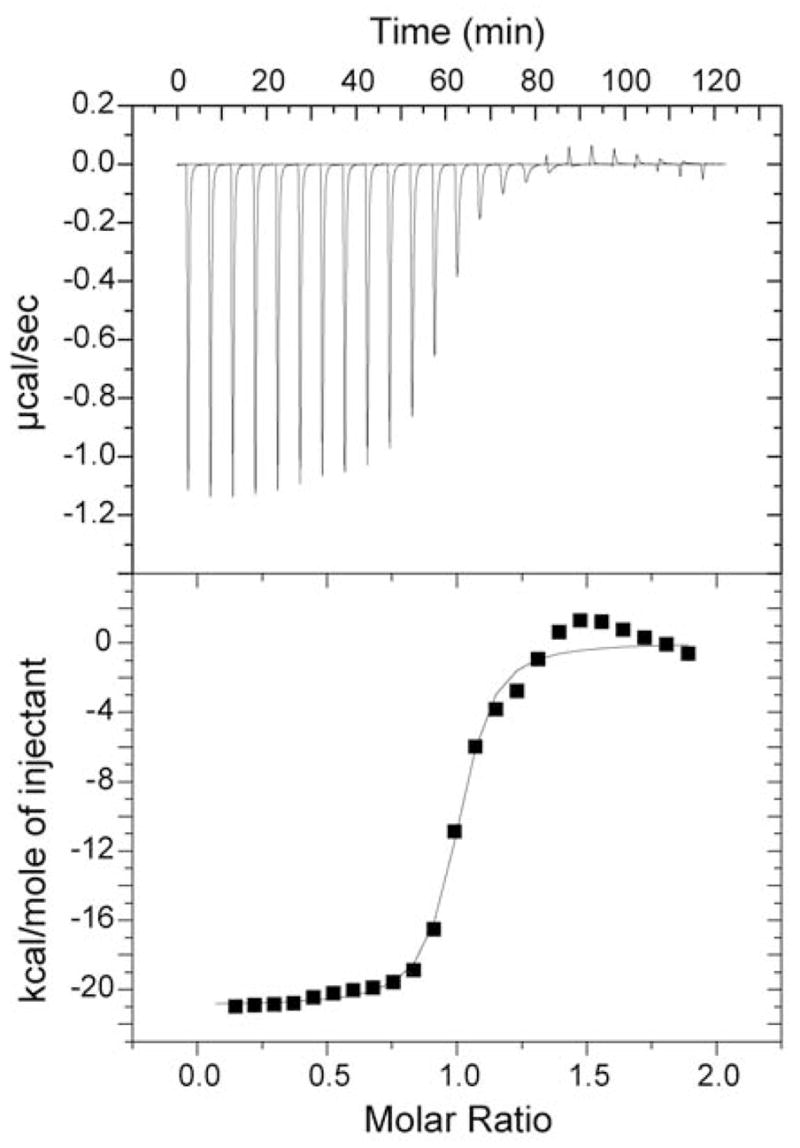
Binding isotherm for titrating CaM into CaMKKαp. Panel A shows the raw heats of titration of CaM into a dilute solution of CaMMαp. Panel B shows the best fit of integrated data to the single binding site model. Best fit values of KD, ΔH (and –TΔS) are given in the text.
Resonance assignments
Sequential assignment of backbone amides and side-chain methyls was necessary, in part, because of the slightly different conditions (+5 °C, −0.2 pH) used here compared to the conditions of the previous structure determination (19). Amides are generally more sensitive to changes in temperature and pH and provide a reasonable measure for the magnitude of chemical shift changes caused by the different sample conditions. The average chemical shift displacement was calculated as where ΔH(N) is the 1H(15N) chemical shift difference between resonances in the two conditions and γN(H) is the gyromagnetic ratio of the indicated nucleus. Approximately 20 residues have a chemical shift change in excess of 0.05; however, only two residues, Thr44 and Ala73, are greater than 0.1. The average chemical shift displacement, Δ̄cs, is 0.03428. Methyl proton and carbon resonances are equally similar. This strongly indicates the structure of the complex is the same in the two conditions.
Backbone dynamics of the CaM:CaMKKαp complex
The primary objective of this study was to determine the sub-nanosecond internal dynamics of CaM in complex with CaMKKαp. The macromolecular correlation time, τm, was determined with T1 and T2 data acquired at 500 and 750 MHz from 42 helical residues, assuming isotropic rotational diffusion. The calculated value of 8.78ns is in excellent agreement with correlation times calculated with HYDROPRO (40) using the crystal structure (20) of CaM:CaMKKp (8.79 ns) and the average NMR model (19) of CaM:CaMKKαp (8.87 ns) after removal of highly dynamic residues. Measurement at two field strengths produced an over-determined data set resulting in high precision of the dynamic parameters. The dynamics of amide bond vectors along the CaM backbone are summarized in Figure 3. The average order parameter of 134 spectrally resolved amide groups is 0.93 (σ = 0.077). In accord with many other protein relaxation studies, the backbone N-H dynamics are generally unrevealing. Aside from the amino and carboxy termini, five residues appear unusually dynamic. A57 and I130 occupy the second position of the second Ca2+ binding loop in their respective EF-hand domains. Low order parameters for these CaM residues and equivalent residues in other calcium binding proteins have been reported previously (41, 42). T79 and S81 are in the domain linker region, and K115 is within a generally flexible region identified in all CaM relaxation studies to date. Indeed, the overall pattern of values is generally consistent between CaM and CaM:CaMKKαp. For 79 amides measured in both states, the average (CaM:CaMKK - CaM) is only 0.0382 (0.053 absolute) with none exceeding 0.225.
Figure 3.
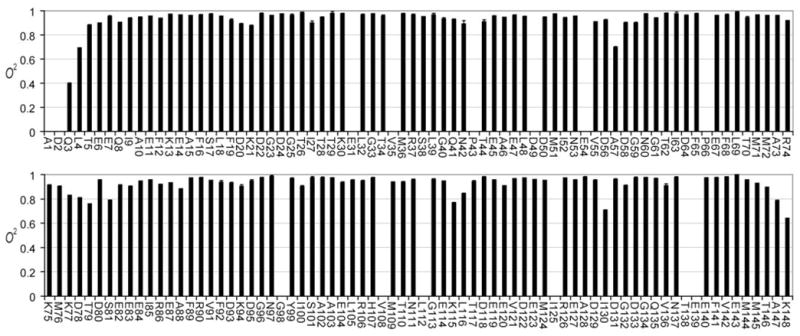
NMR derived backbone N-H bond vector model-free squared generalized order parameters ( ) for CaM in complex with CaMKKap determined with relaxation data obtained at 500 and 750 MHz (1H). The error bars, defined by Monte Carlo simulations, are too small to be resolved from the measured value in most cases. Non-proline residues with no entry were not determined due to spectral overlap.
