Abstract
The accurate formation of cognate aminoacyl-tRNAs (aa-tRNAs) is essential for the fidelity of translation. Most amino acids are esterified onto their cognate tRNA isoacceptors directly by aminoacyl-tRNA synthetases (aaRSs). However, in the case of four amino acids (Gln, Asn, Cys and Sec), aminoacyl-tRNAs are made through indirect pathways in many organisms across all three domains of life. The process begins with the charging of noncognate amino acids to tRNAs by a specialized synthetase in the case of Cys-tRNACys formation or by synthetases with relaxed specificity such as the non-discriminating glutamyl-tRNA synthetase (ND-GluRS), non-discriminating aspartyl-tRNA synthetase (ND-AspRS) and seryl-tRNA synthetase (SerRS). The resulting misacylated tRNAs are then converted to cognate pairs through transformation of the amino acids on the tRNA, which is catalyzed by a group of tRNA-dependent modifying enzymes such as tRNA-dependent amidotransferases, Sep-tRNA:Cys-tRNA synthase (SepCysS), O-phosphoseryl-tRNA kinase (PSTK) and Sep-tRNA:Sec-tRNA synthase (SepSecS). The majority of these indirect pathways are widely spread in all domains of life and thought to be ancient in the course of evolution.
Keywords: aminoacyl-tRNA, indirect pathways, tRNA-dependent amidotransferase, tRNA-dependent cysteine biosynthesis, selenocysteine biosynthesis
In translation, aminoacyl-tRNAs (aa-tRNAs) are employed to convert genetic information stored in mRNA sequence to the three dimensional information manifested in the resulting proteins. Aminoacyl-tRNA synthetases (aaRSs) play a crucial role in maintaining the fidelity of translation by matching each standard amino acid found in proteins to the corresponding tRNA isoacceptors and forming a cognate aa-tRNA pair. The aminoacylation reaction is carried out as a two-step process [1].
ATP+amino acid+AARS → AARS:amino acid~AMP+PPi
tRNA+AARS:amino acid~AMP → AARS+aminoacyl-tRNA+AMP
The first step is the activation of an amino acid with ATP. The aaRSs produce an aminoacyl adenylate by attaching the carboxyl group in the amino acid to the phosphoryl group of AMP. In the second step, the activated amino acid is transferred to the 2′ or 3′ hydroxyl group of the 3′ terminal ribose moiety of tRNA followed by release of the final product, aminoacyl-tRNA. In the classical view, 20 aaRSs catalyze the formation of 20 different aa-tRNA pairs. Each synthetase specifically recognizes a set of tRNA isoacceptors and charges them with the correct amino acid that corresponds to the anticodons of the tRNA molecules.
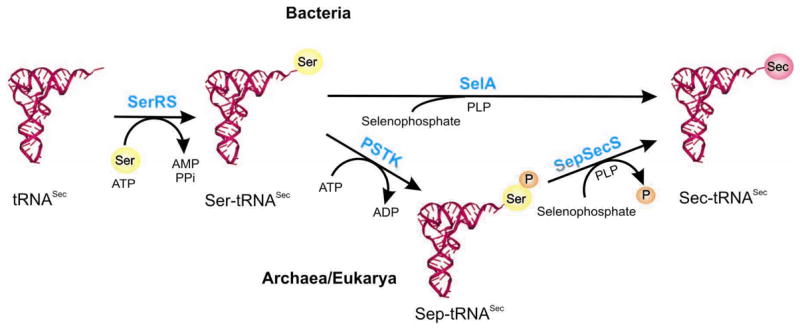
The first exception to this one synthetase/one set of tRNAs/one amino acid rule was discovered 40 years ago [2], when it was shown that in Bacillus Gln-tRNAGln is synthesized from Glu-tRNAGln instead of direct acceptance of Gln on tRNAGln. Thirty years later, the nature of the enzymes catalyzing such tRNA-dependent amino acid transformations was uncovered [3]. With the advance in functional genomics, biochemical and genetic analyses, the indirect pathways for Gln-tRNAGln, Asn-tRNAAsn, Cys-tRNACys and Sec-tRNASec formation have been characterized [4–7]. They all require two types of enzymes. One is aminoacyl-tRNA synthetases which can form misacylated intermediates. The second type is the tRNA-dependent amino acid modifying enzymes which convert the tRNA bound amino acid to form ultimately the cognate aa/tRNA pair. Organisms that posses one or more of these indirect pathways do not have to encode the full set of 20 aaRSs [5, 8–13], once thought to be essential for all living species.
The occurrence of tRNA-dependent amino acid transformations is surprisingly widespread throughout all three domains of life (Table 1). All known archaea [5], most bacteria [8] and chloroplasts [14,15] encode the indirect pathway for Gln-tRNAGln formation. A ND-GluRS aminoacylates tRNAGln with Glu [16,17]. A Glu-tRNAGln amidotransferase (Glu-AdT) then recognizes the mischarged species and transforms it to Gln-tRNAGln in the presence of ATP and an amide donor [Fig. 1(A)] [5]. Likewise, for Asn formation, Asp is first ligated to tRNAAsn by a ND-AspRS [17,18] and then converted to Asn on the tRNA by an Asp-tRNAAsn amidotransferase (Asp-AdT) [Fig. 1(B)] [18,19]. The ND-AspRS/Asp-AdT pathway is present in most bacteria and archaea [5,8,9]. In a large subset of euryarchaeota, a Cys-tRNACys is formed via an O-phosphoseryl-tRNACys (Sep-tRNACys) intermediate catalyzed by a noncanonical synthetase, SepRS. The tRNA bound O-phosphoserine (Sep) is then converted to Cys by SepCysS in the presence of a sulfur donor (Fig. 2) [6]. Selenocysteine formation pathways are exclusively tRNA-dependent and found in all Sec-decoding organisms [13]. In archaea and eukaryotes, it is a multistep process involving SerRS, PSTK and SepSecS (Fig. 3) [7,20]. Ser is esterified onto tRNASec by SerRS and then phosphorylated by PSTK on the tRNA forming Sep, which is further converted to Sec by SepSecS. The archaeal and eukaryotic pathway is different from the bacterial one where the mischarged Ser-tRNASec is directly converted to Sec-tRNASec by selenocysteine synthase (SelA) [13]. In this review, we summarize the latest progress in characterizing the enzymes involved in tRNA-dependent amino acid transformations and the current evolutionary views of these pathways.
Table 1.
| aa-tRNA | Prevalence
|
Reference | |
|---|---|---|---|
| Indirect pathway | Direct Pathway | ||
| Gln-tRNAGln | All known archaea, most bacteria, and chloroplasts | All known eukaryotes and a minority of bacteria | [5,8,14,15] |
| Asn-tRNAAsn | Most bacteria and archaea | All known eukaryotes and a number of bacteria and archaea | [5,8,9,24] |
| Cys-tRNACys | Methanogenic archaea (except M. smithii and M. stadtmanae) and A. fulgidus | All known eukaryotes and bacteria, and most archaea | [6,10–12,61,66–68] |
| Sec-tRNASec | All known Sec-decoding eukaryotes, archaea and bacteria | None known | [98–100,110] |
Prevalence of the indirect and direct pathways for the synthesis of the aa-tRNA species listed in all three domains of life. The table was adapted from [132] and is reprinted with permission from Oxford University Press.
Fig. 1. The tRNA-dependent pathways for Gln-tRNAGln (A) and Asn-tRNAAsn (B) formation.
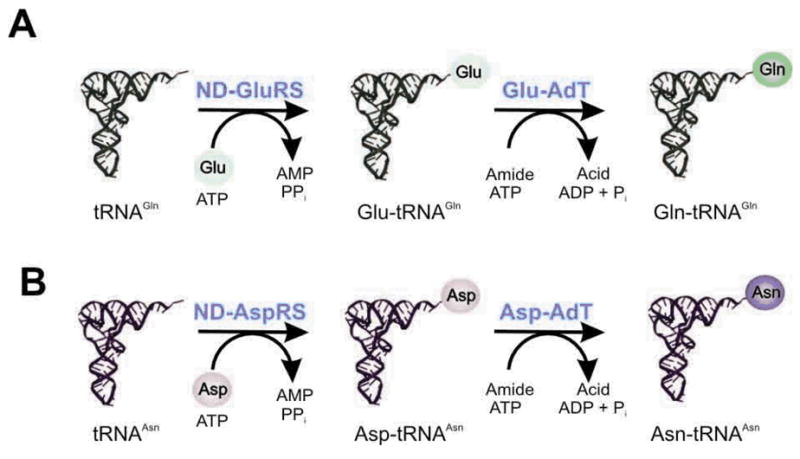
Fig. 2. The tRNA-dependent Cys-tRNACys formation.
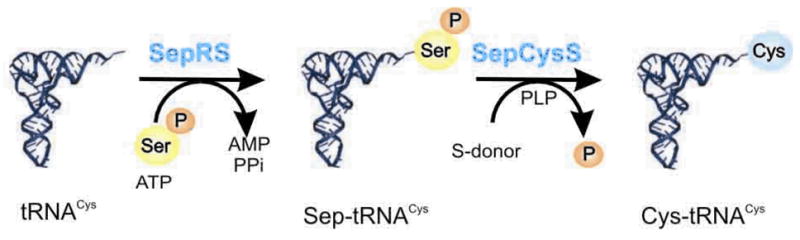
Fig. 3. The selenocysteine biosynthetic pathways in archaea and eukaryotes.
Bacterial tRNA-dependent amidotransferase and tRNA recognition
In bacteria, the tRNA-dependent amidation of Glu and Asp to form Gln-tRNAGln and Asn-tRNAAsn is catalyzed by the same enzyme: the heterotrimeric GatCAB [3]. The exact functional role of the bacterial GatCAB in vivo is determined by the availability of its misacylated substrates Glu-tRNAGln and Asp-tRNAAsn [3,4,21]. In bacteria that only have a ND-GluRS (e.g. Bacillus subtilis) [16], Glu-tRNAGln is generated and GatCAB is exclusively used as a Glu-AdT to transform Glu-tRNAGln to Gln-tRNAGln [3], while in bacteria that only encode a ND-AspRS (e.g. Deinococcus radiodurans, Neisseria meningitides, Pseudomonas aeruginosa and Thermus thermophilus), GatCAB functions as an Asp-AdT, synthesizing Asn on tRNAAsn [4,18,21–26]; however, in bacteria possessing both a ND-GluRS and a ND-AspRS (e.g. Chlamydia trachomatis [27] and Helicobacter pylori [28–30]), GatCAB catalyzes both tRNA-dependent transamidation reactions in vivo [8,27,31].
