Summary
Size specification of macromolecular assemblies in the cytoplasm is poorly understood [1]. In principle, assemblies could scale with cell size, or use intrinsic mechanisms to achieve fixed, but regulated, sizes. For the mitotic spindle, scaling with cell size is expected, since the function of this assembly is to physically move sister chromatids into the center of nascent daughter cells. Anecdotally, spindle length does scale with cell length, but it is not clear if this scaling mechanism could operate at very large cell lengths. Eggs of Xenopus laevis are among the largest cells known that cleave completely during cell division. Cell length in this organism changes by two orders of magnitude (~1200 µm to ~12 µm) while it develops from a fertilized egg into a tadpole [2]. We wondered if, and how, mitotic spindle length and morphology adapt to function at these different length scales. Here, we show that spindle length increases with cell length in small cells, but in very large cells spindle length approaches an upper limit of ~60 µm. To transport the DNA into the center of the daughter cells, the relatively small spindle length is compensated by an enormous anaphase B-like movement. Further evidence for an upper limit to spindle length comes from an embryonic extract system that recapitulates mitotic spindle assembly in a test tube. We conclude that early mitotic spindle length in Xenopus laevis is uncoupled from cell length, reaching an upper bound determined by mechanisms that are intrinsic to the spindle.
Results and Discussion
Spindle length is uncoupled from cell length during first mitoses
We used immunofluorescence to measure spindle size in Xenopus laevis embryos fixed at different stages. Spindle length was measured at metaphase, and cell length was measured in the direction given by the pole-pole axis of the spindle (Fig. 1C). To allow comparison with meiotic spindles, which do not contain centrosomes, we defined spindle length as pole-to-pole distance, where the pole is the position where many microtubules terminate (Fig. 1C). Figure 1E shows a plot of spindle length versus cell length. At stages 8 and 9, spindle length increased with cell length but in earlier stages, and larger cells, appeared to asymptote to an upper limit of ~60 µm. Through mitoses 1 to 7, cell length decreased ~5 fold while spindle length only decreased ~1.2 fold (Fig. 1E).
Figure 1. Spindle size is uncoupled from cell size during first mitoses.
X. laevis embryos at various stages of development were fixed and stained for tubulin (yellow) and DNA (red). A) Embryo at stage 8: animal pole with smaller cells and smaller spindles on top, vegetal pole with larger cells and larger spindles on bottom. B) Embryo at mitosis 7 animal part. C) Second mitotic spindle. White lines define spindle and cell size used throughout this paper. D) Egg arrested at metaphase of meiosis II with arrow pointing at spindle. Bar for upper row = 500 µm. Bar for lower row = 20 µm. E) Plot of cell size versus spindle size at different stages of development. Spindle size increases with cell size but asymptotically reaches an upper limit ~60 µm. Plot on the right is a zoom-in of smaller cells and spindles.
Spindle morphology also changed with development, and cell length. At stages 8 and 9, centrosomes and poles were superimposed at the magnification we used, similar to the case of somatic tissue culture cells (Fig. 1A). At mitosis 7, the centrosomes appeared detached from the spindle poles at metaphase, with a relatively microtubule-sparse region connecting them (Fig. 1B). The distance between centrosomes and poles was even larger in the very early spindles (Fig. 1C) [3]. The partial disconnection of centrosomes might be a strategy of the cell to increase centrosome-to-centrosome distance when spindles reach an upper limit in length. Interestingly, the upper limit to mitotic spindle length was about twice the length of meiotic spindles (Fig. 1D, E).
In smaller cells, where spindle length scales with cell length, we can imagine three spindle length determining mechanisms: I) Spindle length is determined extrinsically via cellular boundaries. II) A factor involved in spindle length determination is provided in limited number. Possible candidates for these factors are tubulin and MAPs that influence microtubule dynamics [4] or microtubule flux properties [5]. III) Length-regulating mechanisms that are intrinsic to the spindle systematically change during development. The independence of spindle length from cell length we observed in very large cells suggests that spindle length is determined via a mechanism that is intrinsic to the spindle, such as microtubules dynamics or DNA content. Alternatively, spindle length may be governed by some internal boundary in the large cells that we were not able to visualize.