Side-chain dynamics of the CaM:CaMKKαp complex
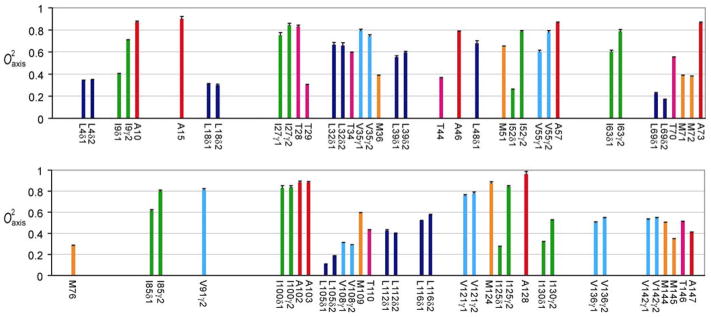
Deuterium relaxation studies of methyl-bearing side-chains indicate that free, calcium-saturated calmodulin itself is an unusually dynamic protein and is characterized by a broad, non-uniform multi-modal distribution of the amplitudes of motion (24). Two-dimensional sampling of deuterium relaxation afforded excellent resolution of methyl 1H-13C cross peaks and allowed relaxation at 71 sites in CaM complexed to the CaMKKαp domain to be quantitated (Figure 4). Unlike amide N-H bond vectors, the symmetry axes of methyl groups in complexed calmodulin are heterogeneously dynamic, even within a specific residue type, with values corresponding to essentially fixed (~1) to nearly isotropic (<0.2) motion (Figure 5). Moreover, methyl-bearing side-chain dynamics are greatly affected by binding of CaMKKαp. Using CaM as a reference state, the average change in methyl order parameter is 0.132 (0.171 absolute) for 47 sites. A few individual changes are quite striking, ranging close to +0.7. It is also noteworthy that a small number of sites (eleven) exhibit an increase in the amplitude of motion (decrease in ) upon binding the CaMKKαp domain. Figure 6A shows the nature of the interface between CaM and the bound CaMKKαp domain. Though nearly completely buried, methyl-bearing side chains at the interface (< 2.5 Å) are heterogeneously dynamic. This response of calmodulin to the binding of the two domains is relatively short range, with the majority of significant changes in occurring within 4 Å of the interface (Figure 6B). In addition, residues at the interfaces in the CaM:smMLCKp and CaM:CaMKKαp complexes respond quite differently to binding (see below).
Figure 4.
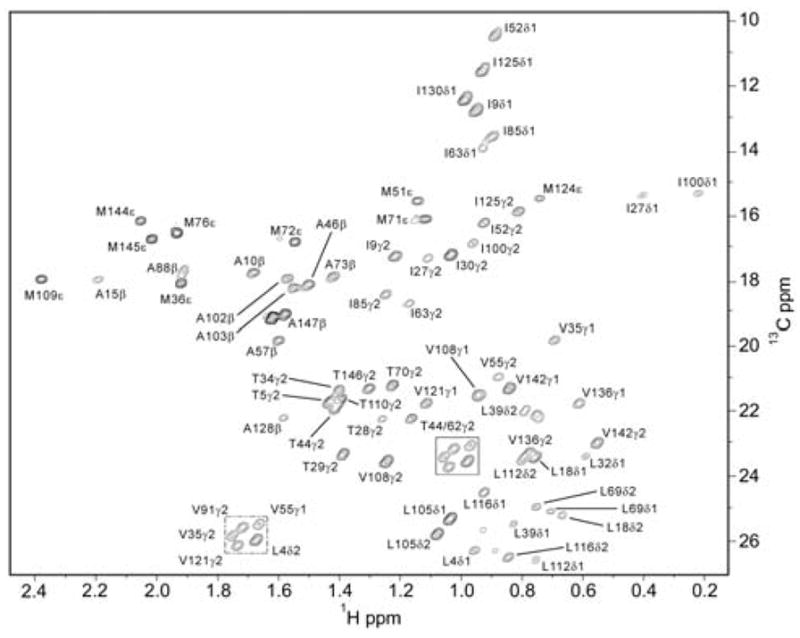
The 1H-13C HSQC spectrum of the CaM:CaMKKap complex. Shown is a contour plot of first time point (7ms) from the IzCzDz experiment acquired at 750 MHz. Seventy-one of 78 methyl groups in calmodulin were adequately resolved for analysis.
Figure 5.
NMR derived side-chain model-free squared generalized order parameters for the symmetry axis ( ) of methyl groups of CaM in complex with CaMKKαp determined with relaxation data obtained at 500 and 750 MHz (1H). Values are color coded by residue.
Figure 6.
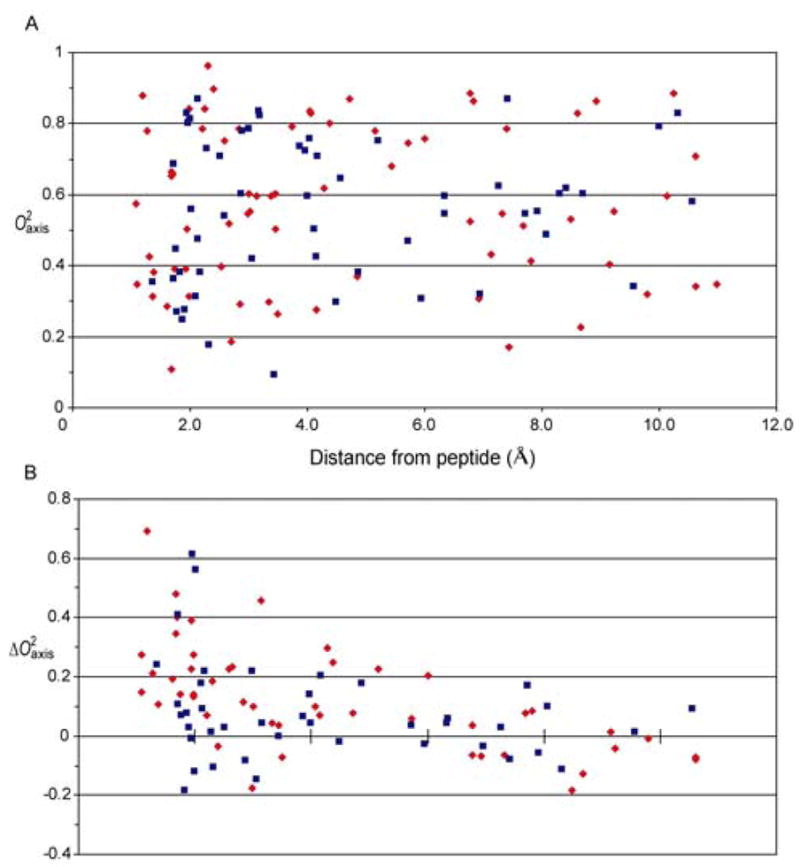
Nature of the interfaces between CaM and CaMKKαp or smMLCKp. Panel A shows the absence of a correlation between distance to the interface between CaM and bound CaMKKαp (red symbols) or smMLCKp (blue symbols) domains and the parameters of methyl groups of CaM. Note the interface is heterogeneously dynamic for both complexes. Panel B shows the correlation between distance to the interface between CaM and bound CaMKKαp (red symbols) or smMLCKp (blue symbols) domains and the parameters (complexed CaM minus free CaM) of methyl groups of CaM. The data for the CaM:smMLCKp complex is from (24).