In general, the anticodon region of the tRNAGln and tRNAAsn is not the major identity element for the recognition by tRNA-dependent amidotransferases [26,32–34]. In order to discriminate against tRNAAsp and tRNAGlu, bacterial GatCAB recognizes the first base pair in the tRNA acceptor stem, which is U1-A72 in tRNAGln [32] and tRNAAsn [26] in contrast to G1-C72 in tRNAAsp and tRNAGlu. Additionally, the supernumerary nucleotide U20A in the D-loop of tRNAAsp and tRNAGlu excludes them as substrates for GatCAB [32]. In N. meningitidis, the mutation of U1-A72 to G1-C72 in tRNAAsn leads to a 100-fold decrease of transamidation activity, while the transplantation of the U1-A72 into tRNAAsp together with the deletion of the U20A converts tRNAAsp into a substrate that behaves similarly to the wild type tRNAAsn for bacterial GatCAB [26]. Sequence comparison of bacterial tRNAGlu, tRNAGln, tRNAAsp and tRNAAsn showed that the first base pair of tRNAAsn and tRNAGln is conserved as U1-A72 in all bacteria encoding tRNA-dependent pathways for Gln and Asn biosynthesis, suggesting that GatCAB utilizes one general mechanism to maintain its substrate specificity [26,32].
Archaeal tRNA-dependent amidotransferases and tRNA recognition
Two types of tRNA-dependent amidotransferases (AdTs) are found in Archaea: the heterodimeric GatDE and the archaeal GatCAB [5]. GatDE is the archaeal Glu-AdT [5]. It exclusively recognizes archaeal tRNAGln and synthesizes glutamine on the tRNA. The archaeal GatCAB only exists in archaea lacking an asparaginyl-tRNA synthetase (AsnRS) [5,9]. In vitro the Methanothermobacter thermautotrophicus GatCAB only acts on Asp-tRNAAsn but not on Glu-tRNAGln suggesting that the archaeal GatCAB has lost its dual function and acts strictly as an Asp-AdT [35]. The advantage of a more specialized archaeal GatCAB compared to its bacterial homologue awaits further investigation.
The tRNA recognition mode of archaeal GatCAB is divergent from the bacterial enzyme. The archaeal GatCAB does not recognize the first base pair of the tRNA substrate; instead, in the case of M. thermautotrophicus GatCAB, it discriminates against tRNAAsp by recognizing U49 and the D-loop as major anti-determinant elements [33]. A mutation of A49 to U49 in the wild-type tRNAAsn leads to more than 99.5% loss of activity in the archaeal GatCAB catalyzed transamidation reaction. Additionally, the length of the variable loop acts as an identity element in tRNAAsn recognition as shown for the M. thermautotrophicus and Methanosarcina barkeri enzymes [26,33]. While the recognition of tRNAGln by the archaeal Glu-AdT, GatDE, relies on the same regions (i.e. the first base pair and the D-loop) as the bacterial GatCAB, the bases recognized are different. GatDE recognizes the first base pair of archaeal tRNAGln conserved as A1-U72, whereas bacterial GatCAB recognizes the U1-A72 base pair of bacterial tRNAGln or tRNAAsn [34]. Mutation of 19 or A20 in the D-loop of M. thermautotrophicus tRNAGln significantly decreases transamidation catalyzed by GatDE [34].
The catalytic mechanism of tRNA-dependent amidotransferases
Transamidation is an ATP-dependent, multistep reaction requiring the presence of an amide donor such as glutamine or asparagine. Despite of the difference in tRNA specificity and natural distribution, GatCAB and GatDE use the same mechanism to catalyze the tRNA-dependent transamidation (Fig. 4). It consists of three sub-reactions: (a) the activation of the amide acceptor (tRNA bound Glu or Asp) at the expense of ATP hydrolysis, forming γ-phosphoryl-Glu-tRNAGln or possibly β-phophoryl-Asp-tRNAAsn as reaction intermediates [15,36–38]. (b) The hydrolysis of an amide donor Gln or Asn to form enzyme captivated ammonia. (c) The transfer of sequestered ammonia to the activated intermediate to form the final product Gln-tRNAGln or Asn-tRNAAsn [38–41]. The kinase (a) and the glutaminase activity (b) of AdTs are tightly coupled upon binding of the misacylated tRNA substrate [8,38,40,42].
Fig. 4. Transamidation reactions to form Gln-tRNAGln (A) and Asn-tRNAAsn (B).
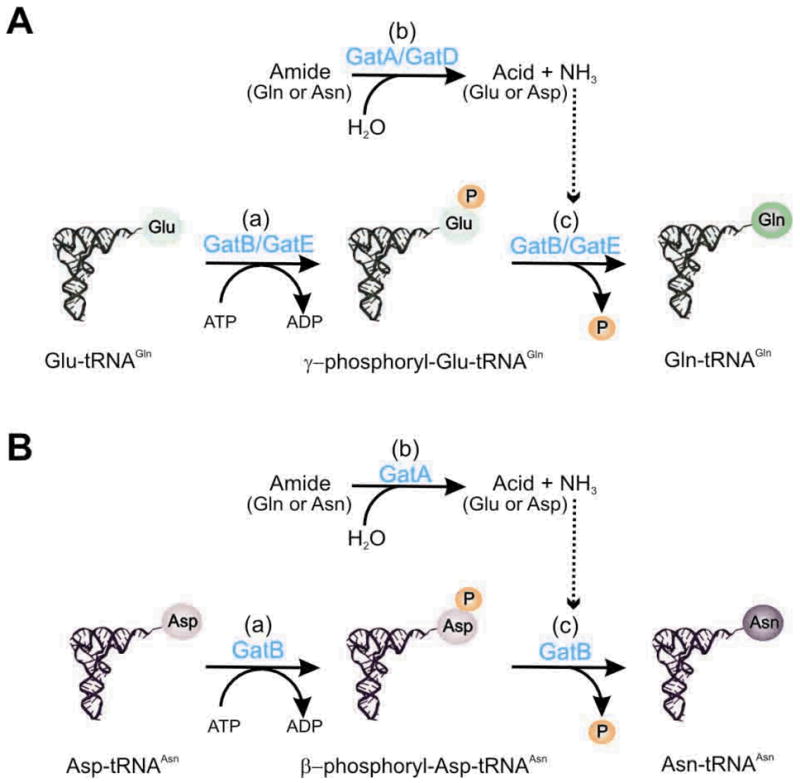
GatCAB and GatDE use the same mechanism to catalyze the tRNA-dependent transamidation. It consists of three sub-reactions: (a) the activation of the amide acceptor (tRNA bound Glu or Asp) at the expense of ATP hydrolysis, forming γ-phosphoryl-Glu-tRNAGln or possibly β-phophoryl-Asp-tRNAAsn as reaction intermediate. (b) The hydrolysis of an amide donor Gln or Asn to form enzyme captivated ammonia. (c) The transfer of sequestered ammonia to the activated intermediate to form the final product Gln-tRNAGln or Asn-tRNAAsn.
GatDE forms an α2β2 tetramer in solution and crystalline condition [Fig. 5(A)] [34,42]. The functional role of each subunit in GatDE is well elucidated. The D subunit carries out the glutaminase activity [5,38,42], which releases ammonia from Gln as well as Asn [8,35]. It consists of three domains. AnsA-like domain 1 and AnsA-like domain 2 connect through a long linker loop to the N-terminal domain that is involved in the binding of the E subunit [34,42]. The D subunit forms a tightly packed dimer with a large surface contact area located at AnsA-like domains between the two protomers. In the dimer interface, two amidase catalytic centers are formed by AnsA-like domain1 and AnsA-like domain 2 from the other subunit [42], where two highly conserved threonine residues, one aspartic acid and a lysine are crucial for the GatD catalyzed glutaminase activity [38]. GatD only hydrolyzes Gln in the presence of the E subunit and Glu-tRNAGln, which couples the hydrolysis of Gln with the activation of tRNA bound Glu, thus preventing an otherwise futile deamination of Gln and the accumulation of free ammonia [38].
Fig. 5. The crystal structure of GatDE from Pyrococcus abyssi (A) and M. thermautotrophicus GatDE complexed to tRNAGln (B).

The ribbon diagram shows GatDE α2β2 dimer with GatD consisting of N-terminal domain (navy), AsnA-like domain 1 (cyan) and AsnA-like domain 2 (royal blue). GatE (A) interacts with tRNA (B) and contains the cradle domain (brown), AspRS-like insertion domain (yellow) and helical domain (copper). The YqeY C-terminal domain in GatE is disordered and therefore not shown in both structures.
GatE interacts with the misacylated tRNA substrate [34,38]. In the presence of ATP, the E subunit alone is able to activate Glu-tRNAGln, forming γ-phosphoryl-Glu-tRNAGln intermediate [38]. Its unique structure consists of a cradle domain, an AspRS-like insertion domain, a helical domain and a C-terminal domain homologous to the YqeY protein family, which may enhance protein affinity towards its tRNA substrate [34,42]. A similar C-terminal YqeY domain appended to the D. radiodurans GlnRS enables the enzyme to bind to tRNAGln productively [43]. The helical domain and the C-terminal domain change their orientation upon tRNA binding and form a concave surface to accommodate the elbow region of the tRNA substrate [Fig. 5(B)] [34]. The lack of tRNAGln-specific and base-specific interactions in this region of the tRNA indicates that a shape-complementary mechanism as the indirect readout of tRNAGln is the main factor for GatE to differentiate tRNAGln from tRNAGlu and tRNAAsn [34]. The cradle domain interacts with the ACCA-terminus of tRNAGln and guides the attached Glu to the catalytic center for the phosphorylation and the subsequent transamidation reaction [34]. The kinase activity requires the presence of Mg2+ ions. Mutations of Mg2+ binding residues in the M. thermautotrophicus GatE subunit drastically affect the enzyme affinity to Mg2+ and abolish the kinase and transamidase activities in vitro [34].
GatCAB and GatDE are evolutionarily related through their kinase domains (GatB and GatE) [5,44]. The GatB subunit containing the catalytic pocket of the kinase and transamidase activities is also involved in tRNA binding [32]. Structurally, bacterial GatB is made of three domains: the cradle domain, the helical domain and the C-terminal YqeY domain, which was shown to participate in tRNA substrate binding [43]. Archaeal GatB is expected to have a different tRNA recognition elements compared to its bacterial homologue as the archaeal GatCAB is a strict Asp-AdT. The detailed interaction map between GatCAB and substrate tRNA is still unknown.
