Abstract
The scaffold protein p62 is involved in internalization and trafficking of TrkA. The receptor is deubiquitinated by the proteasomes prior to degradation by lysosomes. Here we demonstrate that p62 serves as a shuttling protein for interaction of ubiquitinated TrkA with Rpt1, one of the six ATPases of 19S regulatory particle of the 26S proteasome. In p62 −/− mouse brain TrkA failed to interact with the Rpt1. The interaction of TrkA with Rpt1 was reduced in proteasomes isolated from p62 −/− brain, but was restored by addition of p62. The UBA domain of p62 interacts with TrkA and its PB1/UbL domain with AAA-ATPase cassette in the C-terminal region of Rpt1. Last, neurotrophin dependent turnover of TrkA was impaired by reduction in the level of p62. These findings reveal that p62 serves as a shuttling factor for interaction of ubiquitinated substrates with the proteasome and could promote localized protein turnover in neurons.
Keywords: TrkA, p62, Rpt1, proteasome, shuttling protein, degradation
Introduction
Nerve growth factor binding to TrkA receptor triggers activation, ubiquitination, internalization and degradation of TrkA [1,2,3]. Both proteasomes and lysosomes are involved in the degradation of the internalized TrkA receptor, however, TrkA is deubiquitinated by proteasomes prior to its degradation in lysosomes [3]. Previous studies have shown that p62 interacts with polyubiquitinated substrates through its UBA (ubiquitin-associating) domain and interacts with the proteasome through its N-terminal region [4]. The PB1 domain of p62 at its N-terminus region [5] shares structural similarity with the UbL (Ubiquitin Like) domain [6] and may interact with one of the six ATPases of the 19S regulatory particle of the 26S proteasome implicated in binding shuttling factors [7]. In this regard Rpt1 has been implicated as a docking site for interaction with putative shuttling factors with the proteasome [8,9]. Since polyubiquitinated TrkA interacts with UBA domain of p62 [10] and the receptor is deubiquitinated prior to trafficking to lysosomes for degradation [3], this study was undertaken to determine whether p62 may function as a shuttling protein for interaction and deubiquitination of the receptor by proteasomes.
Materials and methods
Materials
The mouse Trk (B-3), myc, HA and rabbit Trk (C-14) antibodies were purchased from Santa Cruz Biotechnology (La Jolla, CA). Rpt1 mouse antibody was obtained from Boston Biochem (Cambridge, MA). Cycloheximide, thrombin and glutathione-agarose beads were from Sigma-Aldrich (St. Louis, MO). 2.5 S nerve growth factor (NGF) was from Bioproducts for Science (Indianapolis, IN).
Animal model
Knock-out mice (p62 −/−) were generated as previously described [11]. All the animals employed were handled according to the Auburn University Institutional Animal Care and Use Committee, which abides the National Institutes of Health guidelines. All experiments were conducted on 6 month–old-mouse brain.
Cell Culture, immunoprecipitation, and western blot analysis
HEK 293 cells and PC12 cells were cultured as previously described [2]. HEK 293 cells were transfected employing the calcium phosphate method using a Mammalian Cell Transfection Kit (Chemicon International, Temecula, CA) and PC12 cells with Lipofectamine (Invitrogen). The cells were lysed with Triton lysis buffer to detect protein-protein interactions (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 10 mM NaF, 0.5% Triton X-100, 1 mM Na3VO4, 1 mM PMSF, 2 µg/ml leupeptin and aprotinin). Protein concentrations were estimated using BSA as a standard by the Bradford procedure (Bio-Rad, Hercules, CA). The lysates were immunoprecipitated and immunoblotted as described [2]. The TrkA C-14 antibody detects both glycosylated and immature forms of TrkA, whereas the B-3 antibody detects the glycosylated form only.
Isolation of the 26S proteasomes
p62 −/− mouse brain was used to isolate the proteasomes as previously described [3,12].
Cortico-hippocampal neurons
Dissociated cultures of cortico-hippocampal neurons were prepared from embryonic day 18 mice, as described [13] and cultured in Neurobasal media with B27 supplements (Invitrogen). At DIV7 the neurons were transfected with GFP-Rpt1 and HATrkA employing Lipofectamine (Invitrogen) in Optimem media (Invitrogen) and processed at DIV9.