Binding of the smMLCKp domain to CaM results in a significant redistribution of the fast side chain dynamics in calmodulin and results in a distribution that is remarkable for its distinct clustering into three apparent classes of motion (24). This is also the case for the CaM:CaMKKαp complex where parameters are distributed in the trimodal manner (Figure 7). Although the distinctive grouping of order parameters seen in the calmodulin complexes is often obscured in other proteins(43), the motional origin of these classes is clear. In the case of calmodulin, two fundamental types of motion occurring on the sub-ns time scale are involved: motion within a rotamer well and motion between rotamer wells of side chain torsion angles (44). It has been shown that the class of motion centered on a squared generalized order parameter value of ~0.35 generally involves a contribution from rotameric interconversion on the nanosecond or faster time scale as it leads to a significant averaging of scalar coupling (J) constants(44). Recent experimental (45) and theoretical simulations (46) suggests this to be general. We will term this group the “J-class”. The distribution of motion at the other extreme is centered on a squared generalized order parameter of ~0.85, which represents highly restricted motion within a rotamer well and is reminiscent of the relative rigidity of the polypeptide backbone. We shall therefore term this class the “ω-class”. The class of motion centered on a squared generalized order parameter of ~0.6 involves little detectable rotamer interconversion and therefore reflects both variation of the amplitude of motion within a single rotamer well and/or the superposition of motion about connected torsion angles. The distribution within this class is generally expected to reflect variance of amplitude of motion within rotamer wells and we therefore term this class the “α-class”. The distributions of the J-, α- and ω-classes in the CaM:CaMKKap complex are satisfactorily described by a sum of three Gaussians (Figure 7). Randomization tests (47) indicate that this distribution is statistically significant and more appropriate than a random distribution or one described by one or two Gaussians.
Figure 7.
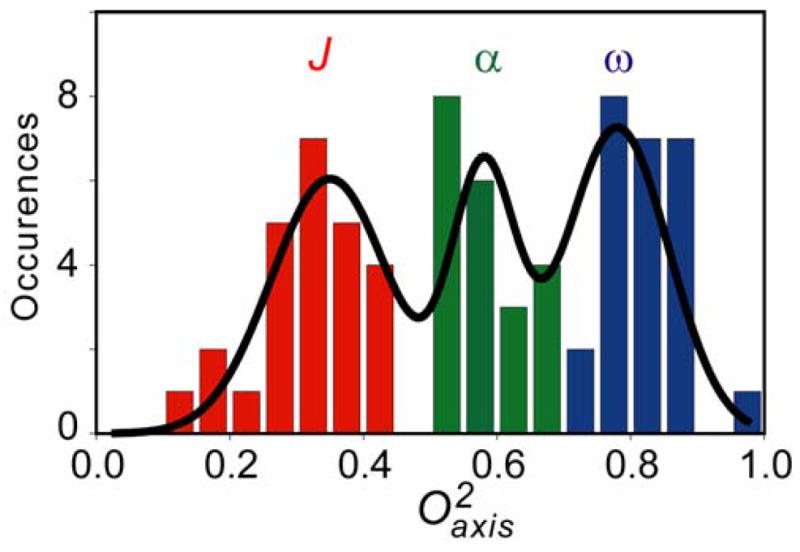
Histogram of the distribution of parameters of methyl groups of CaM in complex with CaMKKαp.
An interesting question is how the various classes of motion in CaM respond to the binding of the CaMKKαp domain. Ten of the eleven methyl groups of CaM whose order parameters decrease upon binding of the CaMKKαp domain remain within the same class of motion (Figure 8A). In contrast, only 15 of the 36 methyl groups whose order parameters increase upon binding the CaMKKαp domain remain within the same class of motion. Ten methyls that are in the J-class in free CaM move to the α-class in the CaM:CaMKKαp complex. One methyl (M124) moves from the J-class to the ω-class (Figure 8A). Ten methyls that are in the α-class in free CaM move to the ω-class in the CaM:CaMKKαp complex. Interestingly, the CaM:smMLCKp complex shows are more limited response (Figure 8B). In this case, the vast majority of methyl groups remain within the class of motion that they occupy in free CaM. Only seven of the 42 methyl groups measured move between adjacent classes and only one, again M124, moves from the J-class to the ω-class (Figure 8B). Above all, these results show the dynamic responses of CaM to binding the smMLCKp and CaMKKαp domains are quite different when examined in detail (Figure 8C).
Figure 8.
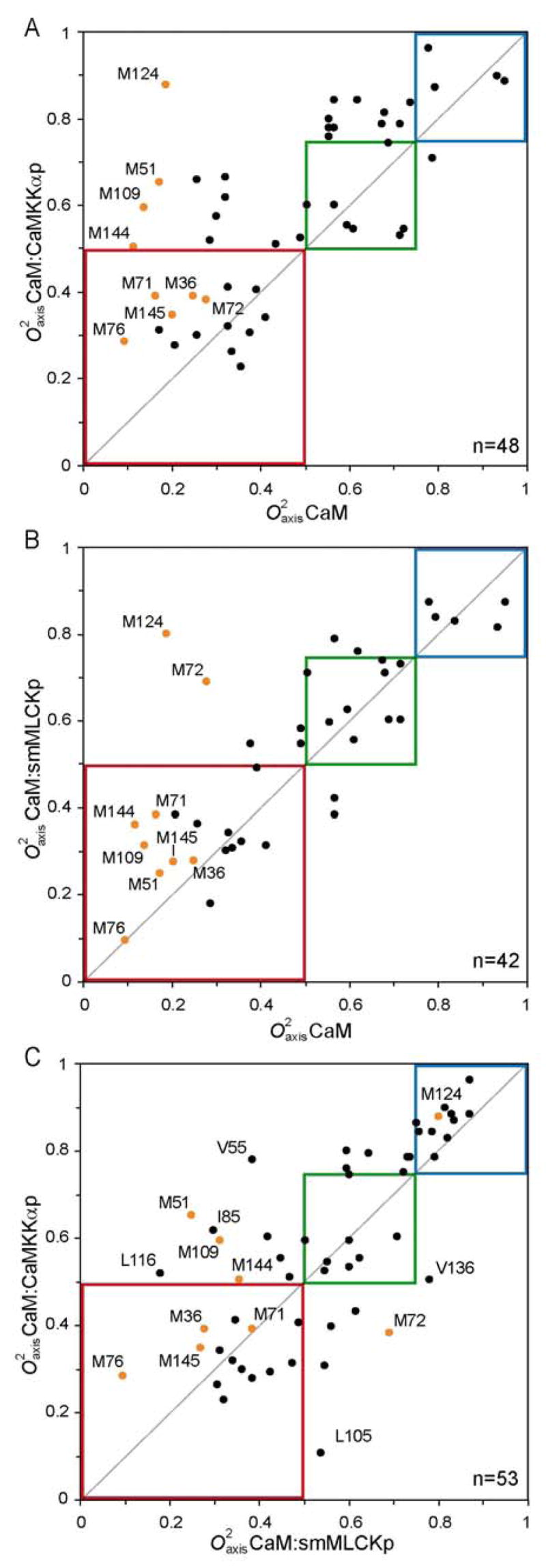
Variation of methyl dynamics between free CaM and its complexes with the CaMKKαp and smMLCKp domains. Panels A and B relate the complexes to CaM and panel C correlates the two complexes with each other. Data points in orange correspond to methionine residues and the boxes indicate the three classes of dynamics: ω-class (blue), α-class (green) and J-class (red).