The bacterial GatCAB alone has a basal glutaminase activity [32,40] which is enhanced significantly in the presence of its mischarged tRNA substrate and ATP [8,40]. GatA belongs to the amidase family. Functionally similar to GatD, it generates active ammonia from an amide donor. The A subunit is a single domain protein possessing Ser-cis-Ser-Lys as the catalytic triad. In the Staphylococcus aureus GatCAB crystal structure, a glutamine molecule located in close proximity to the catalytic triad further proves that the function of subunit A is to liberate ammonia from the amide donor glutamine. Unlike GatD, bacterial GatA prefers Gln as the amide donor to Asn [8,15,45,46]. The shorter side chain in Asn prevents the γ-carboxyl carbon from interacting with the active site Ser and forming a tetrahedral intermediate [32] and thus reduces the deamination efficiency [8,32,46]. The archaeal GatA on the contrary does not seem to have a preference regarding the amide donor as the M. thermautotrophicus enzyme can use Gln or Asn with almost equal efficiency [35], which suggests a possible different arrangement of the active site in archaeal GatA.
GatC is a 10 kDa small protein responsible for stabilizing the GatCAB trimeric protein complex. In the crystal structure of S. aureus GatCAB (Fig. 6), the C subunit shapes as an extended loop with two α helixes at its N-terminus and two β strands at its C-terminus [32]. GatC stabilizes the GatAB complex by extensively interacting with both subunits at the GatA/GatB interface [32]. The proper folding of the GatA subunit also requires the presence of GatC [3].
Fig. 6. The crystal structure of S. aureus GatCAB.
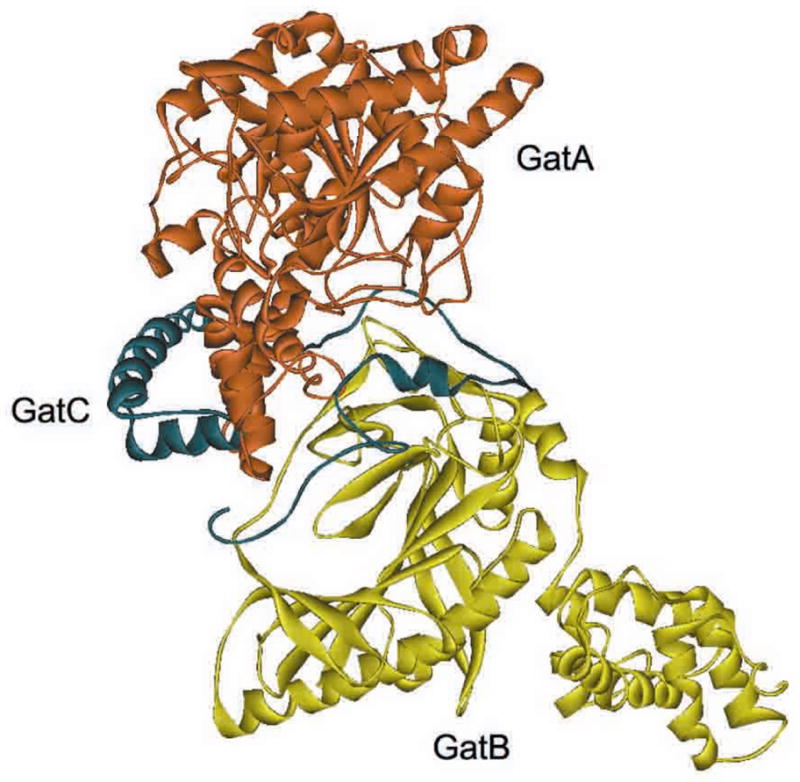
A ribbon presentation of S. aureus GatCAB structure with GatA colored red, GatB colored yellow and GatC colored blue. The YqeY C-terminal domain in GatB is disordered and not shown in the structure.
Gated ammonia channel in tRNA-dependent amidotransferases
A most notable feature in the crystal structures of S. aureus GatCAB and M. thermautotrophicus GatDE is a long protein tunnel (30 Å and 40 Å respectively) connecting the glutaminase activity center in the GatA or GatD subunit to the kinase/transamidase active site in the GatB or GatE subunit [32,34]. The molecular tunnel is made of continuous hydrophilic residues with highly conserved positive and negative residues alternating on the inner surface and surrounded by hydrophobic residues on the outside [32,34]. Ammonia generated in the deaminase center is expected to travel to the transamidase center for the transamidation reaction to occur [32,34,42]. The hydrophilic property of the ammonia tunnel in AdTs suggests that ammonium (NH4+) instead of ammonia (NH3) is transported. The transport is proposed to be carried out through alternating protonation and deprotonation of the ammonium ion [32]. A continuous desolvation of ammonium ions followed by the passage into the tunnel may push ammonium ions to move towards the subsequent reaction center which mimics the mechanism of K+ transport in the potassium channel [34,47].
The coupling of glutaminase and kinase/transamidase activities has been observed in both GatCAB and GatDE [8,38,40,42]. Regarding GatDE, only in the presence of misacylated tRNA substrate does the hydrolysis of Gln or Asn amide donor occur [38,42]. The binding of the tRNA substrate induces a significant conformational change in the D subunit where a catalytic Thr that is 7 Å away from the active site in the apo enzyme moves to the active position [42]. The ammonia tunnel also undergoes conformational changes and it is proposed to switch from a closed to an open state upon binding of the misacylated tRNA [32,34,42]. Unlike GatDE, GatCAB has a less tight coupling between these two subreactions as mentioned earlier [8,32,40]. The lack of a complex structure of GatCAB and tRNA leaves many questions open in this area.
Complex between tRNA, nondiscriminating-aaRS and tRNA-dependent amidotransferase
Several mechanisms have been proposed to maintain the fidelity of translation and to prevent the misacylated tRNAs generated during the described tRNA-dependent amino acid transformations from participating in decoding, such as EF-Tu discriminating against misacylated tRNAs [48] and substrate channeling [14,49]. The first mechanism is based on the diverse binding affinity of EF-Tu towards different tRNA species and the attached amino acids [50]. The cognate aminoacyl-tRNAs bind EF-Tu with similar affinity by thermodynamic compensation, whereas the matching amino acid of a strong binding tRNA is a weak binder to EF-Tu and vice versa [48]. The tRNAGln and tRNAAsn have weaker affinity toward EF-Tu than tRNAGlu and tRNAAsp [48]. Therefore, Glu-tRNAGln and Asp-tRNAAsn as weak/weak combinations are expected to have considerably less affinity for EF-Tu than cognate aminoacyl-tRNAs and thus they are less likely to be involved in translation. Recently, some evidence has been reported to support a substrate channeling mechanism. Structurally, ND-GluRS from T. thermophilus can be docked easily onto M. thermautotrophicus GatDE:tRNAGln, forming a ternary complex [34]. The CCA end of the tRNAGln in the active center of ND-GluRS can move to the kinase/transamidase center in GatE via a simple flip motion resembling the movement of a tRNA and its aaRS with an editing domain [34]. A similar complex of ND-AspRS, GatDE, and tRNA cannot be constructed due to large steric clashes. Biochemically, a stable complex of T. thermophilus ND-AspRS, GatCAB and Asp-tRNAAsn has been observed in vitro and in vivo [51]. The presence of a ND-aaRS reduces the Km of GatCAB for Asp-tRNAAsn and stabilizes Asp-tRNAAsn as well as the final product Asn-tRNAAsn [51,52]. Compared to the free enzymes, complex formation also increases the kcat of ND-AspRS. Substrate channeling couples aminoacylation with transamidation, thus increasing the overall reaction efficiency and preventing the incorporation of misacylated tRNA species in translation.
tRNA-dependent amidotransferase in mitochondria
A number of eukaryotes including Saccharomyces cerevisiae [53] and Homo sapiens [44] encode homologs of AdT subunits in their nuclear genomes. Several lines of evidence suggest that the indirect pathway for Gln-tRNAGln formation may also be used in the mitochondria of these eukaryotes. For example, in yeast mitochondria, the activity of Glu-AdT is present and was first detected nearly three decades ago [54]. Recently, the AdT activity was also found in mammalian (T. Suzuki, unpublished data) and plant mitochondria (A.M. Duchêne and H. Becker, unpublished data). Furthermore, the yeast AdT homologs (Pet112 and YMR293C) are essential for mitochondrial function of [55,56], and Glu-tRNAGln, the substrate of Glu-AdT, was found to be located in the mitochondria of S. cerevisiae [57]. It is intriguing, however, how Glu-tRNAGln is formed as the reaction cannot be catalyzed by the mitochondrial GluRS in vitro [58]. Additionally, cytoplasmic tRNAGln and GlnRS were shown to be imported to the yeast mitochondria as well [58]. The import of tRNAGln was also shown for H. sapiens (J. Alfonzo, unpublished data). It is unclear what may be the reasons (e.g. additional coding functionality) for the presence of dual pathways for Gln-tRNAGln formation in mitochondria.
The evolutionary view of the tRNA-dependent Gln and Asn formation pathways
The indirect pathways for Gln and Asn formation are thought to be ancient and existed in the last universal communal ancestor (LUCA), while the corresponding GlnRS and AsnRS in the direct pathways are later additions during evolution [9,59–62]. The indirect pathway couples amino acid biosynthesis with translation, and the direct pathway requires a de novo synthesis of Gln and Asn independent of tRNA. The indirect pathway for the Asn-tRNA formation can serve as the sole pathway for free Asn formation. In fact, for many organisms encoding the ND-AspRS/Asp-AdT pathway, the enzymes for Asn biosynthesis (AsnA and AsnB) are found to be absent suggesting that the presence of the indirect pathway for Asn formation is essential [8,24]. On the other hand, organisms possessing AsnA, the ammonia-dependent asparagine synthetase, Asn-tRNAAsn is always made using AsnRS through the direct pathway [8,9]. In the case of Gln, bacterial Glu-AdT prefers Gln as the amide donor [8,15,45,46]thus both direct and indirect pathways rely on the de novo biosynthesis of Gln. Archaeal Glu-AdT can use both Gln and Asn as the amide donor [5,35]. Therefore, in archaea possessing AsnA, GatDE could use Asn as the amide donor for Gln-tRNAGln formation catalyzed by the Glu-AdT, adding an additional pathway for Gln biosynthesis.