Immunofluorescence
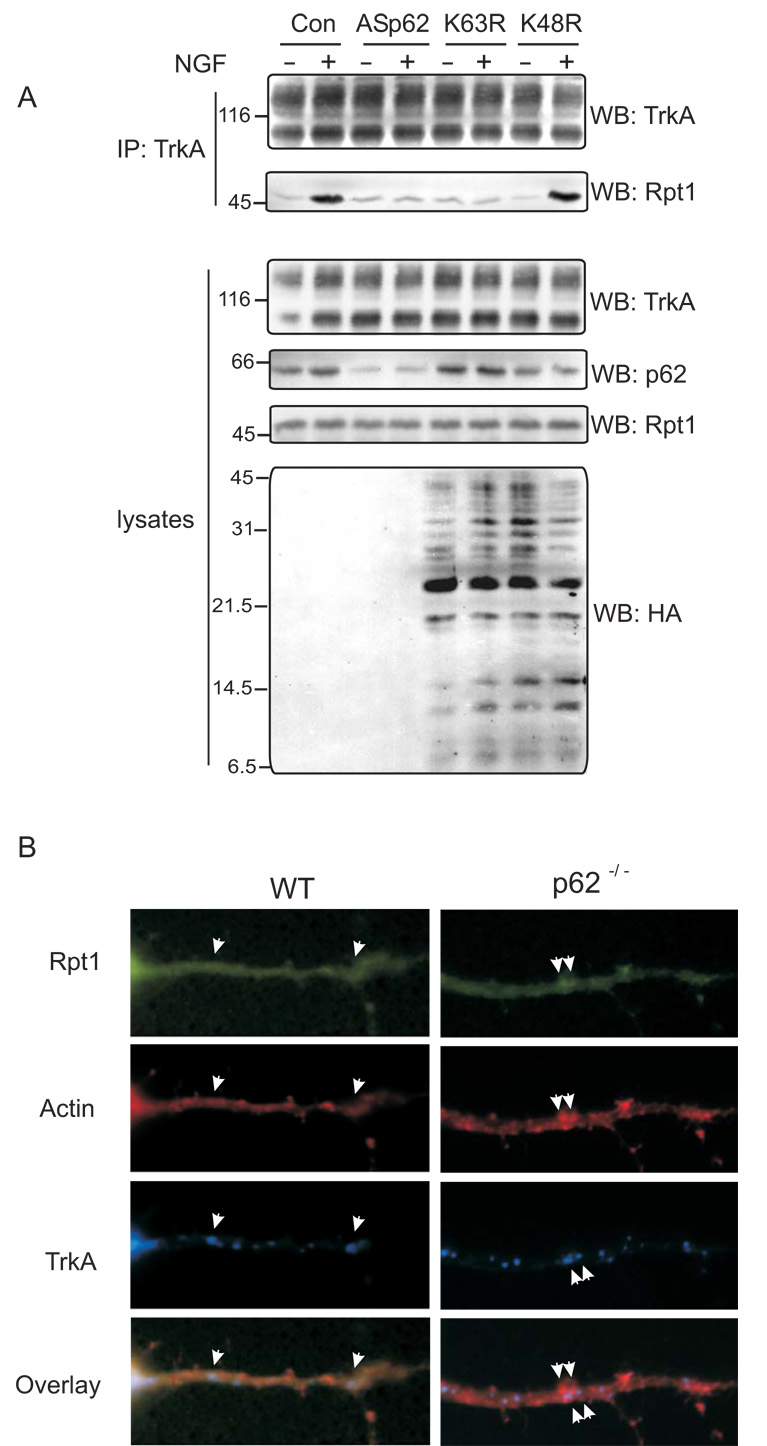
Wild-type and p62 −/− mouse embryonic fibroblast (MEF) cells were treated with 50 ng/ml of NGF at 37°C for 0 and 60 min [3]. MEF cells were washed with phosphate-buffered saline (PBS), followed by fixation with 3% paraformaldehyde and permeabilization in 0.1% Triton X-100 in PBS for 15 min. The cells were blocked in 3% milk in PBS for 1 h and incubated with TrkA (B-3) and Rpt1 antibodies overnight at 4°C and then washed and incubated with secondary antibody coupled to Texas Red or Oregon Green (Molecular Probes, Portland, OR) for 1 h. After washing in PBS, the coverslips were mounted on slides, and were analyzed with a 100 X oil immersion objective on a Bio-Rad MRC 1024 confocal microscope. Images were processed employing Confocal Assistant 4.02 (Bio-Rad) and Adobe Photoshop 6.0 software (Adobe Systems, Mountain View, CA).
GST pull-down assay
Escherichia coli cells were induced with isopropyl 1-thio-β-D-galactopyranoside and lysed in NETN buffer (20 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.1% Nonidet P-40, 2 µg/ml aprotinin, 2 µg/ml leupeptin, and 1 mM PMSF). The GST-tagged proteins were bound to glutathione-agarose overnight at 4°C followed by washing with NETN buffer [4]. The purity was validated by Coomassie blue staining and Western blot with GST antibody. p62 was cleaved from the agarose beads as well as the GST tag by addition of thrombin (1 Unit/100 µg protein) at room temperature for 20 h. The concentration of purified p62 was estimated by mini-Bradford assay. The GST-Rpt captured on agarose beads was washed with binding buffer (20 mM Tris-HCl pH 7.6, 50 mM NaCl, 0.1% Nonidet P-40, 0.5 M dithiothreitol, 1 mM PMSF) prior to pull down assay. Five µg of GST-Rpt was added to 2 µg of purified p62 and rotated for 2 h at 4°C and analyzed by immunoblotting with GST and p62 antibody.