Discussion
Molecular recognition is a complex process composed of several different, often competing, phenomena. In many cases, the free energy of association can be readily measured and decomposed into enthalpic and entropic components using isothermal titration calorimetry. Understanding individual contributions at the atomic level has proven more difficult. In principle, NMR relaxation phenomena can provide access to the protein contribution to binding entropy by quantifying changes in the motions that express residual protein entropy (48). Such an interpretation is model-dependent (38, 39, 49). In an effort to begin to understand how changes in the conformational entropy of proteins can influence high affinity protein-protein interactions, we have undertaken a detailed analysis of the dynamical consequences of calmodulin’s interactions with the calmodulin-binding domains of proteins that it regulates. The heterogeneity of the dynamic response of CaM to the binding of a target domain strongly suggests that the protein is responding as a thermodynamic “particle” and lacks a uniform response with conformational entropy being gathered in an apparently heterogeneous or effectively random fashion. This is most exemplified by the highly varied response of individual side chains even though the binding of both domains results in a general increase of rigidity (Figure 8C). Overall, CaM is slightly more rigid in complex with the CaMKKαp domain than it is in the complex with the smMLCKp domain (average ; n=47). This is consistent with the trend in the change in system entropy upon binding (90 kJ/mol versus 77.5 kJ/mol at 35 °C, respectively) and begins to suggest that the conformational entropy of CaM, inferred from the mobility changes, is acting in concert with the overall change in entropy due to binding of the target domain. This remains to be firmly established by examination of other complexes with varying thermodynamic character for binding. Such studies are on-going.
Here we show that binding of the CaMKKαp domain induces a large scale change in side-chain dynamics involving residues throughout the CaM sequence, an effect observed previously in the CaM:smMLCKp complex. In what follows, we compare the CaM:CaMKKαp backbone and side-chain motional properties with those of the CaM:smMLCKp complex to assess the variability of dynamics among CaM complexes. Seventy eight of 146 backbone amide and 56 of 78 side-chain methyl order parameters were determined for both complexes; including all nine methionine residues lining the interface between CaM and the target domain. This analysis shows potential for revealing binding hot spots of a dynamic (entropic) nature that would be difficult, at best, to detect by structural analysis alone.
No direct backbone amide interactions between calmodulin and target domains are found in the known canonical structures. For the 98 sites measured in both CaM:smMLCKp and the CaM: CaMKKαp complexes, the average change in amide NH order parameter is 0.0106. The average absolute change is 0.0273. This suggests that the backbone dynamics of CaM are similar in CaMKKαp and smMLCKp complexes. One notable exception is a stretch of residues, Lys115-Asp118, located in the carboxy-terminal EF-hand domain with an apparently more rigid backbone (mean ). This area is the site of several charge-charge interactions specific to CaM:CaMKKαp as well as interactions with L17 of the CaMKKαp domain, a residue with no similar counterpart in smMLCKp. As mentioned previously, this is a relatively flexible region of CaM, free and in complex with the smMLCKp domain. Thus motion is apparently restricted by the local spatial restriction specific to CaMKKαp binding. Methyl groups, on the other hand, are featured prominently at the interface between calmodulin and target domains and offer great potential for insight into the dynamical consequences of these high affinity interactions. Many of the generalized order parameters for methyl groups in direct contact or participating in the formation of the hydrophobic binding pockets are significantly altered, some of which we describe in detail below. Subsequently, all values are given as (CaM:CaMKKαp - CaM:smMLCKp), unless indicated otherwise.
Amino-terminal binding EF-hand domain
Sequence alignment of CaM binding domains demonstrates a strong preference for a leucine residue to bind in the amino-terminal EF-hand domain, although methionine and phenylalanine are also sometimes found. CaMKKα is unique in the use of tryptophan in this location; even the CaMKKβ isoform utilizes a leucine residue. The apparent effect of this substitution on the local dynamics is striking (Figure 9). L18(MLCK) contacts the methyl groups of L32, V55, and M71 of CaM. The larger W7(CaMKKαp) contacts these residues as well as I27, M51, and I63. Its side-chain is situated between M51 and V55 and forms close contacts with these residues. Accordingly, the methyl parameters are increased by 0.382 (M51ε), 0.192 (V55γ1), and 0.4 (V55γ2). The methyl groups for Leu32 are not spectroscopically resolved in the CaM:smMLCKp complex; however, the corresponding parameters in the CaM:CaMKKαp complex are approximately 0.25 greater than the average Leu order parameter in the CaM:smMLCKp complex (n=7). I27 and I63 form part of the hydrophobic binding pocket with side-chains oriented parallel to the W7(CaMKKαp) ring structure. parameters for both are similar in the two complexes. Thus, it would appear that the dynamic character of these residues is determined by surrounding CaM residues rather than by the target domain. Interestingly, the side-chain of I52 is in close proximity, but oriented away from the binding pocket, and has very similar order parameter values in both complexes ( and 0.055). Overall, we find that binding of tryptophan decreases the amplitude of motion in the N-terminal binding domain of CaM as compared to binding a leucine. The indole N-H of W7(CaMKKαp) forms a hydrogen bond with the carbonyl of M51 of CaM and the increased number of van der Waals interactions may also compensate for the apparent decrease in local conformational entropy.
Figure 9.