Mitotic spindles in embryo extract
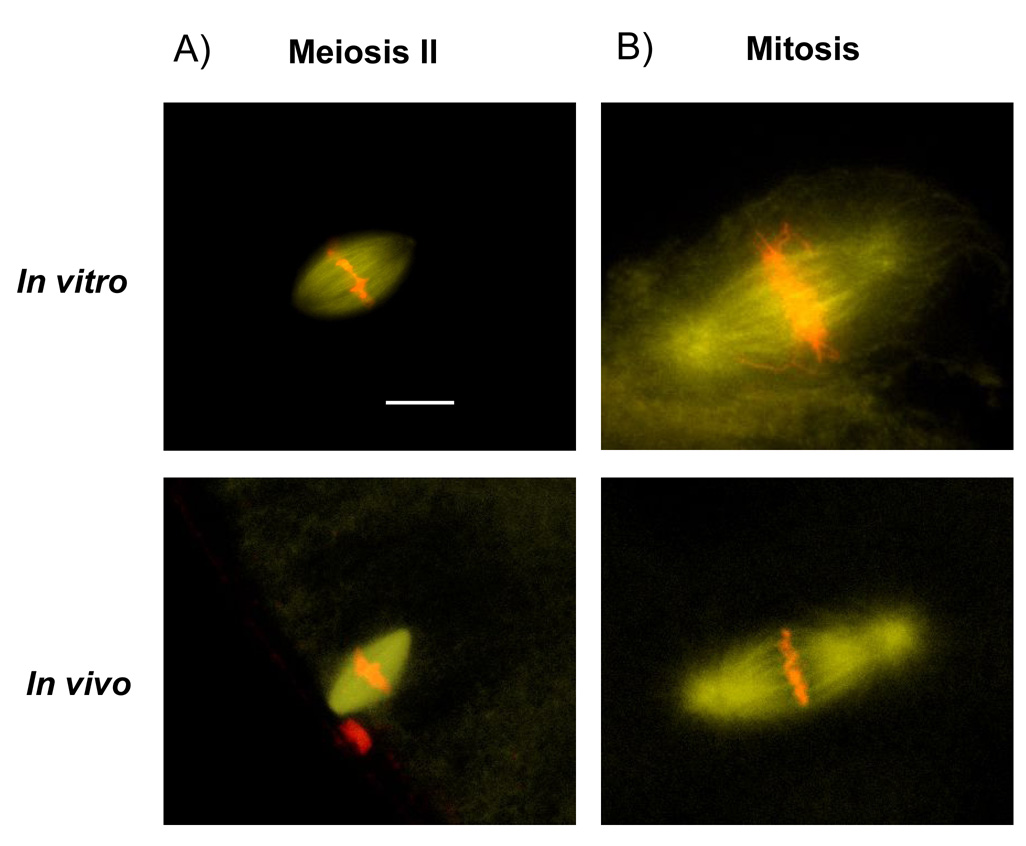
The standard egg extract system for spindle assembly [6] uses cytoplasm from unfertilized eggs that are arrested in meiosis II, and assembles spindles whose length and morphology closely resemble meiosis II spindles in the egg (Fig. 2A)[7]. The length of these spindle cannot be limited by the length of their container (which is a test tube), or by limiting provision of some spindle component, since mean length is insensitive to a wide range of spindle concentrations in the extract [8]. Thus, meiotic spindle length must be limited by a spindle intrinsic mechanism. To test if the same holds true for early mitotic spindles, we developed an embryo extract system that is able to recapitulate their assembly in the test tube. To avoid making a meiotic extract, it is important that the master regulator of meiosis, Mos, be degraded. We made sure that this was the case by preparing the extract from embryos that had already cleaved. By this time Mos is fully degraded [9]. Extract prepared from fertilized eggs is able to go through several cell cycles separated by ~50 minutes [10]. Though sperm nuclei condense during mitosis in this system, we observed no spindles assembling, perhaps because the extract conditions make spindle assembly slow compared to cell cycle progression. We therefore prepared extract from fertilized embryos at the two cell stage, added sperm chromatin, and incubated to allow time in interphase for chromatin assembly and DNA replication. After 80 min, we arrested the extract in mitosis by addition of the C-terminal fragment of EmiI [11] [12]. This fragment is a potent inhibitor of the Anaphase Promoting Complex (APC), which we used rather than the standard mitotic exit inhibitor Cytostatic Factor (CSF), since CSF extract might re-activate meiosis [13]. About 90 min after adding the APC inhibitor, spindles assembled typically with prominent astral microtubules and similar morphology to early mitotic spindles (Fig. 2B). Their length was 48 ± 6 µm (SD, n=28), comparable to mitotic spindles in early blastomeres, and significantly larger than meiotic extract spindles with a length of 32 ± 4 µm (SD) [8]. The length difference of meiotic and mitotic extract spindles appears to reflect the length differences of the in vivo counterparts. To our knowledge this is the first time that truly mitotic spindles could be assembled in a test tube.
Figure 2. Embryonic extract is able to assemble mitotic spindles.