Results and discussion
TrkA interacts with Rpt1 in presence of p62
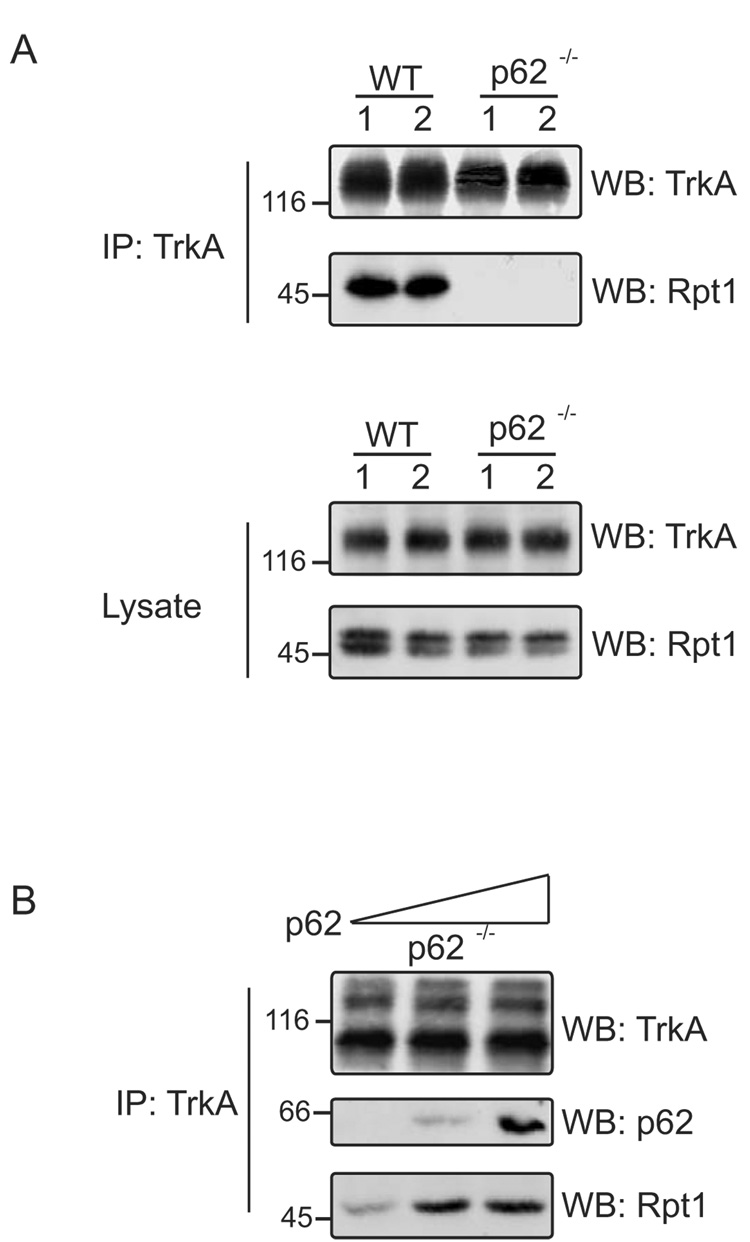
We sought to determine the mechanism whereby ubiquitinated TrkA interacted with the proteasome and set out to examine whether p62 interacted with Rpt1, similar to other UbL/UBA containing shuttling factors. WT and p62 −/− mouse brain homogenates were immunoprecipitated with anti-TrkA and blotted with TrkA and Rpt1 antibody (Fig. 1A); TrkA interacted with Rpt1 in WT lysates but not in lysates from p62 −/− mice. Rpt1 was also immunoprecipitated from WT and p62 −/− mouse brain and Rpt1 associated with TrkA only in the presence of p62 (data not shown). These findings suggest that p62 is needed for TrkA interaction with Rpt1. To further test p62 involvement in directing TrkA interaction, proteasomes were isolated from p62 −/− mouse brain. The interaction of TrkA and Rpt1 was almost nonexistent in proteasomes isolated from p62 −/− mouse brain; however, addition of purified p62 restored the interaction of TrkA with Rpt1 (Fig. 1B).
Fig. 1. Interaction of TrkA with Rpt1 is p62 dependent.
(A) Brain of two mice each, WT and p62 −/− were homogenized in Triton lysis buffer. The homogenate was immunoprecipitated (IP) with TrkA (C-14) antibody and Western blotted (WB) with TrkA (B-3) and Rpt1 antibody. (B) Purified p62 (0, 50, 250 ng) was added to isolated proteasomes from p62 −/− mouse brain and rotated for 1 h at room temperature. TrkA was immunoprecipitated (IP) and Western blotted (WB) with TrkA (B-3), p62 and Rpt1 antibody. (C) WT and p62 −/− MEF cells were stimulated with or without NGF (50 ng/ml) for 1 h, followed by fixation and staining with both anti-TrkA (B-3) and anti-Rpt1. The extent of colocalization (Merge, yellow) was assessed by superimposing red (TrkA, Texas Red) and green (Rpt1, Oregon Green) signals. (D) 50 µg of WT and p62 −/− MEF cells were Western blotted (WB) with TrkA (B-3) and Rpt1 to check their expression.
In addition TrkA colocalization with Rpt1 was examined by immunofluorescence microscopy employing WT and p62 −/− mouse embryo fibroblasts (MEF) (Fig. 1C). The cells were stimulated with NGF and extensive colocalization of the TrkA/Rpt1 was observed in WT MEF cells and to a much lesser extent in p62 −/− cells. As control, the expression of TrkA and Rpt1 in MEF cells was confirmed by immunoblotting (Fig. 1D). Taken together, these results provide compelling evidence for p62 in mediating interaction of TrkA with Rpt1.