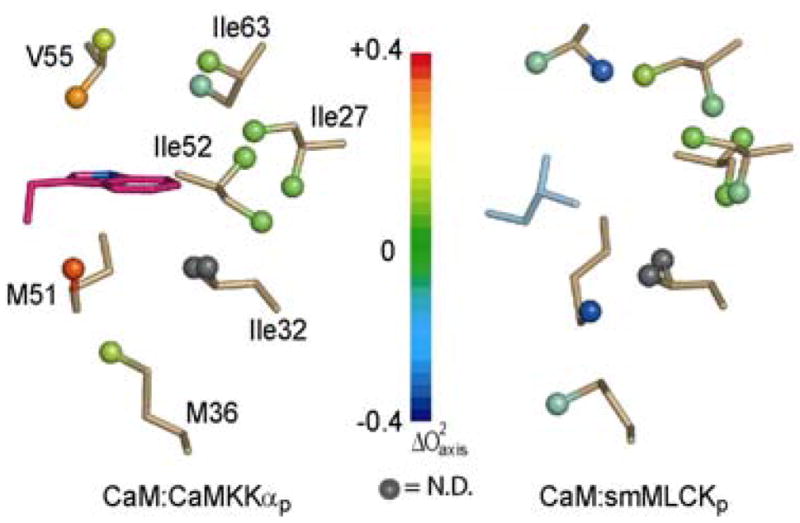
The dynamic effect of different hydrophobic anchors in the amino-terminal EF-hand domain. The two structures were globally superimposed using CaM backbone atoms (2.13Å rmsd). Methyl bearing residues in close contact with the anchor residues are shown in gold sticks with color coded balls for methyl groups. The spatially equivalent residues from each peptide are shown in magenta for W7(CaMKKαp) and cyan for L813(smMLCKp). Color coding reflects changes in values.
Carboxy-terminal EF-hand domain
The opposite situation exists in the carboxy-terminal binding pocket. Here, the preference is for a tryptophan residue; however, CaMKKα/β both utilize phenylalanine. Examination of the structures shows a methionine side-chain from the CaMKK peptide is positioned within the binding pocket, presumably compensating for the smaller side-chain (Figure 10). The dominant effect of the Trp→Phe substitution in this binding pocket is felt by L105δ2 ( ). V136γ1 also experiences a strong decrease (−0.276) in order parameter. The adjacent methyl stereoisomers (L105δ1 and V136γ2) are unfortunately not spectrally resolved. In addition to the residue substitution, there are also structural features which appear to modulate the dynamics of hydrophobic residues in the surrounding area. For example, L116δ1 and V121γ1 experience 0.34 and 0.16 decreases in side-chain mobility, respectively, although neither makes any strong contact with the CaMKKα peptide. The magnitude of change in L116 is due in large part to the fact that it is highly mobile in the CaM:smMLCKp complex, with an nearly 0.1 less than the CaM value. Both of these residues, which are 4.5Å apart, are buried by the β-turn (residues18–22) of CaMKKαp, a feature not found in the smMLCKp domain. Moreover, this sub-structure forms electrostatic interactions with CaM, presumably increasing the overall rigidity of this region. We note that this conclusion is supported by the backbone 15N dynamics. That the effects of the Trp→Phe substitution are not as localized as the N-terminal domain (Leu→Trp) is to some degree a consequence of these charge-charge interactions as well as the β-turn CaMKKαp adopts to properly localize the phenylalanine side-chain. Therefore, a larger number of CaMKKαp residues interact with calmodulin compared to smMLCKp.
Figure 10.
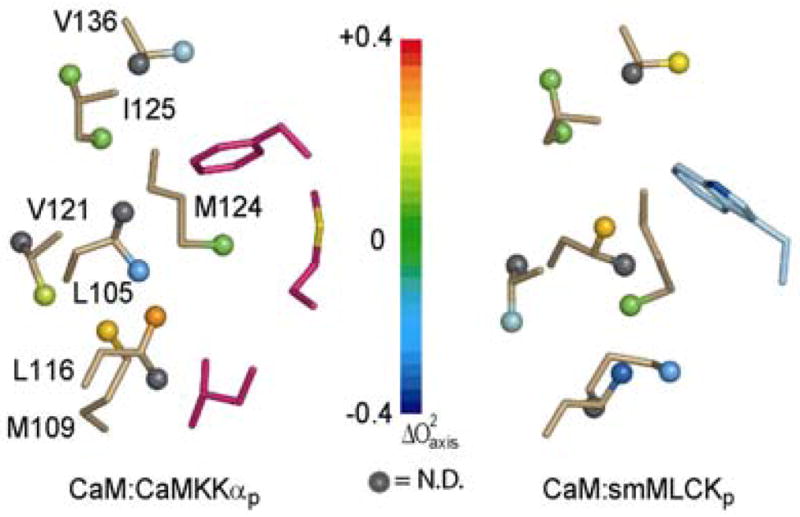
The dynamic effect of different hydrophobic anchors in the carboxy-terminal EF-hand domain. Structure superposition and display are as in Figure 10. Relevant CaMKKαp specific residues, M16(CaMKKαp) and L17(CaMKKαp), are also displayed in magenta.
Methionine Residues
CaM has an unexpectedly high percentage of methionine residues, most of which are localized to the hydrophobic surfaces responsible for binding target domains. Systematic mutagenesis of each methionine residue (Met→Gln) identified three general classes of methionines: those whose mutation affected binding and activation of all, none, or only specific targets (50). A similar analysis can be made using comparative side-chain dynamics (Figure 8A&B, orange data points). M124 and M71 are affected to the same extent in both CaMKKαp and smMLCKp complexes, while M36, M76, and M145 are not strongly perturbed in either. Our analysis leads us to conclude M51, M109 and, to a lesser extent, M144 are preferentially influenced by binding CaMKKαp, whereas M72 is affected by binding smMLCKp. M109 was identified by mutagenesis as a residue involved in binding all of the complexes examined; however, our data suggest this may result from changes in the CaM structure upon mutation. In the CaM/smMLCKp complex, M109 participates in intramolecular hydrophobic interactions between the CaM F and G helices, but does not mediate intermolecular contacts between CaM and smMLCKp. Accordingly, the of M109 is not greatly perturbed by binding smMLCKp. Insertion of a hydrophilic residue in this position may disrupt the proper formation of the C-terminal binding pocket causing a decrease in affinity. In contrast, L17(CaMKKαp) forms close contacts with M109 ( ). Structurally, this appears to stabilize the interactions between M109 and V121/L116 and, therefore, plays a critical role in determining the dynamics of these residues. Another intriguing distinction between smMLCKp and CaMKKαp complexes is the position and dynamics of Met144. In the CaM:CaMKKαp structure, M144 is displaced 6 Å away from the hydrophobic binding pocket by Met16(CaMKKαp). This appears to limit M144 interactions with CaMKKαp, yet its parameter is in fact increased by 0.15.