DNA is shown in red and tubulin in yellow. A) Extract prepared from meiosis II arrested eggs assembles spindles that show similar morphology to meiotic in vivo spindles. B) Spindles in extract prepared from embryos were arrested in mitosis with addition of the APC-inhibitor EmiI. The spindles formed show similar morphology to mitotic in vivo spindles. Bar = 20 µm.
Importantly, the length of these extract mitotic spindles did not scale with the test tube, strongly suggesting that early mitotic spindle length is determined by spindle intrinsic mechanisms, like meiosis II spindles, but not by a cell internal boundary. Mitotic extracts assemble spindles with comparable length to their in vivo counterparts, but the timing of spindle assembly was variable, and we were not able to make this system robust enough for more demanding experiments like immunodepletion or spindle assembly imaging.
The upper limit to spindle length is slightly sensitive to ploidy
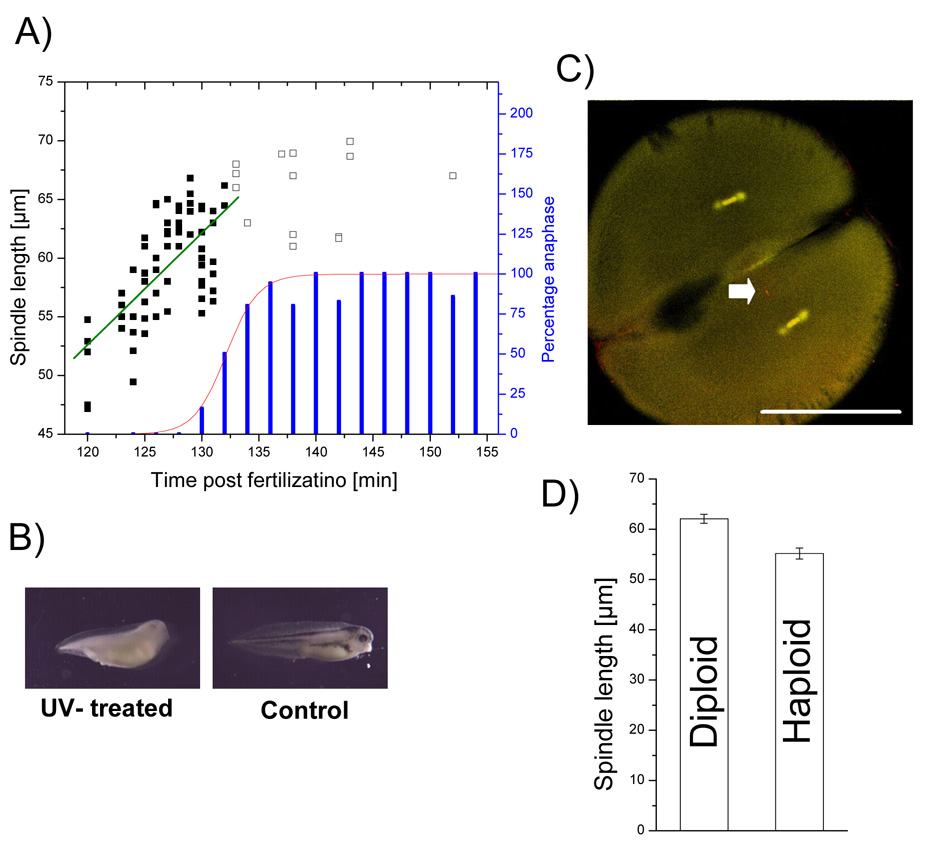
Meiosis II spindles contain only half the number of chromosomes as larger, early mitotic counterparts, and meiotic spindle assembly depends on signals from chromatin [14]. Thus, we wondered if DNA mass plays a role in the spindle intrinsic length determination mechanism in early mitosis [15]. To test this, we compared spindle length in haploid and diploid embryos. We found that spindles lengthen towards the onset of anaphase. To allow more accurate measurement than in Figure 1, we fixed embryos of a synchronously fertilized population between the first and second cytokineses (~112 min and ~160 min post fertilization (pf) respectively) in one minute intervals, and measured spindle length. We chose the two-cell stage because the orientation of the mitotic spindles is clearly defined by the longest cell axis (Fig. 3C), facilitating alignment of spindles in the optical plane for microscopy. By fitting the percentage of cells in anaphase to a cumulative Gaussian distribution, we calculated the most likely time for metaphase-anaphase transition at 132 ± 3 min (SD) (Fig. 3A). A linear fit of spindle length until anaphase onset revealed steady elongation of the spindle during prometaphase-metaphase at ~1.0 µm/min (Fig. 3A). We then defined the maximum metaphase length as the average measured from embryos fixed during a 5 minute window before the peak of anaphase transition. For the diploid population in Fig. 3A this value was 61.6 ± 3.1 µm (SD) (n=28). We then produced haploid embryos by fertilizing albino eggs with UV treated sperm from a pigmented male [16], and compared their spindle length to diploid embryos derived from the same parents, fertilized at nearly the same time (Fig. 3A) [17]. The large majority of UV-sperm fertilized tadpoles showed no pigment (>97 %), and a phenotype typical of haploids (Fig. 4A) [18, 19]. Haploidy was further confirmed by counting chromosomes (data not shown). At the two-cell stage, the UV-treated sperm nucleus, with few microtubule associated, was typically observed away from the spindle (Arrow Fig. 3C). Only one free nucleus was observed, indicating that UV treatment inhibited replication.