Interaction of TrkA and p62 with Rpt1
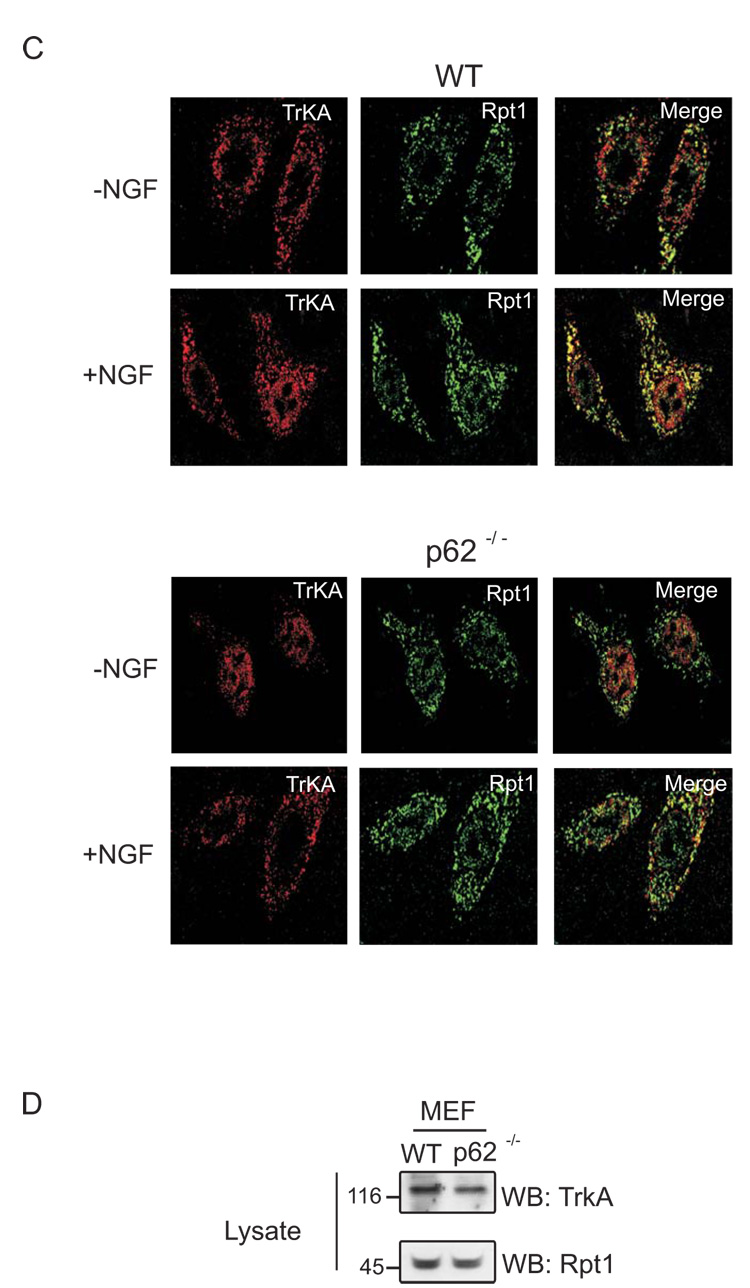
Several proteins containing UBA and UbL domains interact with ubiquitinated substrates and proteasome, thereby targeting delivery of substrates for degradation [8,14,15]. We next set out to examine which domain of p62 was involved in mediating TrkA interaction with Rpt1. HEK cells were cotransfected with HA-TrkA, myc-p62 and either ΔN-terminal or ΔUBA mutants of p62. The cells were stimulated with nerve growth factor (NGF) to promote TrkA ubiquitination [10] and evaluated for the cointeraction of TrkA, p62 and Rpt1 (Fig. 2A). TrkA was able to interact with the delta N-term of p62 but not the delta UBA domain, however an absence of p62’s UBA domain impaired TrkA interaction with Rpt1. On the other hand, p62 interacted with TrkA and Rpt1 but the interaction with TrkA was absent if p62’s UBA domain was deleted. Full-length and delta UBA mutant of p62 interacted with Rpt1 (Fig. 2B). These findings reveal that the N-term of p62 interacts with Rpt1, whereas the C-term of p62 interacts with TrkA. This observation is consistent with previous findings revealing the ubiquitinated TrkA interacts with p62’s UBA domain [10].
Fig. 2. The site of interaction between TrkA/p62 and Rpt1.