The retention of the indirect pathway for Gln formation in Archaea may be due to the unique archaeal tRNAGln [44], which cannot be recognized and aminoacylated by GlnRS from Escherichia coli or S. cerevisiae [5]. On the other hand, bacterial tRNAGln from B. subtilis, an organism that encodes the indirect pathway, is a good substrate for E. coli GlnRS [16]. In all free-living organisms, ammonium is fixed mainly through the conversion of Glu to Gln by glutamine synthetase. The free amino acid Gln also serves as an amide donor for several other biosynthetic pathways as well as a signaling molecule for the nitrogen metabolism [63,64]. A number of bacteria encoding a Glu-AdT have increased amounts of free Glu in their cells, which favors the indirect pathway for Gln-tRNAGln formation suggesting another possible reason to maintain the ancient indirect pathway. The reason why the indirect pathways have not been replaced by the direct pathways in these bacteria may include nitrogen, carbon regulation and translation fidelity. The details await further investigation.
tRNA-dependent cysteine synthesis in Archaea
In a large subset of Euryarchaeota [65], Cys is synthesized in a tRNA-dependent manner (Table 1) [6]. This indirect pathway for Cys-tRNACys formation utilizes two enzymes, O-phosphoseryl-tRNA synthetase (SepRS) and O-phosphoseryl-tRNA:cysteinyl-tRNA synthase (SepCysS) (Fig. 2) [6]. SepRS aminoacylates tRNACys with O-phosphoserine (Sep) [6]. The Sep moiety is then converted to Cys by SepCysS in the presence of an unknown sulfur donor to form Cys-tRNACys [6].
Genomic analyses revealed that SepRS and SepCysS are both encoded in the sulfate reducing archaeon Archaeoglobus fulgidus [66] and all known methanogenic archaea [65] except Methanosphaera stadtmanae [67] and Methanobrevibacter smithii [68]. In most of these archaea, CysRS is also coded for though it may not be essential [65,69]; for example, in Methanococcus maripaludis CysRS is dispensable [70].
In many of these euryarchaeal genomes the enzymes required for the formation of free Cys (i.e. tRNA-independent) biosynthesis are not encoded [6,65]. The indirect route for Cys-tRNACys formation using SepRS and SepCysS is likely the sole means for Cys biosynthesis in these organisms [6]. This is consistent with an earlier report demonstrating that Sep is a precursor for Cys biosynthesis in M. jannaschii [71]. Further studies have shown that an archaeal Sep biosynthetic pathway can provide sufficient Sep levels for Ser, cystathionine and tRNA-dependent Cys production [72]. Furthermore, knocking out sepS in M. maripaludis resulted in a Cys auxotroph [6]. Therefore, the use of SepRS and SepCysS for the tRNA-dependent Cys synthesis in these organisms likely enables coupling of protein synthesis with Cys production.
SepRS directly aminoacylates tRNACys with O-phosphoserine
SepRS is a subclass IIc aaRS like PheRS and PylRS [6,73], sharing a common ancestor with the α-subunit of PheRS [65,73]. Both biophysical and structural analyses demonstrate that SepRS is a homotetramer [74,75]. The core of this α4 assembly resembles that of PheRS and consists mostly of the four catalytic domains [74]. While the active site of SepRS is structurally similar to that of PheRS, SepRS uniquely recognizes its amino acid substrate, Sep [75]. The phosphate group of the latter is highly recognized by SepRS with each of the three non-bridging oxygen atoms forming two hydrogen bonds to residues in the enzyme’s binding pocket. Mutation of these residues in the M. maripaludis SepRS resulted in inactive mutant enzymes [74]. The recognition of the phosphate moiety includes hydrogen bonding between two non-bridging oxygens and the α-amino group of active site residues, which is unique amongst aaRSs to SepRS. Structural results suggest that dipole interactions between Sep and a central α-helix in the active site of SepRS stabilize the polar side chain of the substrate, another feature not observed in other aaRSs [75].
Each monomer of SepRS can recognize and aminoacylate tRNACys in cis [75], in contrast to PheRS where the tRNA anticodon recognition site and aminoacylation active site are found on different subunits. However, in the co-crystal structure of the A. fulgidus SepRS with tRNACys only two tRNA molecules bound to the tetramer [75]. Computer modeling though does suggest that four tRNAs could be accommodated by the complex [75]. The stoichiometry in solution of SepRS to tRNACys is not clear and awaits further investigation.
Biochemical studies revealed that M. maripaludis SepRS recognizes the same major identity elements in tRNACys (G34, C35, and A36) as the homologous CysRS [76]. Both aaRSs also use G15 and A47 in tRNACys as minor identity elements. However the base pairs G1:C72 and G10:C25, and nucleotides G37 and A59 serve as minor identity elements for only SepRS [76]. The use of similar identity elements in tRNACys recognition by both SepRS and CysRS, both of which were present in LUCA [65,73], suggests that the genetic code predates the modern aminoacylation machinery [76]. Given that SepRS, like other class II aaRSs, approaches the tRNA from the major groove side while CysRS, a class I aaRS, approaches it from the minor groove side has lead to speculation that a complex between SepRS, tRNACys, and CysRS is possible [75], though the in vivo role of such a complex is currently not clear.
Sep-tRNA:Cys-tRNA synthase catalyzed formation of Cys-tRNACys
The pyridoxal phosphate (PLP)-dependent enzyme SepCysS modifies the Sep bound to tRNACys to form Cys-tRNACys [6]. The sulfur donor for this enzyme is unknown though in vitro sulfide is sufficient [6]. The A. fulgidus SepCysS crystal structure (2.4 Å resolution) [77] revealed that it belongs to the fold type I family [78] with its large N-terminal domain being comprised of a characteristic seven stranded β-sheet which typifies this family of enzymes. In addition, the structure showed that the enzyme forms a homodimer [77]. The active site of the enzyme is formed in a large basic cleft in the dimer interface and is comprised of conserved residues from both monomers [77]. Modeling a SepCysS:Sep-tRNACys complex suggests that a conserved Arg79, His103, and Tyr 104 (A. fulgidus numbering) recognize the phosphate group of the Sep moiety of the tRNA substrate [77]. The same work implicates one of the three conserved Cys residues (39, 42 or 247, A. fulgidus numbering) in the SepCysS active site as the persulfide sulfur carrier essential for catalysis (Fig. 7) [77] though this awaits further study.
Fig. 7. The crystal structure of the active sites in A. fulgidus SepCysS and M. maripaludis SepSecS.
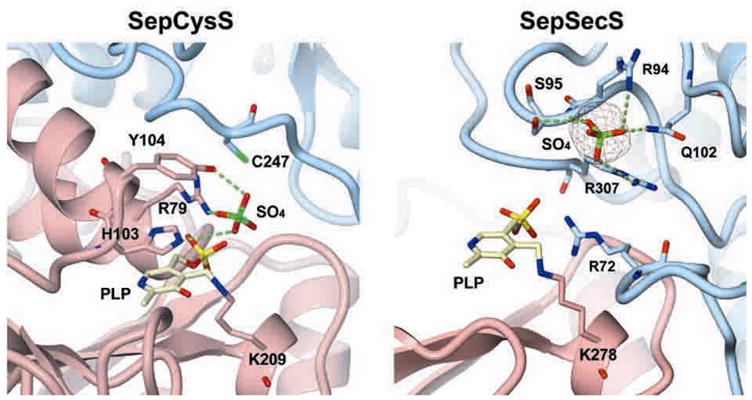
The protein active sites are presented as ribbon diagrams with essential residues highlighted as stick models. Monomers in each protein are colored pink and blue. The figure was adapted from [132] and is reprinted with permission from Oxford University Press.
The crystal structure of SepCysS with PLP alone revealed that the co-factor formed a Schiff-base linkage with the conserved Lys209 (A. fulgidus numbering) in the active site of SepCysS [77]. A hydrogen bond between SepCysS and the nitrogen atom of the ring structure of PLP is achieved through the side chain of a conserved Asn and not an Asp as is found in most PLP-dependent enzymes [77]. Nevertheless, like in other PLP-dependent enzymes, the co-factor in SepCysS is thought to stabilize the negatively charged transition state formed during catalysis of the β-replacement reaction [79].
M. maripaludis encodes both the direct and indirect paths for Cys-tRNACys synthesis. As noted above, the sole route for Cys formation is tRNACys dependent. Intriguingly while sepS (encoding SepRS) can be deleted when the organism is grown in the presence of Cys, pscS (encoding SepCysS) cannot (T. Major, M. Hohn, D. Su, W.B. Whitman, unpublished data), raising the question whether SepCysS possesses an additional function in M. maripaludis that is essential.
Cysteine synthesis in Archaea
Four different routes for Cys formation have been discovered in Archaea: the eukaryotic pathway in which the precursor is cystathionine [80], the bacterial pathway where O-acetylserine serves as the precursor [81–83], a modified bacterial pathway with free Sep as the precursor [84,85], and the tRNA-dependent route with Sep-tRNACys as the precursor [6]. Cys is implicated as the major sulfur source for a variety of biosynthetic pathways including Fe-S cluster formation, tRNA modification, and biosynthesis of co-factors in bacteria (reviewed in [86]). Fe-S cluster proteins are highly encoded in the genomes of methanogenic archaea [87]. Whether Cys generated through the tRNA-dependent pathway is used as the sulfur source is an open area of investigation. It is thought that these euryarchaea must balance the need for Cys in protein synthesis and other biosynthetic pathways by controlling the deacylation of Cys-tRNA, thus regulating the level of free Cys relative to Cys-tRNACys; however, it may well be that these methanogens which grow in environments rich in reduced sulfur compounds may be able to use inorganic sulfur directly.
Evolution of the two Cys-tRNACys biosynthetic pathways
It has been speculated that the indirect pathways for aa-tRNA formation predate the direct ones [88]. While phylogenetics supports this speculation for amide aa-tRNA synthesis (discussed above), analyses using structure-based amino acid alignments [65,73] suggest that both pathways (SepRS/SepCysS and CysRS) for Cys-tRNACys formation were present in LUCA. The phylogenetic data suggest that only early bacteria retained CysRS while the ancestral archaea possessed SepRS and that CysRS was later horizontally transferred to Archaea. In some archaeal lineages the bacterial CysRS replaced the indirect pathway for Cys-tRNACys synthesis while in many euryarchaea CysRS either coexisted with SepRS/SepCysS or was not retained [65]. Why the indirect pathway for Cys-tRNACys formation has been retained in these euryarchaea remains an open question. It is speculated that a link between Cys formation, sulfur metabolism and methanogenesis may exist that would favor retention of the tRNA-dependent route for Cys biosynthesis [65,89]though this awaits further experimental inquiry.
tRNA-dependent selenocysteine formation
Sec is the major biological form of selenium, an essential dietary trace element in humans implicated in cancer prevention [90,91]. Sec is coded as the 21st amino acid in a number of species across all three domains of life (reviewed in [13,92]). Under physiological conditions (pH 7), Sec is more stable in its ionized form than Cys due to the lower redox potential which thus lowers the pKa of the selenol group of Sec compared to the thiol group of Cys (5.2 and 8.5, respectively) [93]. Sec thus serves as an excellent nucleophile in the active sites of proteins involved in oxidation-reduction reactions.