Figure 3. Halving the DNA content reduces spindle length by ~10%.
A) Percentage of embryos (synchronously fertilized) in anaphase (blue bars) was fitted to a cumulative Gaussian distribution (red line), calculating the time for metaphase-anaphase transition at 132 ± 3 min (SD). Spindle length before peak of anaphase onset (full squares) was fitted linearly (green line) revealing spindle growth of 1.0 µm/min. Delayed spindles (shown as empty squares) were ignored for growth measurement as this would have systematically underestimated growth rate. B) Albino eggs were fertilized with UV-treated sperm from a pigmented male resulting in tadpoles with no pigments but haploid phenotype. Control developed with pigments and diploid phenotype C) Sperm derived DNA (arrow) is separate from spindles at two-cell stage of haploid embryo. Bar = 500 µm. D) Spindle mean length for haploids is 55.2 µm and therefore ~10 % shorter than diploids with 62.1 µm. Standard errors are 0.9 µm and 1.1 µm, respectively, with a statistically significant p-value of 0.005 %.
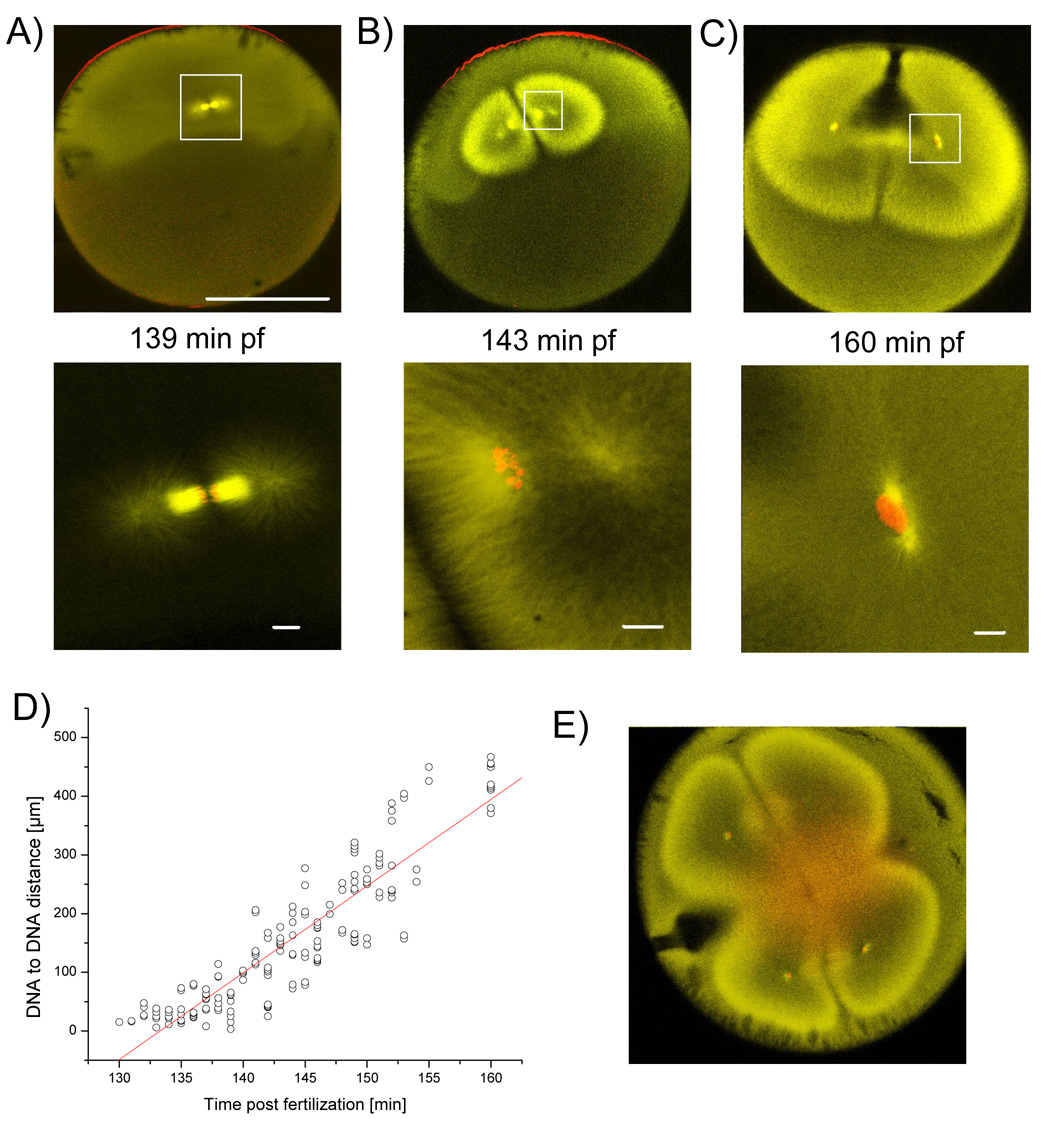
Figure 4. Relatively small spindle is compensated by enormous anaphase B like movement.