(A) HA-tagged TrkA along with myc-tagged WT p62, ΔN-term, or ΔUBA (as shown) was expressed in HEK 293 cells followed by NGF stimulation (50 ng/ml) for 60 min. The cells were lysed with Triton lysis buffer, and the association of TrkA with p62 and Rpt1 was determined by immunoprecipitation of the lysates with antibody to TrkA (C-14) and Western blotting with antibody to HA, myc and Rpt1. (B) The same transfected HEK lysates from above was also immunoprecipitated with anti-myc and Western blotted with HA, myc and Rpt1 antibody. Expression of myc-tagged p62 constructs is shown with an arrow.
p62 interacts with C-terminal region of Rpt1
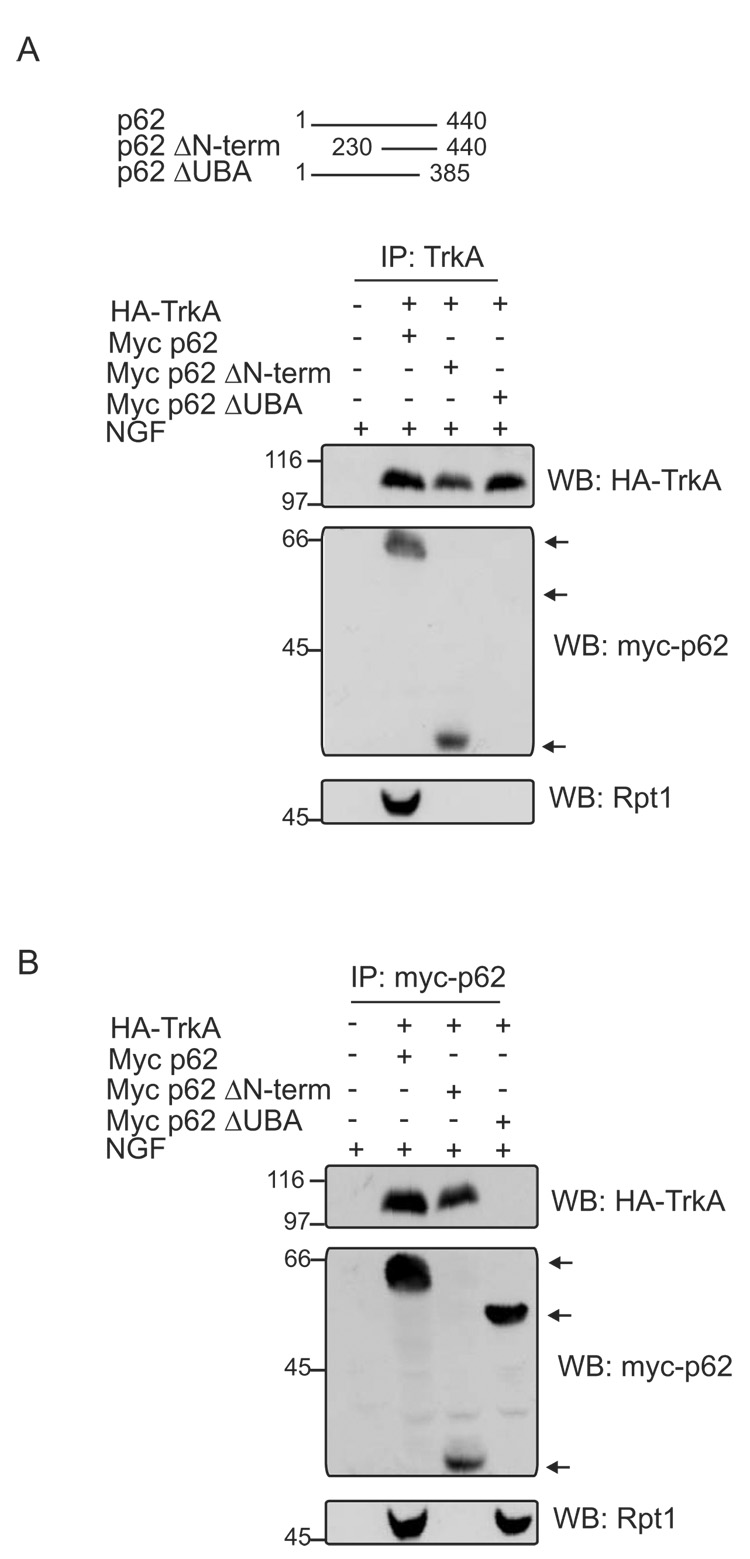
The six Rpt subunits (Rpt1-6) present in the base of the regulatory particle (RP) of the 26S proteasome contain a coiled coil domain in the N-terminal region and an AAA-ATPase (ATPases Associated with a variety of cellular Activities) cassette in the C-terminus [16,17]. As p62 interacts with Rpt1, additional experiments were undertaken to map the region of Rpt1 that interacts with p62. GST-tagged Rpt1 or N-term, C-term mutant proteins captured on agarose beads were employed in a pull-down assay along with GST-Rpt6 to test for specificity (Fig. 3A). p62 interacted with full-length Rpt1 and C-term but not with the mutant N-term Rpt1 or Rpt6. Increasing concentrations of p62 included in the GST-Rpt1 pull down assay, resulted in capture of increasing amounts of p62 (Fig 3B). By contrast, GST-Rpt6 failed to capture p62, even when higher concentrations were included in the assay (Fig. 3C). Therefore, p62 interacts directly with C-terminal region of Rpt1 and not with Rpt6.
Fig. 3. Interaction of p62 with the C-terminal region of Rpt1.