Selenoprotein synthesis requires the formation of selenocysteinyl-tRNASec (Sec-tRNASec). No aaRS, [i.e. a selenocysteinyl-tRNA synthetase (SecRS)], has been identified in Sec-decoding organisms that can carry out the task (Table 1) and instead Sec is synthesized on tRNASec (Fig. 3). Why Sec-tRNA is only formed via indirect paths remains unknown but it may be due to selective pressures to maintain translational fidelity. CysRSs from Escherichia coli [94] and Phaseolus aureus [95] have the ability to aminoacylate tRNACys with Sec. Therefore, given the fact that misincorporation of Sec in place of Cys can be detrimental to protein function [96], the levels of free Sec are likely well regulated and kept low to prevent misacylation of tRNACys with Sec. It is also worth noting that Sec to Cys mutations lead to mutant enzymes with significantly reduced activities [97]. Synthesizing Sec on tRNASec enables formation of Sec-tRNASec while potentially minimizing the free Sec levels in vivo, preventing misacylation of tRNACys with Sec; thus the retention of the tRNA-dependent Sec pathway may be a mechanism of ensuring the accurate decoding of Cys and Sec [98].
In all known Sec-decoding organisms [99–101], tRNASec is first serylated by seryl-tRNA synthetase (SerRS) to form Ser-tRNASec [102–104]. Work in the 1990s revealed in bacteria that selenocyteine synthase (SelA) transforms Ser bound to tRNASec to Sec (Fig. 3) [13]. The Sec-tRNASec formed is then used in protein synthesis to decode UGA (usually a stop codon) when an RNA element, the selenocysteine insertion sequence element, is present in the mRNA [13]. A unique elongation factor, SelB, brings the Sec-tRNASec to the ribosome [13]. The mischarged species, Ser-tRNASec, in Sec-decoding archaea and eukaryotes (Fig. 3) is not directly modified to Sec-tRNASec, but rather the Ser moiety on the tRNA is phosphorylated by O-phosphoseryl-tRNA kinase (PSTK) to form Sep-tRNASec [105,106]. The tRNA-bound Sep is then converted to Sec by the PLP-dependent enzyme Sep-tRNA:Sec-tRNA synthase (SepSecS) [7,20].
Like SelA, SepSecS uses selenophosphate as the selenium donor to produce Sec-tRNASec in vitro and in vivo [7,20,107–109]. SepSecS is unable to use Ser-tRNASec as a substrate, recognizing only Sep-tRNASec [7,20]. However, in vitro bacterial SelA in addition to Ser-tRNASec, can convert Sep-tRNASec to Sec-tRNASec [20], though the biological relevance of this is not clear as PSTK is not encoded in bacterial genomes [7]. In all known Sec-decoding archaeal and eukaryal genomes, PSTK and SepSecS are always both encoded [110].
It is unknown why archaea and eukaryotes use Sep-tRNASec as an intermediate in tRNA-dependent Sec biosynthesis. The carboxyl ester bond between Sep and tRNASec is more stable than Ser and the tRNASec [105]. In addition, the phosphate moiety of Sep is likely a better leaving group than the hydroxyl moiety of Ser. Thus, Sep-tRNASec may serve as a better precursor for Sec-tRNASec formation than Ser-tRNASec (7).
tRNASec-dependent serine phosphorylation
Work with extracts from rat and rooster liver and lactating bovine mammary gland in the 1970s first demonstrated that Ser-tRNA could be phosphorylated [111,112]. While it was later shown with partially purified enzyme from bovine liver that the enzyme had a high affinity for tRNASec, it was only in 2004 that the protein (PSTK) was identified from mouse [105], and soon after the archaeal homolog [106].
PSTK phosphorylates Ser-tRNASec by transferring the γ-phosphate of ATP onto the Ser moiety in an Mg2+-dependent manner [105,110]. The enzyme belongs to the P-loop kinase superfamily [113], possessing a phosphate-binding loop (P-loop), a Walker B motif, and an RxxxR motif in its N-terminal domain [110]. Mutation of conserved residues in these motifs in the M. jannaschii PSTK resulted in mutant enzymes with significantly reduced activity [110]. While PSTK prefers ATP, in vitro the enzyme is able to use other NTPs (GTP, CTP, UTP and dATP) as substrates, like T4 polynucleotide kinase [110]. Similar to other members of the kinase superfamily [113], the ATPase activity of PSTK is activated in the presence of its other substrate Ser-tRNASec [110]. Interestingly, the activity is also enhanced when unacylated tRNASec is provided [110].
PSTK recognition of Sep-tRNASec
It does not appear that PSTK uses the Sep moiety of tRNASec as a major recognition element as the enzyme has a similar Kd for unacylated tRNASec as Ser-tRNASec (39 nM and 53 nM, respectively) [110]. In vivo, the concentration of tRNASec is approximately 10% of that of tRNASer [114,115], the other tRNA substrate of SerRS. It may well be that PSTK serves as a tRNASec scavenger for SerRS [110]. PSTK may also assist in maintaining translation fidelity by preventing the misacylated tRNASec intermediates from being used in protein synthesis and channeling Sep-tRNASec to SepSecS [110].
Surprisingly, it appears that archaeal and eukaryotic PSTK enzymes recognize different elements in Ser-tRNASec. While tRNASec possesses an extended variable loop like tRNASer, a major identity element for SerRS recognition of tRNA [116], it is distinct from other tRNA isoacceptors by possessing an elongated acceptor stem and D-stem [117,118]. For tRNASec recognition by eukaryotic PSTK, the major element is the length and conformation of the elongated D-stem [119]. For archaeal PSTK, the D-stem is a minor identity element and the G2-C71 and C3-G70 base pairs in the acceptor stem of archaeal tRNASec serve as the major recognition elements in the tRNA [120]. Given the deep phylogenetic divide between archaeal and eukaryotic PSTK [110], this may be a strong indication of co-evolution of PSTK and tRNASec [120].
Interestingly, while bacterial tRNASec has an 8 bp acceptor stem and a 5 bp T-stem, and archaeal and eukaryotic tRNASec have a 9 bp acceptor stem and 4 bp T-stem arrangement [117,121–123], PSTK and SepSecS can use E. coli tRNASec in vivo [7]. In turn, E. coli can use the human tRNASec in place of its own tRNASec both in vivo and in vitro [124]. It may well be that tRNASec is functionally conserved between the different domains of life despite bacteria using a different tRNA-dependent route for Sec formation than Sec-decoding archaea and eukaryotes.
Sep-tRNA:Sec-tRNA synthase catalyzed Sec-tRNASec formation
While SepSecS catalyzes a similar reaction as SepCysS, a tRNA-dependent β-replacement of Sep, a structural phylogeny revealed that SepSecS is not closely related to SepCysS nor other PLP-dependent enzymes [125]. The recently completed crystal structures of the SepSecS from M. maripaludis [125] and mouse [126] to high resolution have enabled insight into how the enzyme catalyzes the tRNA-dependent formation of Sec. SepSecS forms an (α2)2 homotetramer, mediated by an N-terminal extension in SepSecS. Each dimer has two active sites, each formed by conserved residues from both subunits in the dimer interface (Fig. 7) [125,126]. Interestingly deleting the N-terminal extension, thus apparently disrupting dimerization, gives rise to inactive SepSecS [125]. Tetramerization is speculated to also enable formation of large patches of positive electronic potential on the surface of the tetramer, which are predicted to be tRNASec binding sites [126].
As in the SepCysS active site, a conserved Asn in the SepSecS (247, M. maripaludis numbering) active site binds to the nitrogen of the ring structure of the PLP and similar to other PLP-dependent enzymes a conserved Lys (278, M. maripaludis numbering) forms a Schiff base linkage with the co-factor [125]. The phosphate group(s) of Sep-tRNASec and/or selenophosphate are proposed to interact with conserved Arg, Gln, and Ser in the active site of SepSecS [125]. Mutations to those residues results in mutant enzymes with significantly reduced activities both in vitro and in vivo [125]. SepSecS may exclude free amino acids including Sep from its active site by use of a conserved Glu which could repel the carboxyl group of free amino acids [126]. Unlike SepCysS, a conserved Cys residue is not found in the active site of SepSecS, suggesting formation of a perselenide intermediate is unlikely [125].
Relationship between tRNA-dependent cysteine and selenocysteine biosynthesis
The tRNA-dependent route for Cys biosynthesis is similar to that for Sec-tRNASec formation in archaea and eukaryotes, since in both of them Sep-tRNA serves as the final precursor prior to product formation. Both pathways were present in LUCA [7,65]. Interestingly, SepSecS can use thiophosphate in vitro to form Cys-tRNASec [125] instead of selenophosphate to synthesize Sec-tRNASec. Numerous homologs of selenoproteins are found in nature, which possess Cys in place of Sec. Given that and the similarity between Sec and Cys codons (UGA and UGY, respectively), it is interesting to speculate that a dynamic relationship exists between the two amino acids over the course of evolution [127].
Outlook
The tRNA-dependent pathways forming Gln-tRNAGln and Asn-tRNAAsn by amino acid transformations are thought to have evolved earlier than the direct aminoacylation of the tRNAs with their cognate amino acids [9,59–62]. In the case of Cys-tRNACys formation, both the direct and the indirect pathways are shown to be present in the time of LUCA [65,74,76]. Regarding to selenocysteine, Sec-tRNASec is formed only through the indirect tRNA-dependent amino acid transformations in all domains of life [99–101]. Even though it cannot be generalized as that indirect pathways are ancient pathways, it appears that indirect pathways have unique features as discussed above and are retained during evolution. For example, the tRNA-dependent Sec-tRNASec formation may provide a solution to discriminate against Cys, an extremely similar amino acid, and maintain a faithful translation.
A common feature among these indirect pathways is the existence of misacylated intermediates, which would drastically decrease the fidelity of translation if participating in decoding. Even though elongation factors bind misacylated intermediates with too low or too high affinity in vitro [21,48,50,128–130], it is still ambiguous whether the discrimination by the elongation factor alone could ensure the accuracy of translation in vivo [51,131]. Substrate channeling provides an additional mechanism to prevent misincorporation. Interestingly, complexes consist of enzymes in the same tRNA-dependent pathway and the corresponding tRNA molecules have either been observed or proposed based on computer modeling [34,51,77]. Furthermore, complex formation increases the overall reaction efficiency as well as the stability of the end product [51,52]. The tRNA-dependent amino acid transformations couple translation with the biosynthesis of amino acids that may be involved in other biological pathways [63]. To better understand the connection and regulation among different biological processes, systems biology is likely to be a very useful approach which may also lead to discovery of other exciting aspects of tRNA-dependent amino acid transformation pathways.