Embryos of a synchronously fertilized population were fixed between first and second cytokineses and stained for tubulin (yellow) and DNA (red). A) At anaphase the astral microtubules start to elongate. B) Up to a DNA to DNA distance of ~180 µm, DNA is still condensed and surrounded by high staining of microtubules. Astral microtubules form a hollow structure. C) Nuclear envelope has reformed and finally the nuclei have been separated by ~400 µm, astral microtubules reach the cell cortex and cytokinesis starts. A–C) Bar for upper row = 500 µm, Bars in lower row = 20 µm. D) Plot of DNA to DNA distance versus time. Linear fit estimates speed of DNA separation at ~15 µm/min. E) Cytokinesis, but not separation of DNA, is inhibited by addition of 33 µg/ml of actin depolymerazing Cytochalasin B.
Average spindle length was measured as 55.2 ± 3.9 µm (SD, n=14) for haploids and 62.1 ± 3.1 µm (SD, n=12) for diploids (Fig. 3D). A t-test resulted in a p-value of 0.005 % making the small difference statistically significant. We conclude that the upper limit to mitotic spindle size can be reduced by ~10 % by halving the amount of DNA. This difference is similar to the DNA-dependent length difference observed in meiotic extract spindles [20]. Thus, signaling from chromatin may contribute to spindle length control in meiotic and mitotic spindles, but it does not appear to be a major factor governing length. Haploid mitotic spindles were about two-fold longer than meiosis II spindles (Fig. 1C, D, E) that contain the same amount of DNA, showing that ploidy alone cannot account for length differences between meiosis and mitosis.
Relatively small spindles undergo long, fast anaphase B-like movement
How can a spindle that is only 1/20th of the cell length (Fig. 1E) segregate chromosomes to the center of the daughter cells? To find out, we fixed synchronously fertilized populations at different time intervals at the two cell stage, and observed the distribution of DNA and microtubules. At the onset of anaphase, astral microtubules started to extend (Fig. 4A,B), rapidly forming a hollow structure, where (presumed) plus ends move out towards the cortex, but many minus ends apparently move out at roughly the same rate. Chromosomes stayed condensed, and were surrounded by bright stain for tubulin, until they had separated by ~180 µm (Fig. 4B). At approximately this distance the nuclear envelope reformed, but the DNA continued to separate to a final distance of ~400µm. By this time, astral microtubule plus ends were touching the cell cortex, and the second cytokinesis is initiated (Fig. 4C). Sister DNA separation during anaphase was plotted versus time (Fig. 2D). A linear fit showed a distance increase of ~15 µm/minute (Fig. 2D), with no obvious difference in separation rate for condensed or uncondensed DNA. This is fast compared to ~4 µm/min observed for anaphase-B movement in HeLa cells [21].
To test whether actin is involved in the separation of the DNA [22], we observed fixed embryos that had been incubated with the F-actin capping drug Cytochalasin B (33 µg/ml) [23]. This resulted in inhibition of cytokinesis, but spindle assembly and separation of DNA were not measurably perturbed (Fig. 4E). However, while the drug did block cleavage, it is possible that its concentration was insufficient to block actin dependent processes deep in the embryo, so our conclusion that F actin is not required for DNA separation is provisional.
Our measurements in large cells, and extract experiments, suggest that Xenopus early mitotic spindle length is determined via an intrinsic mechanism that sets an upper length of ~60 µm. This limit was reduced by ~10 % in haploid spindles, suggesting signals from DNA contribute to setting length, but are not a major factor. Recently, we proposed a model for meiotic spindle length regulation in which length depends primarily on a balance between microtubule nucleation-loss and transport by motors [5]. Perhaps a spindle intrinsic mechanism of this kind also operates in mitotic spindles.