(A) GST-tagged full length Rpt1, its mutant N-term, C-term or GST-tagged Rpt6 captured on glutathione agarose beads were allowed to interact with purified p62 in a pull-down assay. The pull downs were Western blotted with anti-GST. The blot was stripped and blotted with p62 antibody. (B) GST-Rpt1 pull down assay was carried out with increasing concentrations of p62 (0, 1, 2, 4 µg) and immunoblotted with anti-GST and anti-p62. (C) GST-Rpt6 pull down was conducted same as in B. The assay was Western blotted with anti-GST. The blot was stripped and probed with p62 antibody.
Turnover of TrkA
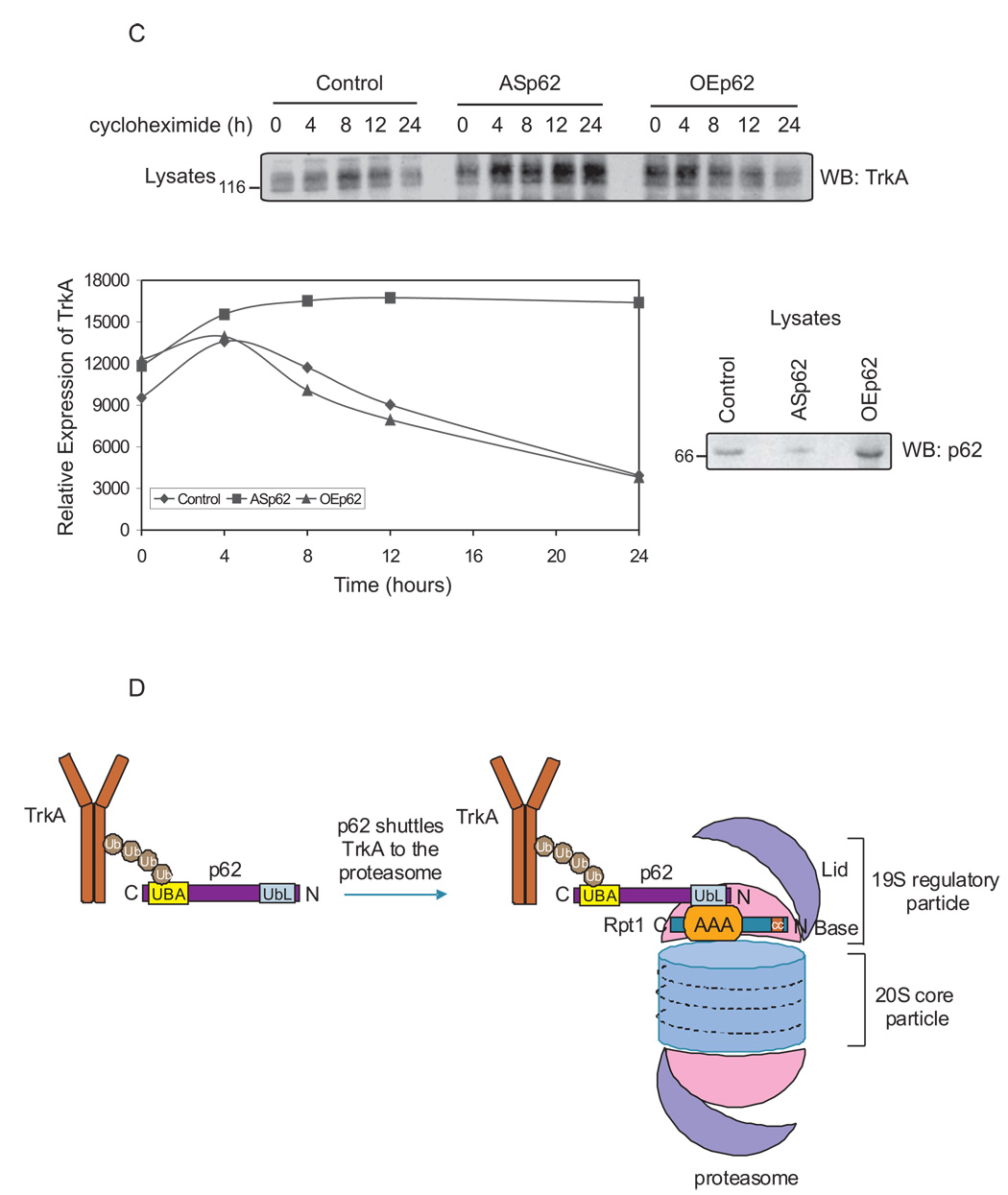
TrkA is deubiquitinated post-internalization by proteasomes prior to degradation by lysosomes [3]. As p62 interacts with the proteasomal subunit, Rpt1, it is possible that the TrkA receptor may employ p62 as a shuttling protein to regulate its turnover. To explore this possibility, PC12 cells were transfected with full-length antisense p62 (ASp62) to reduce the endogenous levels of p62 [4] followed by stimulation with NGF. TrkA was immunoprecipitated and Western blotted with anti-TrkA and anti-Rpt1 to detect the interaction of TrkA with proteasomes (Fig. 4A). Control cells treated with NGF revealed TrkA interaction with Rpt1. However, in lysates with diminished p62 expression TrkA failed to interact with Rpt1, consistent with earlier findings. Furthermore, to examine whether Rpt1 is indeed capturing ubiquitinated TrkA, K63R and K48R ubiquitin mutants were expressed prior to treatment with NGF. TrkA is K63 polyubiquitinated [2,10]; expression of K63R ubiquitin mutant but not K48R ubiquitin mutant blocked TrkA interaction with Rpt1 (Fig. 4A). Collectively, these findings reveal that Rpt1 is interacting with the ubiquitinated form of TrkA. To further examine the role of p62 in TrkA/Rpt1 interaction, cortico-hippocampal neurons from WT and p62 −/− mice were transfected with GFP-Rpt1 and HA-TrkA, stimulated, stained and examined by confocal microscopy. In WT neurons, the colocalization of TrkA, actin and Rpt1 was visualized; however, the interaction of TrkA and Rpt1 was absent in p62 −/− neurons (Fig. 4B). Rpt1 is also known to function in the binding of nascent misfolded proteins [9, 18]. We next examined whether p62 played a significant role in regulating the turnover of the TrkA receptor. PC12 cells were transfected with ASp62 or p62 constructs to over express (OE) the protein followed by treatment with the translation inhibitor cycloheximide (Fig. 4C). Cell lysates were prepared and the expression of TrkA and p62 was analyzed by immunoblotting. Diminished p62 expression enhanced the stability of TrkA compared to control or cells overexpressing p62. Likewise, expression of the K63R but not the K48R mutant ubiquitin construct impaired TrkA turnover (data not shown). Altogether, these results reveal that p62 plays a role in the presentation of ubiquitinated TrkA to the proteasome for deubiquitination and therefore regulates TrkA degradation.