Biological implications
Site-specific variability in response to binding different ligands is a key indicator of the functional relevance of residual protein entropy in modulating CaM activity (Figure 8C). Local entropy differences between the two complexes are clearly evident by the greater breadth of data points within the 0–0.75 range (red and blue boxes). This contrasts with the tight cluster of thirteen residues found to be generally rigid in both complexes. This is not surprising given the similar CaM structure and binding interface in the two complexes. Taking into account the highly conserved nature of CaM, it appears that the target domain sequences have evolved to exploit the reservoir of CaM internal motional freedom at sites specific to each complex. Furthermore, CaM clearly retains a significant number of dynamic residues in either complex, indicating additional modulation may be possible; although this may produce unexpected effects on overall protein and/or complex stability. The majority of differentially perturbed CaM:CaMKKαp residues discussed above are readily identified in Figure 8C, with the exception of Ile85. Examination of the NMR and x-ray structures does not immediately provide a rational explanation for the increased at this site.
We have shown that on a per residue basis the side-chain dynamics of CaM respond differentially to the binding of peptides derived from two kinases, but cumulatively in a manner consistent with the overall change in system entropy for each complex. The formation of all CaM/kinase complexes examined to date by ITC are enthalpically-driven, high affinity associations with significant negative entropic components; complexes formed with CaMKKαp, smMLCKp, and CaMKIp are entropically unfavorable by 120–140 kJ/mol. However, the fact that high affinity for CaM is not necessarily accompanied by a strong entropic penalty [e.g. the complex with the CaM-binding domain of neural nitric oxide synthase (23)] suggests the large entropic penalty consistently present for CaM/kinase formation is significant and fulfills some functional role. It is interesting to speculate that the significant unfavorable entropic contribution to binding may be involved with the deactivation of CaM-dependant kinases; whereby phosphorylation of residues within or in proximity to the CaM binding domain would induce electrostatic repulsion with the highly acidic CaM altering the balance of enthalpic and entropic terms. Phosphorylation of smMLCK and CaMKII has been demonstrated to greatly reduce or abolish CaM binding, respectively (54, 55). In line with this model, autophosphorylation and/or exogenous phosphorylation sites, such as PKA, are consistently found in CaM-dependent kinases many CaM binding domains (50–53). Experiments are currently underway with complexes involving the CaM-binding domains of CaMKII or CaMKIV to test this hypothesis. Interestingly, although a number of CaM residues experience similar changes in dynamics (conformational entropy) across a set of complexes studied in our lab (this work; Lee et al. (24); unpublished results), there are many additional residues that do not. Thus, it is possible that the kinase sequences that have evolved to bind with high affinity while also utilizing entropy to prime the complexes for rapid deactivation. This clearly exploits the ability of CaM to absorb a variety of discrete, heterogeneously distributed entropic penalties. It will be highly informative to compare the present results with the dynamic behavior of CaM in complex with peptides whose association is entropically driven. Such studies are on-going and will be reported elsewhere.
Acknowledgments
We thank Professor Mark Greene for access to the isothermal titration calorimeter and Dr. Kathy Valentine for assistance with NMR relaxation experiments.
Footnotes
Supported by NIH research grant DK 39806.
Abbreviations used: CaM, calcium-saturated calmodulin; CaMKK, calcium/calmodulin-dependent protein kinase kinase; CaMKKαp, peptide based on the CaMKKα calmodulin-binding domain with sequence CaMKKα sequence: GSVKLIPSWTTVILVKSMLRKRSFGNPF; MALDI-TOF, matrix-assisted laser desorption ionization-time of flight; NOE, {1H}-15N nuclear Overhauser effect; and , Lipari-Szabo squared generalized order parameters for the methyl group symmetry axis and amide N-H bond, respectively; PCR, polymerase chain reaction; rmsd, root mean squared deviation; smMLCKp, peptide based on the calmodulin-binding domain of the smooth muscle myosin light chain kinase with sequence: GSARRKWQKTGHAVRAIGRLS; T1, longitudinal relaxation time; T2, transverse relaxation time.
References
- 1.Chin D, Means AR. Calmodulin: a prototypical calcium sensor. Trends Cell Biol. 2000;10:322–8. doi: 10.1016/s0962-8924(00)01800-6. [DOI] [PubMed] [Google Scholar]
- 2.Crivici A, Ikura M. Molecular and structural basis of target recognition by calmodulin. Annu Rev Biophys Biomol Struct. 1995;24:85–116. doi: 10.1146/annurev.bb.24.060195.000505. [DOI] [PubMed] [Google Scholar]
- 3.Shen X, Valencia CA, Szostak JW, Dong B, Liu R. Scanning the human proteome for calmodulin-binding proteins. Proc Natl Acad Sci U S A. 2005;102:5969–74. doi: 10.1073/pnas.0407928102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Eldick LJ, Watterson DM. Calmodulin and Signal Transduction. Academic Press; San Diego: 1998. [Google Scholar]
- 5.Selbert MA, Anderson KA, Huang QH, Goldstein EG, Means AR, Edelman AM. Phosphorylation and activation of Ca(2+)-calmodulin-dependent protein kinase IV by Ca(2+)-calmodulin-dependent protein kinase Ia kinase. Phosphorylation of threonine 196 is essential for activation. J Biol Chem. 1995;270:17616–21. doi: 10.1074/jbc.270.29.17616. [DOI] [PubMed] [Google Scholar]
- 6.Tokumitsu H, Brickey DA, Glod J, Hidaka H, Sikela J, Soderling TR. Activation mechanisms for Ca2+/calmodulin-dependent protein kinase IV. Identification of a brain CaM-kinase IV kinase. J Biol Chem. 1994;269:28640–7. [PubMed] [Google Scholar]