Relatively small spindle size in large cells requires adaptation of the mitotic process, which includes an unusually long and fast anaphase B, and perhaps also partial separation of centrosomes from the spindle. One question puzzles us greatly: how can the relatively small spindle orient itself in the large cell to specify the next cleavage plane perpendicular to the longest cell axis (Fig. 3C) [24],[25],[26]? In more ordinary sized cells, spindle orientation is thought to require contact of astral microtubules with the cortex [27],[28]. Perhaps some microtubules are long enough to reach cortex during prometaphase-metaphase of early Xenopus mitosis, but this seems unlikely because these microtubules would have to be much longer than the spindle microtubules, and they would have to elongate to the cortex much faster than the astral microtubules that grow out at anaphase (~15 µm/min, estimated from images like those in Figure 4). Rather, we suspect some uncharacterized spindle orientation mechanism must exist. Perhaps the astral microtubules at late anaphase can sense the longest cell axis and determine centrosome orientation for the next spindle.
Experimental Procedures
Immunofluorescence of embryos
Embryos were raised at 16° C. Previous protocols [29] were modified as follows. Embryos were fixed in 50 mM EGTA, 10 % H2O, 90 % methanol for at least 12 h. Pigmented embryos were bleached in 10 % H2O2, 20 % H20, and 70 % methanol under illumination for 24 hours. Specimen were dehydrated with a series of 20 %, 40 %, 80 % 100 % TBS/Methanol. For hemisection embryos were cut in TBS on an agarose cushion with a scalpel. Specimen were incubated with directly labeled a-tubulin antibody (T6074 (sigma) 4.6/AB labeling ratio, Alexa 547 (Invitrogen)) 1:100 for at least 12 h at 4° C in TBSN (TBS + 0.1 % Nonidet P40 + 0.1 % sodium azide, 2 % BSA, 1 % FCS). Embryos were washed in TBSN for at least 24 hours. DNA was stained with YoPro3 (Invitrogen) (5mM) or To-Pro-1 (Invitrogen) (5mM) in TBSN for 30 min and washed in TBSN for 1h. After one wash in TBS and two changes of methanol embryos were cleared in Murray’s clear (benzyl benzoate, part benzyl alcohol 2:1)and mounted in metal slides with a hole (thickness of 1.2 mm for whole mount or 0.8 mm for hemisected). The hole was closed on the bottom with parafilm attached coverslip. Microscopy was performed on an upright Biorad Radiance 2000 or inverted Zeiss Meta 550 with 10 × (0.3 NA) or 20 × (0.75 NA) objectives.
Comparison of haploid and diploid spindle length
To generate haploid embryos half a testis was macerated with an Eppendorf pestle in 1ml of MMR and pressed with a syringe through cheese cloth to remove tissue junks. The suspension was placed on a Petri dish with 7 cm diameter and irradiated 2 times at 30,000 microjoules/cm2 with swirling in between in a UV Stratalinker 2400 [30]. Embryos were fertilized with this suspension and fixed at the two cell stage, hemisected along the first cleavage plane and prepared for immunofluorescence as described above. Spindle size within one embryo is more similar than spindles in the whole population. Therefore for the t-test (ttest2 function in Matlab (Mathworks)) the average spindle length per embryo was used. Karyotyping was performed as described [31]. Curve fitting was performed with cftool in Matlab (Mathworks).
Embryo extract spindles
Published protocols [32],[6] for meiotic extract spindles were modified as follows to give mitotic extract spindles. Females were squeezed, eggs fertilized and dejellied. Embryos from different animals were kept separate and only if fertilization rate was close to 100% embryos were used. After the first cleavage non-fertilized eggs were sorted out because of dominant effect of CSF. Embryos were washed in XB (100 mM KCl, 0.1mM CaCl2, 1mM MgCl2, 10 mM Hepes, 50 mM Sucrose, pH 7.8 (KOH)). 0.75 ml of silicon oil AP100 (Fluka) were added to a 50Ultra-Clear Tube (11×34mm) (Beckmann), embryos were transferred to top, incubated for 15 min on ice, and spun at 2000 rpm in a JS4.2 (Beckmann) for 4 min at 4° C. Buffer and oil were removed. Embryos were crushed at 12000rpm in a TLS-55 (Beckmann) for 15 min at 4 ° C. The cyotplasmic fraction was removed with a syringe. A clearing spin in a tabletop centrifuge at 4° C, 4 min, 12000g followed to remove residual oil. Cytochalasin D (10µg/ml), LPC (10 µg/ml each of leupeptin, pepstatin, chymostatin) and Energy Mix (7.5 mM creatine phosphate,1 mM ATP, 0.1 mM EGTA, 1 mM MgCl2) were added. Demembrenated sperm was added and extract allowed to cycle at RT. After ~80 min bacterially expressed C-terminal fragment of EmiI (23mg/ml) was added at 1:200. Spindles typically formed after an additional 90 minutes. C-terminal fragment of EmiI was purified via a His-tag and frozen in XB + 200 mM KCl.