Fig. 4. TrkA turnover is p62 dependent.
(A) PC12 cells were transfected with antisense (AS) p62, HA-tag K63R or K48R mutant ubiquitin constructs. The cells were treated with NGF (50 ng/ml) for 60 min or not before lysing the cells in Triton lysis buffer. Proteins (750 µg) were immunoprecipitated (IP) with anti-TrkA (C-14) and analysed by Western blotting (WB) with TrkA (B-3) and Rpt1 antibody. The expression of TrkA, p62 and ubiquitin constructs in the lysates (50 µg) was determined by Western blotting. (B) Cortico-hippocampal neurons transfected with GFP-Rpt1, HA-TrkA were stimulated with 60 mM K+ for 15 min followed by fixation and staining for actin with phallodin (red), or HA for TrkA. Secondary antibody tagged with CY5 (Molecular Probes, Eugene, OR) was employed to detect HA-TrkA. The merged image is shown. When all three proteins colocalize (red-green-blue) a lavender image appears, whereas miscolocalization of GFP-Rpt1 results in red-blue that produces a violet image. (C) PC12 cells were transfected either with ASp62 or p62 (OEp62). Thirty-six hours post-transfection, the cells were treated with 20 µg of cycloheximide at 37°C for the indicated time periods and lysed with Triton lysis buffer. Equal amounts of cell lysate (50 µg) were Western blotted (WB) with anti-TrkA (B-3) to assess the turnover of endogenous TrkA. The TrkA blot was scanned and the relative expression of TrkA is represented in the graphical plot. The lysates were analyzed for protein expression of p62. (D) Schematic representation of TrkA/p62 proteasomal shuttling. Ubiquitinated TrkA [2] interacts with p62's UBA domain [10] and is targeted by the UbL domain of p62 to the proteasome by interacting with Rpt1. These findings (A–C) are representative of three other independent experiments.
Our findings with TrkA are in contrast with recent reports on centrin/Cdc31 a newly identified shuttling factor which interact with both the proteasome and multiubiquitinated proteins [7]. Cdc31 binds to Rpt1, Rpt4 and Rpt6 subunits of 19S regulatory particle but does not require prior attachment to a multiubiquitin chain. In contrast, p62 binds Rpt1 when loaded with K63 ubiquitinated cargo such as TrkA (Fig. 4D). In this regard, the function of p62 is similar to other proteins containing UBA and UbL domains such as: Ddi1, Rad23 and Dsk2 in shuttling of substrates to the proteasome [8,19,20]. There is growing evidence for local degradation of proteins in neuronal spines to fine tune synaptic architecture [21]. Given that p62 can also regulate delivery of proteins to autophagosomes through a separate LC3 interacting region (LIR) [22], reveals that p62 could thereby preserve protein degradation capabilities in circumstances where activity of the proteasome is impaired. Such conditions are common in numerous neurodegenerative diseases. Our results reveal that deubiquitination and degradation [3] of a defined K63-polyubiquitinated substrate, TrkA [10], is dependent upon p62.