- 7.Haribabu B, Hook SS, Selbert MA, Goldstein EG, Tomhave ED, Edelman AM, Snyderman R, Means AR. Human calcium-calmodulin dependent protein kinase I: cDNA cloning, domain structure and activation by phosphorylation at threonine-177 by calcium-calmodulin dependent protein kinase I kinase. EMBO J. 1995;14:3679–86. doi: 10.1002/j.1460-2075.1995.tb00037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmitt JM, Wayman GA, Nozaki N, Soderling TR. Calcium activation of ERK mediated by calmodulin kinase I. J Biol Chem. 2004;279:24064–72. doi: 10.1074/jbc.M401501200. [DOI] [PubMed] [Google Scholar]
- 9.Enslen H, Sun P, Brickey D, Soderling SH, Klamo E, Soderling TR. Characterization of Ca2+/calmodulin-dependent protein kinase IV. Role in transcriptional regulation. J Biol Chem. 1994;269:15520–7. [PubMed] [Google Scholar]
- 10.Matthews RP, Guthrie CR, Wailes LM, Zhao X, Means AR, McKnight GS. Calcium/calmodulin-dependent protein kinase types II and IV differentially regulate CREB-dependent gene expression. Mol Cell Biol. 1994;14:6107–16. doi: 10.1128/mcb.14.9.6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun P, Enslen H, Myung PS, Maurer RA. Differential activation of CREB by Ca2+/calmodulin-dependent protein kinases type II and type IV involves phosphorylation of a site that negatively regulates activity. Genes Dev. 1994;8:2527–39. doi: 10.1101/gad.8.21.2527. [DOI] [PubMed] [Google Scholar]
- 12.Edelman AM, Mitchelhill KI, Selbert MA, Anderson KA, Hook SS, Stapleton D, Goldstein EG, Means AR, Kemp BE. Multiple Ca(2+)-calmodulin-dependent protein kinase kinases from rat brain. Purification, regulation by Ca(2+)-calmodulin, and partial amino acid sequence. J Biol Chem. 1996;271:10806–10. doi: 10.1074/jbc.271.18.10806. [DOI] [PubMed] [Google Scholar]
- 13.Anderson KA, Means RL, Huang QH, Kemp BE, Goldstein EG, Selbert MA, Edelman AM, Fremeau RT, Means AR. Components of a calmodulin-dependent protein kinase cascade. Molecular cloning, functional characterization and cellular localization of Ca2+/calmodulin-dependent protein kinase kinase beta. J Biol Chem. 1998;273:31880–9. doi: 10.1074/jbc.273.48.31880. [DOI] [PubMed] [Google Scholar]
- 14.Picciotto MR, Czernik AJ, Nairn AC. Calcium/calmodulin-dependent protein kinase I. cDNA cloning and identification of autophosphorylation site. J Biol Chem. 1993;268:26512–21. [PubMed] [Google Scholar]
- 15.Sakagami H, Saito S, Kitani T, Okuno S, Fujisawa H, Kondo H. Localization of the mRNAs for two isoforms of Ca2+/calmodulin-dependent protein kinase kinases in the adult rat brain. Brain Res Mol Brain Res. 1998;54:311–5. doi: 10.1016/s0169-328x(97)00362-8. [DOI] [PubMed] [Google Scholar]
- 16.Peters M, Mizuno K, Ris L, Angelo M, Godaux E, Giese KP. Loss of Ca2+/calmodulin kinase kinase beta affects the formation of some, but not all, types of hippocampus-dependent long-term memory. J Neurosci. 2003;23:9752–60. doi: 10.1523/JNEUROSCI.23-30-09752.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizuno K, Giese KP. Hippocampus-dependent memory formation: do memory type-specific mechanisms exist? J Pharmacol Sci. 2005;98:191–7. doi: 10.1254/jphs.crj05005x. [DOI] [PubMed] [Google Scholar]
- 18.Rhoads AR, Friedberg F. Sequence motifs for calmodulin recognition. FASEB J. 1997;11:331–40. doi: 10.1096/fasebj.11.5.9141499. [DOI] [PubMed] [Google Scholar]
- 19.Osawa M, Tokumitsu H, Swindells MB, Kurihara H, Orita M, Shibanuma T, Furuya T, Ikura M. A novel target recognition revealed by calmodulin in complex with Ca2+-calmodulin-dependent kinase kinase. Nat Struct Biol. 1999;6:819–24. doi: 10.1038/12271. [DOI] [PubMed] [Google Scholar]
- 20.Kurokawa H, Osawa M, Kurihara H, Katayama N, Tokumitsu H, Swindells MB, Kainosho M, Ikura M. Target-induced conformational adaptation of calmodulin revealed by the crystal structure of a complex with nematode Ca(2+)/calmodulin-dependent kinase kinase peptide. J Mol Biol. 2001;312:59–68. doi: 10.1006/jmbi.2001.4822. [DOI] [PubMed] [Google Scholar]
- 21.Meador WE, Means AR, Quiocho FA. Target enzyme recognition by calmodulin: 2.4 A structure of a calmodulin-peptide complex. Science. 1992;257:1251–5. doi: 10.1126/science.1519061. [DOI] [PubMed] [Google Scholar]
- 22.Meador WE, Means AR, Quiocho FA. Modulation of calmodulin plasticity in molecular recognition on the basis of x-ray structures. Science. 1993;262:1718–21. doi: 10.1126/science.8259515. [DOI] [PubMed] [Google Scholar]
- 23.Brokx RD, Lopez MM, Vogel HJ, Makhatadze GI. Energetics of target peptide binding by calmodulin reveals different modes of binding. J Biol Chem. 2001;276:14083–91. doi: 10.1074/jbc.M011026200. [DOI] [PubMed] [Google Scholar]
- 24.Lee AL, Kinnear SA, Wand AJ. Redistribution and loss of side chain entropy upon formation of a calmodulin-peptide complex. Nat Struct Biol. 2000;7:72–7. doi: 10.1038/71280. [DOI] [PubMed] [Google Scholar]
- 25.Urbauer JL, Short JH, Dow LK, Wand AJ. Structural analysis of a novel interaction by calmodulin: high-affinity binding of a peptide in the absence of calcium. Biochemistry. 1995;34:8099–109. doi: 10.1021/bi00025a016. [DOI] [PubMed] [Google Scholar]
- 26.Grzesiek S, Bax A. An efficient experiment for sequential backbone assignment of medium sized isotopically enriched proteins. J Magn Reson. 1992;99:201–207. [Google Scholar]
- 27.Muhandiram DR, Kay LE. Gradient-enhanced triple-resonance three-dimensional NMR experiments with improved sensitivity. J Magn Reson Ser B. 1994;103:203–216. [Google Scholar]
- 28.Wittekind M, Mueller L. HNCACB, a High-Sensitivity 3D NMR Experiment to Correlate Amide-Proton and Nitrogen Resonances with the Alpha- and Beta-Carbon Resonances in Proteins. J Mag Reson B. 1993;101:201–205. [Google Scholar]
- 29.Uhrin D, Uhrinova S, Leadbeater C, Nairn J, Price NC, Barlow PN. 3D HCCH(3)-TOCSY for resonance assignment of methyl-containing side chains in (13)C-labeled proteins. J Magn Reson Ser B. 2000;142:288–93. doi: 10.1006/jmre.1999.1951. [DOI] [PubMed] [Google Scholar]
- 30.Bax A, Delaglio F, Grzesiek S, Vuister GW. Resonance assignment of methionine methyl groups and chi 3 angular information from long-range proton-carbon and carbon-carbon J correlation in a calmodulin-peptide complex. J Biomol NMR. 1994;4:787–97. doi: 10.1007/BF00398409. [DOI] [PubMed] [Google Scholar]