Acknowledgements
We would like to thank Jagesh Shah, Andrew Murray, Marc Kirschner, Rebecca Ward, Yifat Merbl, Tom Maresca, Jay Gatlin, Cell division group Woods Hole, Eva Kiermaier and people in the Mitchison lab for helpful suggestions and discussion, Olaf Stemmann for training, Michael Rape for plasmid of EmiI, Jim Horn for technical assistance, Nikon Imaging Center (HMS) and Zeiss Woods Hole for imaging assistance. DJN was supported by the Life Sciences Research Foundation, sponsored by Novartis. This work was supported by the National Institutes of Health (NIH) grants GM39565 and P50 GM068763-1.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Marshall WF. Cellular length control systems. Annu Rev Cell Dev Biol. 2004;20:677–693. doi: 10.1146/annurev.cellbio.20.012103.094437. [DOI] [PubMed] [Google Scholar]
- 2.Montorzi M, Burgos MH, Falchuk KH. Xenopus laevis embryo development: arrest of epidermal cell differentiation by the chelating agent 1,10-phenanthroline. Mol Reprod Dev. 2000;55:75–82. doi: 10.1002/(SICI)1098-2795(200001)55:1<75::AID-MRD10>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 3.Gard DL, Cha BJ, Schroeder MM. Confocal immunofluorescence microscopy of microtubules, microtubule-associated proteins, and microtubule-organizing centers during amphibian oogenesis and early development. Curr Top Dev Biol. 1995;31:383–431. doi: 10.1016/s0070-2153(08)60234-3. [DOI] [PubMed] [Google Scholar]
- 4.Andersen SS, Ashford AJ, Tournebize R, Gavet O, Sobel A, Hyman AA, Karsenti E. Mitotic chromatin regulates phosphorylation of Stathmin/Op18. Nature. 1997;389:640–643. doi: 10.1038/39382. [DOI] [PubMed] [Google Scholar]
- 5.Burbank KS, Mitchison TJ, Fisher DS. Slide-and-cluster models for spindle assembly. Curr Biol. 2007;17:1373–1383. doi: 10.1016/j.cub.2007.07.058. [DOI] [PubMed] [Google Scholar]
- 6.Sawin KE, Mitchison TJ. Mitotic spindle assembly by two different pathways in vitro. J Cell Biol. 1991;112:925–940. doi: 10.1083/jcb.112.5.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lohka MJ, Maller JL. Induction of nuclear envelope breakdown, chromosome condensation, and spindle formation in cell-free extracts. J Cell Biol. 1985;101:518–523. doi: 10.1083/jcb.101.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitchison TJ, Maddox P, Gaetz J, Groen A, Shirasu M, Desai A, Salmon ED, Kapoor TM. Roles of polymerization dynamics, opposed motors, and a tensile element in governing the length of Xenopus extract meiotic spindles. Mol Biol Cell. 2005;16:3064–3076. doi: 10.1091/mbc.E05-02-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sagata N, Watanabe N, Vande Woude GF, Ikawa Y. The c-mos protooncogene product is a cytostatic factor responsible for meiotic arrest in vertebrate eggs. Nature. 1989;342:512–518. doi: 10.1038/342512a0. [DOI] [PubMed] [Google Scholar]
- 10.Murray AW, Kirschner MW. Cyclin synthesis drives the early embryonic cell cycle. Nature. 1989;339:275–280. doi: 10.1038/339275a0. [DOI] [PubMed] [Google Scholar]
- 11.Reimann JD, Freed E, Hsu JY, Kramer ER, Peters JM, Jackson PK. Emi1 is a mitotic regulator that interacts with Cdc20 and inhibits the anaphase promoting complex. Cell. 2001;105:645–655. doi: 10.1016/s0092-8674(01)00361-0. [DOI] [PubMed] [Google Scholar]
- 12.Schmidt A, Rauh NR, Nigg EA, Mayer TU. Cytostatic factor: an activity that puts the cell cycle on hold. J Cell Sci. 2006;119:1213–1218. doi: 10.1242/jcs.02919. [DOI] [PubMed] [Google Scholar]
- 13.Moses RM, Masui Y. Cytostatic Factor (Csf) in the Eggs of Xenopus-Laevis. Experimental Cell Research. 1989;185:271–276. doi: 10.1016/0014-4827(89)90055-4. [DOI] [PubMed] [Google Scholar]