Acknowledgements
This study was funded by NIH Grants NS33661 (MWW) and CA83875 (KM). We thank Dr. Erin M. Schuman, Cal. Tech. for the GFP-Rpt1 construct.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Grimes ML, Zhou J, Beattie EC, Yuen EC, Hall DE, Valletta JS, Topp KS, LaVail JH, Bunnett NW, Mobley WC. Endocytosis of activated TrkA: evidence that nerve growth factor induces formation of signaling endosomes. J. Neurosci. 1996;16:7950–7964. doi: 10.1523/JNEUROSCI.16-24-07950.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geetha T, Jiang J, Wooten MW. Lysine 63 polyubiquitination of the nerve growth factor receptor TrkA directs internalization and signaling. Mol. Cell. 2005;20:301–312. doi: 10.1016/j.molcel.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 3.Geetha T, Wooten MW. TrkA receptor endolysosomal degradation is both ubiquitin and proteasome dependent. Traffic. 2008;9:1–11. doi: 10.1111/j.1600-0854.2008.00751.x. [DOI] [PubMed] [Google Scholar]
- 4.Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR, Wooten MW. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol. Cell. Biol. 2004;24:8055–8068. doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamark T, Perander M, Outzen H, Kristiansen K, Overvatn A, Michaelsen E, Bjorkoy G, Johansen T. Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. J. Biol. Chem. 2003;278:34568–34581. doi: 10.1074/jbc.M303221200. [DOI] [PubMed] [Google Scholar]
- 6.Hirano Y, Yoshinaga S, Ogura K, Yokochi M, Noda Y, Sumimoto H, Inagaki F. Solution structure of atypical protein kinase C PB1 domain and its mode of interaction with ZIP/p62 and MEK5. J. Biol. Chem. 2004;279:31883–31890. doi: 10.1074/jbc.M403092200. [DOI] [PubMed] [Google Scholar]
- 7.Chen L, Madura K. Centrin/Cdc31 is a novel regulator of protein degradation. Mol. Cell. Biol. 2008;28:1829–1840. doi: 10.1128/MCB.01256-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L, Madura K. Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol. Cell. Biol. 2002;22:4902–4913. doi: 10.1128/MCB.22.13.4902-4913.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chuang SM, Madura K. Saccharomyces cerevisiae Ub-conjugating enzyme Ubc4 binds the proteasome in the presence of translationally damaged proteins. Genetics. 2005;171:1477–1484. doi: 10.1534/genetics.105.046888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wooten MW, Geetha T, Babu JR, Seibenhener ML, Peng J, Cox N, Diaz-Meco MT, Moscat J. Essential role of SQSTM1/p62 in trafficking of K63-ubiquitinated proteins. J. Biol. Chem. 2008;283:6783–6789. doi: 10.1074/jbc.M709496200. [DOI] [PubMed] [Google Scholar]
- 11.Rodriguez A, Duran A, Selloum M, Champy M, Diez-Guerra F, Flores J, Serrano M, Auwerx H, Diaz-Meco MT, Moscat J. Mature-onset obesity and insulin resistance in mice deficient in the signaling adapter p62. Cell Metab. 2006;3:211–222. doi: 10.1016/j.cmet.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 12.Bardag-Gorce F, Venkatesh R, Li J, French BA, French SW. Hyperphosphorylation of rat liver proteasome subunits; the effects of ethanol and okadaic acid are compared. Life Sci. 2004;75:585–597. doi: 10.1016/j.lfs.2003.12.027. [DOI] [PubMed] [Google Scholar]
- 13.Matsuda S, Miura E, Matsuda K, Kakegawa K, Kohda K, Watanabe M, Yuzaki M. Accumulation of AMPA receptors in autophagosomes in neuronal axons lacking adaptor protein AP-4. Neuron. 2008;57:730–745. doi: 10.1016/j.neuron.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 14.Funakoshi M, Sasaki T, Nishimoto T, Kobayashi H. Budding yeast Dsk2p is a polyubiquitin-binding protein that can interact with the proteasome. Proc. Natl. Acad. Sci. USA. 2002;99:745–750. doi: 10.1073/pnas.012585199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verma R, Oania R, Graumann J, Deshaies RJ. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell. 2004;118:99–110. doi: 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 16.Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried VA, Finley D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- 17.Rubin DM, Glickman MH, Larsen CN, Dhruvakumar S, Finley D. Active site mutants in the six regulatory particle ATPases reveal multiple roles for ATP in the proteasome. EMBO J. 1998;17:4909–4919. doi: 10.1093/emboj/17.17.4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chuang SM, Chen L, Lambertson D, Anand M, Kinzy TG, Madura K. Proteasome-mediated degradation of cotranslationally damaged proteins involves translation elongation factor 1A. Mol. Cell. Biol. 2005;25:403–413. doi: 10.1128/MCB.25.1.403-413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clarke DJ, Mondesert G, Segal M, Bertolaet BL, Jensen S, Wolff M, Henze M, Reed SI. Dosage suppressors of pds1 implicate ubiquitin-associated domains in checkpoint control. Mol. Cell. Biol. 2001;21:1997–2007. doi: 10.1128/MCB.21.6.1997-2007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rao H, Sastry A. Recognition of specific ubiquitin conjugates is important for the proteolytic functions of the ubiquitin-associated domain proteins Dsk2 and Rad23. J. Biol. Chem. 2002;277:11691–11695. doi: 10.1074/jbc.M200245200. [DOI] [PubMed] [Google Scholar]
- 21.Bingol B, Schuman EM. Activity-dependent dynamics and sequestration of proteasomes in dendritic spines. Nature. 2006;44:1144–1148. doi: 10.1038/nature04769. [DOI] [PubMed] [Google Scholar]
- 22.Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 2005;171:601–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]