Acknowledgments
We thank all the current members of Dr. Dieter Söll’s laboratory for their discussion and revision of the manuscript. We thank Drs. A.M. Duchêne, T. Suzuki, H. Becker, J. Alfonzo and W.B. Whitman for communicating some of their unpublished results. Work from Dr. Dieter Söll’s laboratory reviewed in this manuscript was supported by the grants from the Department of Energy, the National Institute of General Medical Sciences, and the National Science Foundation.
References
- 1.Ibba M, Söll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 2.Wilcox M, Nirenberg M. Transfer RNA as a cofactor coupling amino acid synthesis with that of protein. Proc Natl Acad Sci USA. 1968;61:229–236. doi: 10.1073/pnas.61.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Curnow AW, Hong K, Yuan R, Kim S, Martins O, Winkler W, Henkin TM, et al. Glu-tRNAGln amidotransferase: a novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc Natl Acad Sci USA. 1997;94:11819–11826. doi: 10.1073/pnas.94.22.11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Curnow AW, Tumbula DL, Pelaschier JT, Min B, Söll D. Glutamyl-tRNAGln amidotransferase in Deinococcus radiodurans may be confined to asparagine biosynthesis. Proc Natl Acad Sci USA. 1998;95:12838–12843. doi: 10.1073/pnas.95.22.12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tumbula DL, Becker HD, Chang WZ, Söll D. Domain-specific recruitment of amide amino acids for protein synthesis. Nature. 2000;407:106–110. doi: 10.1038/35024120. [DOI] [PubMed] [Google Scholar]
- 6.Sauerwald A, Zhu W, Major TA, Roy H, Palioura S, Jahn D, Whitman WB, et al. RNA-dependent cysteine biosynthesis in archaea. Science. 2005;307:1969–1972. doi: 10.1126/science.1108329. [DOI] [PubMed] [Google Scholar]
- 7.Yuan J, Palioura S, Salazar JC, Su D, O’Donoghue P, Hohn MJ, Cardoso AM, et al. RNA-dependent conversion of phosphoserine forms selenocysteine in eukaryotes and archaea. Proc Natl Acad Sci USA. 2006;103:18923–18927. doi: 10.1073/pnas.0609703104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sheppard K, Akochy PM, Salazar JC, Söll D. The Helicobacter pylori amidotransferase GatCAB is equally efficient in glutamine-dependent transamidation of Asp-tRNAAsn and Glu-tRNAGln. J Biol Chem. 2007;282:11866–11873. doi: 10.1074/jbc.M700398200. [DOI] [PubMed] [Google Scholar]
- 9.Roy H, Becker HD, Reinbolt J, Kern D. When contemporary aminoacyl-tRNA synthetases invent their cognate amino acid metabolism. Proc Natl Acad Sci USA. 2003;100:9837–9842. doi: 10.1073/pnas.1632156100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bult CJ, White O, Olsen GJ, Zhou L, Fleischmann RD, Sutton GG, Blake JA, et al. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 11.Smith DR, Doucette-Stamm LA, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, et al. Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slesarev AI, Mezhevaya KV, Makarova KS, Polushin NN, Shcherbinina OV, Shakhova VV, Belova GI, et al. The complete genome of hyperthermophile Methanopyrus kandleri AV19 and monophyly of archaeal methanogens. Proc Natl Acad Sci USA. 2002;99:4644–4649. doi: 10.1073/pnas.032671499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Böck A, Thanbichler M, Rotherand M, Resch A. The Aminoacyl-tRNA Synthetases. Georgetown: Landes Bioscience; 2005. [Google Scholar]
- 14.Schön A, Kannangara CG, Gough S, Söll D. Protein biosynthesis in organelles requires misaminoacylation of tRNA. Nature. 1988;331:187–190. doi: 10.1038/331187a0. [DOI] [PubMed] [Google Scholar]
- 15.Jahn D, Kim YC, Ishino Y, Chen MW, Söll D. Purification and functional characterization of the Glu-tRNAGln amidotransferase from Chlamydomonas reinhardtii. J Biol Chem. 1990;265:8059–8064. [PubMed] [Google Scholar]
- 16.Lapointe J, Duplain L, Proulx M. A single glutamyl-tRNA synthetase aminoacylates tRNAGlu and tRNAGln in Bacillus subtilis and efficiently misacylates Escherichia coli tRNAGln1in vitro. J Bacteriol. 1986;165:88–93. doi: 10.1128/jb.165.1.88-93.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cathopoulis T, Chuawong P, Hendrickson TL. Novel tRNA aminoacylation mechanisms. Mol Biosyst. 2007;3:408–418. doi: 10.1039/b618899k. [DOI] [PubMed] [Google Scholar]
- 18.Becker HD, Reinbolt J, Kreutzer R, Giege R, Kern D. Existence of two distinct aspartyl-tRNA synthetases in Thermus thermophilus. Structural and biochemical properties of the two enzymes. Biochemistry. 1997;36:8785–8797. doi: 10.1021/bi970392v. [DOI] [PubMed] [Google Scholar]
- 19.Curnow AW, Ibba M, Söll D. tRNA-dependent asparagine formation. Nature. 1996;382:589–590. doi: 10.1038/382589b0. [DOI] [PubMed] [Google Scholar]
- 20.Xu XM, Carlson BA, Mix H, Zhang Y, Kazima S, Glass RS, Berry MJ, et al. Biosynthesis of Seloncystein on its tRNA in eukaryotes. PLoS Biology. 2007;5:96–105. doi: 10.1371/journal.pbio.0050004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Becker HD, Kern D. Thermus thermophilus: a link in evolution of the tRNA-dependent amino acid amidation pathways. Proc Natl Acad Sci USA. 1998;95:12832–12837. doi: 10.1073/pnas.95.22.12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Becker HD, Min B, Jacobi C, Raczniak G, Pelaschier J, Roy H, Klein S, et al. The heterotrimeric Thermus thermophilus Asp-tRNAAsn amidotransferase can also generate Gln-tRNAGln. FEBS Lett. 2000;476:140–144. doi: 10.1016/s0014-5793(00)01697-5. [DOI] [PubMed] [Google Scholar]
- 23.Becker HD, Roy H, Moulinier L, Mazauric MH, Keith G, Kern D. Thermus thermophilus contains an eubacterial and an archaebacterial aspartyl-tRNA synthetase. Biochemistry. 2000;39:3216–3230. doi: 10.1021/bi992573y. [DOI] [PubMed] [Google Scholar]
- 24.Min B, Pelaschier JT, Graham DE, Tumbula-Hansen D, Söll D. Transfer RNA-dependent amino acid biosynthesis: an essential route to asparagine formation. Proc Natl Acad Sci USA. 2002;99:2678–2683. doi: 10.1073/pnas.012027399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akochy PM, Bernard D, Roy PH, Lapointe J. Direct glutaminyl-tRNA biosynthesis and indirect asparaginyl-tRNA biosynthesis in Pseudomonas aeruginosa PAO1. J Bacteriol. 2004;186:767–776. doi: 10.1128/JB.186.3.767-776.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bailly M, Giannouli S, Blaise M, Stathopoulos C, Kern D, Becker HD. A single tRNA base pair mediates bacterial tRNA-dependent biosynthesis of asparagine. Nucleic Acids Res. 2006;34:6083–6094. doi: 10.1093/nar/gkl622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raczniak G, Becker HD, Min B, Söll D. A single amidotransferase forms asparaginyl-tRNA and glutaminyl-tRNA in Chlamydia trachomatis. J Biol Chem. 2001;276:45862–45867. doi: 10.1074/jbc.M109494200. [DOI] [PubMed] [Google Scholar]
- 28.Skouloubris S, Ribas de Pouplana L, De Reuse H, Hendrickson TL. A noncognate aminoacyl-tRNA synthetase that may resolve a missing link in protein evolution. Proc Natl Acad Sci USA. 2003;100:11297–11302. doi: 10.1073/pnas.1932482100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salazar JC, Ahel I, Orellana O, Tumbula-Hansen D, Krieger R, Daniels L, Söll D. Coevolution of an aminoacyl-tRNA synthetase with its tRNA substrates. Proc Natl Acad Sci USA. 2003;100:13863–13868. doi: 10.1073/pnas.1936123100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chuawong P, Hendrickson TL. The nondiscriminating aspartyl-tRNA synthetase from Helicobacter pylori: anticodon-binding domain mutations that impact tRNA specificity and heterologous toxicity. Biochemistry. 2006;45:8079–8087. doi: 10.1021/bi060189c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cathopoulis TJ, Chuawong P, Hendrickson TL. A thin-layer electrophoretic assay for Asp-tRNAAsn/Glu-tRNAGln amidotransferase. Anal Biochem. 2007;360:151–153. doi: 10.1016/j.ab.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakamura A, Yao M, Chimnaronk S, Sakai N, Tanaka I. Ammonia channel couples glutaminase with transamidase reactions in GatCAB. Science. 2006;312:1954–1958. doi: 10.1126/science.1127156. [DOI] [PubMed] [Google Scholar]
- 33.Namgoong S, Sheppard K, Sherrer RL, Söll D. Co-evolution of the archaeal tRNA-dependent amidotransferase GatCAB with tRNAAsn. FEBS Lett. 2007;581:309–314. doi: 10.1016/j.febslet.2006.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oshikane H, Sheppard K, Fukai S, Nakamura Y, Ishitani R, Numata T, Sherrer RL, et al. Structural basis of RNA-dependent recruitment of glutamine to the genetic code. Science. 2006;312:1950–1954. doi: 10.1126/science.1128470. [DOI] [PubMed] [Google Scholar]
- 35.Sheppard K, Sherrer RL, Söll D. Archaeal tRNAGln confines the amidotransferase GatCAB to Asn-tRNA formation. Journal of Molecular Biology. 2008;377:845–853. doi: 10.1016/j.jmb.2008.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilcox M. Gamma-phosphoryl ester of Glu-tRNAGln as an intermediate in Bacillus subtilis glutaminyl-tRNA synthesis. Cold Spring Harb Symp Quant Biol. 1969;34:521–528. doi: 10.1101/sqb.1969.034.01.059. [DOI] [PubMed] [Google Scholar]
- 37.Wilcox M. Gamma-glutamyl phosphate attached to glutamine-specific tRNA. A precursor of glutaminyl-tRNA in Bacillus subtilis. Eur J Biochem. 1969;11:405–412. doi: 10.1111/j.1432-1033.1969.tb00788.x. [DOI] [PubMed] [Google Scholar]
- 38.Feng L, Sheppard K, Tumbula-Hansen D, Söll D. Gln-tRNAGln formation from Glu-tRNAGln requires cooperation of an asparaginase and a Glu-tRNAGln kinase. J Biol Chem. 2005;280:8150–8155. doi: 10.1074/jbc.M411098200. [DOI] [PubMed] [Google Scholar]