- 31.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 32.Goddard T, Kneller D. SPARKY 3. University of California; San Francisco: 2004. [Google Scholar]
- 33.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 34.Muhandiram DR, Yamazaki T, Sykes BD, Kay LE. Measurement of 2H T1 and T1.rho. Relaxation Times in Uniformly 13C-Labeled and Fractionally 2H-Labeled Proteins in Solution. J Am Chem Soc. 1995;117 [Google Scholar]
- 35.Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. J Am Chem Soc. 1982;104:4546–4559. [Google Scholar]
- 36.Dellwo MJ, Wand AJ. Model-Independent and Model-Dependent Analysis of the Global and Internal Dynamics of Cyclosporine-A. J Am Chem Soc. 1989;111:4571–4578. [Google Scholar]
- 37.Kneller JM, Lu M, Bracken C. An effective method for the discrimination of motional anisotropy and chemical exchange. J Am Chem Soc. 2002;124:1852–3. doi: 10.1021/ja017461k. [DOI] [PubMed] [Google Scholar]
- 38.Li Z, Raychaudhuri S, Wand AJ. Insights into the local residual entropy of proteins provided by NMR relaxation. Protein Sci. 1996;5:2647–50. doi: 10.1002/pro.5560051228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang DW, Kay LE. Contributions to conformational entropy arising from bond vector fluctuations measured from NMR-derived order parameters: Application to protein folding. J Mol Biol. 1996;263:369–382. doi: 10.1006/jmbi.1996.0581. [DOI] [PubMed] [Google Scholar]
- 40.Garcia De La Torre J, Huertas ML, Carrasco B. Calculation of hydrodynamic properties of globular proteins from their atomic-level structure. Biophys J. 2000;78:719–30. doi: 10.1016/S0006-3495(00)76630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baldellon C, Alattia JR, Strub MP, Pauls T, Berchtold MW, Cave A, Padilla A. 15N NMR relaxation studies of calcium-loaded parvalbumin show tight dynamics compared to those of other EF-hand proteins. Biochemistry. 1998;37:9964–75. doi: 10.1021/bi980334p. [DOI] [PubMed] [Google Scholar]
- 42.Barbato G, Ikura M, Kay LE, Pastor RW, Bax A. Backbone dynamics of calmodulin studied by 15N relaxation using inverse detected two-dimensional NMR spectroscopy: the central helix is flexible. Biochemistry. 1992;31:5269–78. doi: 10.1021/bi00138a005. [DOI] [PubMed] [Google Scholar]
- 43.Best RB, Clarke J, Karplus M. The origin of protein sidechain order parameter distributions. J Am Chem Soc. 2004;126:7734–5. doi: 10.1021/ja049078w. [DOI] [PubMed] [Google Scholar]
- 44.Lee AL, Sharp KA, Kranz JK, Song XJ, Wand AJ. Temperature dependence of the internal dynamics of a calmodulin-peptide complex. Biochemistry. 2002;41:13814–13825. doi: 10.1021/bi026380d. [DOI] [PubMed] [Google Scholar]
- 45.Chou JJ, Case DA, Bax A. Insights into the mobility of methyl-bearing side chains in proteins from (3)J(CC) and (3)J(CN) couplings. J Am Chem Soc. 2003;125:8959–66. doi: 10.1021/ja029972s. [DOI] [PubMed] [Google Scholar]
- 46.Best RB, Clarke J, Karplus M. What contributions to protein side-chain dynamics are probed by NMR experiments? A molecular dynamics simulation analysis. J Mol Biol. 2005;349:185–203. doi: 10.1016/j.jmb.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 47.Edgington E. Randomization Tests. 3. Marcel Dekker; New York: 1995. [Google Scholar]
- 48.Wand AJ. Dynamic activation of protein function: a view emerging from NMR spectroscopy. Nat Struct Biol. 2001;8:926–31. doi: 10.1038/nsb1101-926. [DOI] [PubMed] [Google Scholar]
- 49.Akke M, Bruschweiler R, Palmer AG. NMR Order Parameters and Free-Energy - an Analytical Approach and Its Application to Cooperative Ca2+ Binding by Calbindin-D(9k) J Am Chem Soc. 1993;115:9832–9833. [Google Scholar]
- 50.Chin D, Means AR. Methionine to glutamine substitutions in the C-terminal domain of calmodulin impair the activation of three protein kinases. J Biol Chem. 1996;271:30465–71. doi: 10.1074/jbc.271.48.30465. [DOI] [PubMed] [Google Scholar]
- 51.Conti MA, Adelstein RS. The relationship between calmodulin binding and phosphorylation of smooth muscle myosin kinase by the catalytic subunit of 3′:5′ cAMP-dependent protein kinase. J Biol Chem. 1981;256:3178–81. [PubMed] [Google Scholar]
- 52.Colbran RJ, Soderling TR. Calcium/calmodulin-independent autophosphorylation sites of calcium/calmodulin-dependent protein kinase II. Studies on the effect of phosphorylation of threonine 305/306 and serine 314 on calmodulin binding using synthetic peptides. J Biol Chem. 1990;265:11213–9. [PubMed] [Google Scholar]
- 53.Abe M, Hasegawa K, Hosoya H. Activation of chicken gizzard myosin light chain kinase by Ca2+/calmodulin is inhibited by autophosphorylation. Cell Struct Funct. 1996;21:183–8. doi: 10.1247/csf.21.183. [DOI] [PubMed] [Google Scholar]
- 54.Okuno S, Kitani T, Fujisawa H. Regulation of Ca(2+)/calmodulin-dependent protein kinase kinase alpha by cAMP-dependent protein kinase: I. Biochemical analysis. J Biochem (Tokyo) 2001;130:503–13. doi: 10.1093/oxfordjournals.jbchem.a003013. [DOI] [PubMed] [Google Scholar]
- 55.Lukas TJ, Burgess WH, Prendergast FG, Lau W, Watterson DM. Calmodulin binding domains: characterization of a phosphorylation and calmodulin binding site from myosin light chain kinase. Biochemistry. 1986;25:1458–64. doi: 10.1021/bi00354a041. [DOI] [PubMed] [Google Scholar]
- 56.Hashimoto Y, Schworer CM, Colbran RJ, Soderling TR. Autophosphorylation of Ca2+/calmodulin-dependent protein kinase II. Effects on total and Ca2+-independent activities and kinetic parameters. J Biol Chem. 1987;262:8051–5. [PubMed] [Google Scholar]
- 57.Okada S, Pessin JE. Insulin and epidermal growth factor stimulate a conformational change in Rap1 and dissociation of the CrkII-C3G complex. J Biol Chem. 1997;272:28179–82. doi: 10.1074/jbc.272.45.28179. [DOI] [PubMed] [Google Scholar]
- 58.Yi W, Lee TH, Tompkins JD, Zhu F, Wu X, Her C. Physical and Functional Interaction between hMSH5 and c-Abl. Cancer Res. 2006;66:151–8. doi: 10.1158/0008-5472.CAN-05-3019. [DOI] [PubMed] [Google Scholar]
- 59.DerMardirossian C, Schnelzer A, Bokoch GM. Phosphorylation of RhoGDI by Pak1 mediates dissociation of Rac GTPase. Mol Cell. 2004;15:117–27. doi: 10.1016/j.molcel.2004.05.019. [DOI] [PubMed] [Google Scholar]