- 14.Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- 15.Nicklas RB, Gordon GW. The total length of spindle microtubules depends on the number of chromosomes present. J Cell Biol. 1985;100:1–7. doi: 10.1083/jcb.100.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reinschmidt DC, Simon SJ, Volpe EP, Tompkins R. Production of Tetraploid and Homozygous Diploid Amphibians by Suppression of 1st Cleavage. Journal of Experimental Zoology. 1979;210:137–143. [Google Scholar]
- 17.Noramly S, Zimmerman L, Cox A, Aloise R, Fisher M, Grainger RM. A gynogenetic screen to isolate naturally occurring recessive mutations in Xenopus tropicalis. Mech Dev. 2005;122:273–287. doi: 10.1016/j.mod.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 18.Hamilton L. An Experimental Analysis of Development of Haploid Syndrome in Embryos of Xenopus Laevis. Journal of Embryology and Experimental Morphology. 1963;11:267-&. [PubMed] [Google Scholar]
- 19.Porter KR. Androgenetic development of the egg of Rana pipiens. Biological Bulletin. 1939;77:233–257. [Google Scholar]
- 20.Brown KS, Blower MD, Maresca TJ, Grammer TC, Harland RM, Heald R. Xenopus tropicalis egg extracts provide insight into scaling of the mitotic spindle. J Cell Biol. 2007;176:765–770. doi: 10.1083/jcb.200610043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayr MI, Hummer S, Bormann J, Gruner T, Adio S, Woehlke G, Mayer TU. The human kinesin Kif18A is a motile microtubule depolymerase essential for chromosome congression. Curr Biol. 2007;17:488–498. doi: 10.1016/j.cub.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 22.Verlhac MH, Lefebvre C, Guillaud P, Rassinier P, Maro B. Asymmetric division in mouse oocytes: with or without Mos. Curr Biol. 2000;10:1303–1306. doi: 10.1016/s0960-9822(00)00753-3. [DOI] [PubMed] [Google Scholar]
- 23.Gard DL, Cha BJ, Roeder AD. F-actin is required for spindle anchoring and rotation in Xenopus oocytes: a re-examination of the effects of cytochalasin B on oocyte maturation. Zygote. 1995;3:17–26. doi: 10.1017/s0967199400002331. [DOI] [PubMed] [Google Scholar]
- 24.Hertwig O. Ueber den Werth der ersten Furchungszellen f ur die Organbildung des Embryo. Experimentelle Studien am Frosch- und Tritonei. Arch. mikr. Anat. 1893;xlii:662–807. [Google Scholar]
- 25.Born G. Ueber Druckversuche an Froscheiern. Anat. Anz. 1893;viii:609–627. [Google Scholar]
- 26.Sawai T, Yomota A. Cleavage plane determination in amphibian eggs. Ann N Y Acad Sci. 1990;582:40–49. doi: 10.1111/j.1749-6632.1990.tb21666.x. [DOI] [PubMed] [Google Scholar]
- 27.Strauss B, Adams RJ, Papalopulu N. A default mechanism of spindle orientation based on cell shape is sufficient to generate cell fate diversity in polarised Xenopus blastomeres. Development. 2006;133:3883–3893. doi: 10.1242/dev.02578. [DOI] [PubMed] [Google Scholar]
- 28.Thery M, Jimenez-Dalmaroni A, Racine V, Bornens M, Julicher F. Experimental and theoretical study of mitotic spindle orientation. Nature. 2007;447:493–496. doi: 10.1038/nature05786. [DOI] [PubMed] [Google Scholar]
- 29.Becker BE, Gard DL. Visualization of the cytoskeleton in Xenopus oocytes and eggs by confocal immunofluorescence microscopy. Methods Mol Biol. 2006;322:69–86. doi: 10.1007/978-1-59745-000-3_6. [DOI] [PubMed] [Google Scholar]
- 30.Tompkins R. Triploid and Gynogenetic Diploid Xenopus-Laevis. Journal of Experimental Zoology. 1978;203:251–256. [Google Scholar]
- 31.Hirsch N, Zimmerman LB, Gray J, Chae J, Curran KL, Fisher M, Ogino H, Grainger RM. Xenopus tropicalis transgenic lines and their use in the study of embryonic induction. Dev Dyn. 2002;225:522–535. doi: 10.1002/dvdy.10188. [DOI] [PubMed] [Google Scholar]
- 32.Desai A, Murray A, Mitchison TJ, Walczak CE. The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods Cell Biol. 1999;61:385–412. doi: 10.1016/s0091-679x(08)61991-3. [DOI] [PubMed] [Google Scholar]