- 39.Harpel MR, Horiuchi KY, Luo Y, Shen L, Jiang W, Nelson DJ, Rogers KC, et al. Mutagenesis and mechanism-based inhibition of Streptococcus pyogenes Glu-tRNAGln amidotransferase implicate a serine-based glutaminase site. Biochemistry. 2002;41:6398–6407. doi: 10.1021/bi012126u. [DOI] [PubMed] [Google Scholar]
- 40.Horiuchi KY, Harpel MR, Shen L, Luo Y, Rogers KC, Copeland RA. Mechanistic studies of reaction coupling in Glu-tRNAGln amidotransferase. Biochemistry. 2001;40:6450–6457. doi: 10.1021/bi002599l. [DOI] [PubMed] [Google Scholar]
- 41.Shin S, Yun YS, Koo HM, Kim YS, Choi KY, Oh BH. Characterization of a novel Ser-cisSer-Lys catalytic triad in comparison with the classical Ser-His-Asp triad. J Biol Chem. 2003;278:24937–24943. doi: 10.1074/jbc.M302156200. [DOI] [PubMed] [Google Scholar]
- 42.Schmitt E, Panvert M, Blanquet S, Mechulam Y. Structural basis for tRNA-dependent amidotransferase function. Structure. 2005;13:1421–1433. doi: 10.1016/j.str.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 43.Deniziak M, Sauter C, Becker HD, Paulus CA, Giege R, Kern D. Deinococcus glutaminyl-tRNA synthetase is a chimer between proteins from an ancient and the modern pathways of aminoacyl-tRNA formation. Nucleic Acids Res. 2007;35:1421–1431. doi: 10.1093/nar/gkl1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sheppard K, Söll D. On the evolution of the tRNA-dependent amidotransferases, GatCAB and GatDE. Journal of Molecular Biology. 2008;377:831–844. doi: 10.1016/j.jmb.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strauch MA, Zalkin H, Aronson AI. Characterization of the glutamyl-tRNAGln-to-glutaminyl-tRNAGln amidotransferase reaction of Bacillus subtilis. J Bacteriol. 1988;170:916–920. doi: 10.1128/jb.170.2.916-920.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bailly M, Blaise M, Roy H, Deniziak M, Lorber B, Birck C, Becker HD, et al. tRNA-dependent asparagine formation in prokaryotes: Characterization, isolation and structural and functional analysis of a ribonucleoprotein particle generating Asn-tRNA(Asn) Methods. 2008;44:146–163. doi: 10.1016/j.ymeth.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 47.Morais-Cabral JH, Zhou Y, MacKinnon R. Energetic optimization of ion conduction rate by the K+ selectivity filter. Nature. 2001;414:37–42. doi: 10.1038/35102000. [DOI] [PubMed] [Google Scholar]
- 48.LaRiviere FJ, Wolfson AD, Uhlenbeck OC. Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science. 2001;294:165–168. doi: 10.1126/science.1064242. [DOI] [PubMed] [Google Scholar]
- 49.Srivastava DK, Bernhard SA. Metabolite transfer via enzyme-enzyme complexes. Science. 1986;234:1081–1086. doi: 10.1126/science.3775377. [DOI] [PubMed] [Google Scholar]
- 50.Asahara H, Uhlenbeck OC. The tRNA specificity of Thermus thermophilus EF-Tu. Proc Natl Acad Sci USA. 2002;99:3499–3504. doi: 10.1073/pnas.052028599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bailly M, Blaise M, Lorber B, Becker HD, Kern D. The transamidosome: a dynamic ribonucleoprotein particle dedicated to prokaryotic tRNA-dependent asparagine biosynthesis. Molecular Cell. 2007;28:228–239. doi: 10.1016/j.molcel.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 52.Huot JL, Balg C, Jahn D, Moser J, Emond A, Blais SP, Chenevert R, Lapointe J. Mechanism of a GatCAB amidotransferase: aspartyl-tRNA synthetase increases its affinity for Asp-tRNAAsn and novel aminoacyl-tRNA analogues are competitive inhibitors. Biochemistry. 2007;46:13190–13198. doi: 10.1021/bi700602n. [DOI] [PubMed] [Google Scholar]
- 53.Kim SI, Stange-Thomann N, Martins O, Hong KW, Söll D, Fox TD. A nuclear genetic lesion affecting Saccharomyces cerevisiae mitochondrial translation is complemented by a homologous Bacillus gene. J Bacteriol. 1997;179:5625–5627. doi: 10.1128/jb.179.17.5625-5627.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dirheimer D, Keith G, Sibler AP, Martin RP. Transfer RNA: structure, properties, and recognition. 1. New York: Cold Spring Harbor; 1979. [Google Scholar]
- 55.Mulero JJ, Rosenthal JK, Fox TL. PET112, a Saccharomyces cerevisiae nuclear gene required to maintain rho+ mitochondrial DNA. Current Genetics. 1994;25:299–304. doi: 10.1007/BF00351481. [DOI] [PubMed] [Google Scholar]
- 56.Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 57.Martin NC, Rabinowitz M, Fukuhara H. Yeast mitochondrial DNA specifies tRNA for 19 amino acids. Deletion mapping of the tRNA genes. Biochemistry. 1977;16:4672–4677. doi: 10.1021/bi00640a022. [DOI] [PubMed] [Google Scholar]
- 58.Rinehart J, Krett B, Rubio MA, Alfonzo JD, Söll D. Saccharomyces cerevisiae imports the cytosolic pathway for Gln-tRNA synthesis into the mitochondrion. Genes Dev. 2005;19:583–592. doi: 10.1101/gad.1269305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lamour V, Quevillon S, Diriong S, N’Guyen VC, Lipinski M, Mirande M. Evolution of the Glx-tRNA synthetase family: the glutaminyl enzyme as a case of horizontal gene transfer. Proc Natl Acad Sci USA. 1994;91:8670–8674. doi: 10.1073/pnas.91.18.8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Woese CR, Olsen GJ, Ibba M, Söll D. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol Mol Biol Rev. 2000;64:202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O’Donoghue P, Luthey-Schulten Z. On the evolution of structure in aminoacyl-tRNA synthetases. Microbiol Mol Biol Rev. 2003;67:550–573. doi: 10.1128/MMBR.67.4.550-573.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O’Donoghue P, Luthey-Schulten Z. Evolutionary profiles derived from the QR factorization of multiple structural alignments gives an economy of information. J Mol Biol. 2005;346:875–894. doi: 10.1016/j.jmb.2004.11.053. [DOI] [PubMed] [Google Scholar]
- 63.Forchhammer K. Glutamine signalling in bacteria. Front Biosci. 2007;12:358–370. doi: 10.2741/2069. [DOI] [PubMed] [Google Scholar]
- 64.Sonenshein AL. Control of key metabolic intersections in Bacillus subtilis. Nat Rev Microbiol. 2007;5:917–927. doi: 10.1038/nrmicro1772. [DOI] [PubMed] [Google Scholar]
- 65.O’Donoghue P, Sethi A, Woese CR, Luthey-Schulten ZA. The evolutionary history of Cys-tRNACys formation. Proc Natl Acad Sci USA. 2005;102:19003–19008. doi: 10.1073/pnas.0509617102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Klenk HP, Clayton RA, Tomb JF, White O, Nelson KE, Ketchum KA, Dodson RJ, et al. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 67.Fricke WF, Seedorf H, Henne A, Kruer M, Liesegang H, Hedderich R, Gottschalk G, et al. The genome sequence of Methanosphaera stadtmanae reveals why this human intestinal archaeon is restricted to methanol and H2 for methane formation and ATP synthesis. J Bacteriol. 2006;188:642–658. doi: 10.1128/JB.188.2.642-658.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Samuel BS, Hansen EE, Manchester JK, Coutinho PM, Henrissat B, Fulton R, Latreille P, et al. Genomic and metabolic adaptations of Methanobrevibacter smithii to the human gut. Proc Natl Acad Sci USA. 2007;104:10643–10648. doi: 10.1073/pnas.0704189104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li T, Graham DE, Stathopoulos C, Haney PJ, Kim HS, Vothknecht U, Kitabatake M, et al. Cysteinyl-tRNA formation: the last puzzle of aminoacyl-tRNA synthesis. FEBS Lett. 1999;462:302–306. doi: 10.1016/s0014-5793(99)01550-1. [DOI] [PubMed] [Google Scholar]
- 70.Stathopoulos C, Kim W, Li T, Anderson I, Deutsch B, Palioura S, Whitman W, et al. Cysteinyl-tRNA synthetase is not essential for viability of the archaeon Methanococcus maripaludis. Proc Natl Acad Sci USA. 2001;98:14292–14297. doi: 10.1073/pnas.201540498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.White RH. The biosynthesis of cysteine and homocysteine in Methanococcus jannaschii. Biochim Biophys Acta. 2003;1624:46–53. doi: 10.1016/j.bbagen.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 72.Helgadottir S, Rosas-Sandoval G, Söll D, Graham DE. Biosynthesis of phosphoserine in the Methanococcales. J Bacteriol. 2007;189:575–582. doi: 10.1128/JB.01269-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kavran JM, Gundllapalli S, O’Donoghue P, Englert M, Söll D, Steitz TA. Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation. Proc Natl Acad Sci USA. 2007;104:11268–11273. doi: 10.1073/pnas.0704769104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kamtekar S, Hohn MJ, Park HS, Schnitzbauer M, Sauerwald A, Söll D, Steitz TA. Toward understanding phosphoseryl-tRNACys formation: the crystal structure of Methanococcus maripaludis phosphoseryl-tRNA synthetase. Proc Natl Acad Sci USA. 2007;104:2620–2625. doi: 10.1073/pnas.0611504104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fukunaga R, Yokoyama S. Structural insights into the first step of RNA-dependent cysteine biosynthesis in archaea. Nat Struct Mol Biol. 2007;14:272–279. doi: 10.1038/nsmb1219. [DOI] [PubMed] [Google Scholar]
- 76.Hohn MJ, Park HS, O’Donoghue P, Schnitzbauer M, Söll D. Emergence of the universal genetic code imprinted in an RNA record. Proc Natl Acad Sci USA. 2006;103:18095–18100. doi: 10.1073/pnas.0608762103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fukunaga R, Yokoyama S. Structural insights into the second step of RNA-dependent cysteine biosynthesis in archaea: crystal structure of Sep-tRNA:Cys-tRNA synthase from Archaeoglobus fulgidus. J Mol Biol. 2007;370:128–141. doi: 10.1016/j.jmb.2007.04.050. [DOI] [PubMed] [Google Scholar]
- 78.Schneider G, Kack H, Lindqvist Y. The manifold of vitamin B6 dependent enzymes. Structure. 2000;8:R1–6. doi: 10.1016/s0969-2126(00)00085-x. [DOI] [PubMed] [Google Scholar]
- 79.Eliot AC, Kirsch JF. Pyridoxal phosphate enzymes: mechanistic, structural, and evolutionary considerations. Annu Rev Biochem. 2004;73:383–415. doi: 10.1146/annurev.biochem.73.011303.074021. [DOI] [PubMed] [Google Scholar]
- 80.Zhou D, White RH. Transsulfuration in archaebacteria. J Bacteriol. 1991;173:3250–3251. doi: 10.1128/jb.173.10.3250-3251.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kitabatake M, So MW, Tumbula DL, Söll D. Cysteine biosynthesis pathway in the archaeon Methanosarcina barkeri encoded by acquired bacterial genes? J Bacteriol. 2000;182:143–145. doi: 10.1128/jb.182.1.143-145.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Borup B, Ferry JG. Cysteine biosynthesis in the Archaea: Methanosarcina thermophila utilizes O-acetylserine sulfhydrylase. FEMS Microbiol Lett. 2000;189:205–210. doi: 10.1111/j.1574-6968.2000.tb09231.x. [DOI] [PubMed] [Google Scholar]
- 83.Borup B, Ferry JG. O-Acetylserine sulfhydrylase from Methanosarcina thermophila. J Bacteriol. 2000;182:45–50. doi: 10.1128/jb.182.1.45-50.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mino K, Ishikawa K. Characterization of a novel thermostable O-acetylserine sulfhydrylase from Aeropyrum pernix K1. J Bacteriol. 2003;185:2277–2284. doi: 10.1128/JB.185.7.2277-2284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mino K, Ishikawa K. A novel O-phospho-L-serine sulfhydrylation reaction catalyzed by O-acetylserine sulfhydrylase from Aeropyrum pernix K1. FEBS Lett. 2003;551:133–138. doi: 10.1016/s0014-5793(03)00913-x. [DOI] [PubMed] [Google Scholar]
- 86.Mihara H, Esaki N. Bacterial cysteine desulfurases: their function and mechanisms. Appl Microbiol Biotechnol. 2002;60:12–23. doi: 10.1007/s00253-002-1107-4. [DOI] [PubMed] [Google Scholar]
- 87.Major TA, Burd H, Whitman WB. Abundance of 4Fe-4S motifs in the genomes of methanogens and other prokaryotes. FEMS Microbiol Lett. 2004;239:117–123. doi: 10.1016/j.femsle.2004.08.027. [DOI] [PubMed] [Google Scholar]
- 88.Di Giulio M. The origin of the genetic code: theories and their relationships, a review. Biosystems. 2005;80:175–184. doi: 10.1016/j.biosystems.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 89.Klipcan L, Frenkel-Morgenstern M, Safro MG. Presence of tRNA-dependent pathways correlates with high cysteine content in methanogenic Archaea. Trends Genet. 2008;24:59–63. doi: 10.1016/j.tig.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 90.Rayman MP. The importance of selenium to human health. Lancet. 2000;356:233–241. doi: 10.1016/S0140-6736(00)02490-9. [DOI] [PubMed] [Google Scholar]
- 91.Moghadaszadeh B, Beggs AH. Selenoproteins and their impact on human health through diverse physiological pathways. Physiology. 2006;21:307–315. doi: 10.1152/physiol.00021.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hatfield DL, Carlson BA, Xu XM, Mix H, Gladyshev VN. Selenocysteine incorporation machinery and the role of selenoproteins in development and health. Prog Nucleic Acid Res Mol Biol. 2006;81:97–142. doi: 10.1016/S0079-6603(06)81003-2. [DOI] [PubMed] [Google Scholar]
- 93.Huber RE, Criddle RS. Comparison of the chemical properties of selenocysteine and selenocystine with their sulfur analogs. Arch Biochem Biophys. 1967;122:164–173. doi: 10.1016/0003-9861(67)90136-1. [DOI] [PubMed] [Google Scholar]
- 94.Young PA, Kaiser II. Aminoacylation of Escherichia coli cysteine tRNA by selenocysteine. Arch Biochem Biophys. 1975;171:483–489. doi: 10.1016/0003-9861(75)90057-0. [DOI] [PubMed] [Google Scholar]
- 95.Shrift A, Bechard D, Harcup C. Utilization of selenocysteine by a cysteinyl-tRNA synthetase from Phaseolus aureus. Plant Physiol. 1976;58:248–252. doi: 10.1104/pp.58.3.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Müller S, Senn H, Gsell B, Vetter W, Baron C, Böck A. The formation of diselenide bridges in proteins by incorporation of selenocysteine residues: biosynthesis and characterization of (Se)2-thioredoxin. Biochemistry. 1994;33:3404–3412. doi: 10.1021/bi00177a034. [DOI] [PubMed] [Google Scholar]
- 97.Johansson L, Gafvelin G, Arner ES. Selenocysteine in proteins-properties and biotechnological use. Biochim Biophys Acta. 2005;1726:1–13. doi: 10.1016/j.bbagen.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 98.Böck A, Forchhammer K, Heider J, Leinfelder W, Sawers G, Veprek B, Zinoni F. Selenocysteine: the 21st amino acid. Mol Microbiol. 1991;5:515–520. doi: 10.1111/j.1365-2958.1991.tb00722.x. [DOI] [PubMed] [Google Scholar]
- 99.Gladyshev VN, Hatfield DL. Selenocysteine-containing proteins in mammals. J Biomed Sci. 1999;6:151–160. doi: 10.1007/BF02255899. [DOI] [PubMed] [Google Scholar]
- 100.Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, Gladyshev VN. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 101.Kryukov GV, Gladyshev VN. The prokaryotic selenoproteome. EMBO Rep. 2004;5:538–543. doi: 10.1038/sj.embor.7400126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bilokapic S, Korencic D, Söll D, Weygand-Durasevic I. The unusual methanogenic seryl-tRNA synthetase recognizes tRNASer species from all three kingdoms of life. Eur J Biochem. 2004;271:694–702. doi: 10.1111/j.1432-1033.2003.03971.x. [DOI] [PubMed] [Google Scholar]
- 103.Mizutani T, Narihara T, Hashimoto A. Purification and properties of bovine liver seryl-tRNA synthetase. Eur J Biochem. 1984;143:9–13. doi: 10.1111/j.1432-1033.1984.tb08331.x. [DOI] [PubMed] [Google Scholar]
- 104.Leinfelder W, Zehelein E, Mandrand-Berthelot MA, Böck A. Gene for a novel tRNA species that accepts L-serine and cotranslationally inserts selenocysteine. Nature. 1988;331:723–725. doi: 10.1038/331723a0. [DOI] [PubMed] [Google Scholar]
- 105.Carlson BA, Xu XM, Kryukov GV, Rao M, Berry MJ, Gladyshev VN, Hatfield DL. Identification and characterization of phosphoseryl-tRNA[Ser]Sec kinase. Proc Natl Acad Sci USA. 2004;101:12848–12853. doi: 10.1073/pnas.0402636101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kaiser JT, Gromadski K, Rother M, Engelhardt H, Rodnina MV, Wahl MC. Structural and functional investigation of a putative archaeal selenocysteine synthase. Biochemistry. 2005;44:13315–13327. doi: 10.1021/bi051110r. [DOI] [PubMed] [Google Scholar]
- 107.Leinfelder W, Forchhammer K, Veprek B, Zehelein E, Bock A. In vitro synthesis of selenocysteinyl-tRNAUCA from seryl-tRNAUCA: involvement and characterization of the selD gene product. Proc Natl Acad Sci USA. 1990;87:543–547. doi: 10.1073/pnas.87.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Leinfelder W, Forchhammer K, Zinoni F, Sawers G, Mandrand-Berthelot MA, Böck A. Escherichia coli genes whose products are involved in selenium metabolism. J Bacteriol. 1988;170:540–546. doi: 10.1128/jb.170.2.540-546.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Xu XM, Carlson BA, Irons R, Mix H, Zhong N, Gladyshev VN, Hatfield DL. Selenophosphate synthetase 2 is essential for selenoprotein biosynthesis. Biochem J. 2007;404:115–120. doi: 10.1042/BJ20070165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sherrer RL, O’Donoghue P, Söll D. Characterization and evolutionary history of an archaeal kinase involved in selenocysteinyl-tRNA formation. Nucleic Acids Res. 2008;36:1247–1259. doi: 10.1093/nar/gkm1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mäenpää PH, Bernfield MR. A specific hepatic transfer RNA for phosphoserine. Proc Natl Acad Sci USA. 1970;67:688–695. doi: 10.1073/pnas.67.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sharp SJ, Stewart TS. The characterization of phosphoseryl tRNA from lactating bovine mammary gland. Nucleic Acids Res. 1977;4:2123–2136. doi: 10.1093/nar/4.7.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Leipe DD, Koonin EV, Aravind L. Evolution and classification of P-loop kinases and related proteins. J Mol Biol. 2003;333:781–815. doi: 10.1016/j.jmb.2003.08.040. [DOI] [PubMed] [Google Scholar]
- 114.Hatfield D, Lee BJ, Hampton L, Diamond AM. Selenium induces changes in the selenocysteine tRNA[Ser]Sec population in mammalian cells. Nucleic Acids Res. 1991;19:939–943. doi: 10.1093/nar/19.4.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dong H, Nilsson L, Kurland CG. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J Mol Biol. 1996;260:649–663. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- 116.Wu XQ, Gross HJ. The long extra arms of human tRNA(Ser)Sec and tRNASer function as major identify elements for serylation in an orientation-dependent, but not sequence-specific manner. Nucleic Acids Res. 1993;21:5589–5594. doi: 10.1093/nar/21.24.5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sturchler C, Westhof E, Carbon P, Krol A. Unique secondary and tertiary structural features of the eucaryotic selenocysteine tRNASec. Nucleic Acids Res. 1993;21:1073–1079. doi: 10.1093/nar/21.5.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schön A, Bock A, Ott G, Sprinzl M, Söll D. The selenocysteine-inserting opal suppressor serine tRNA from E. coli is highly unusual in structure and modification. Nucleic Acids Res. 1989;17:7159–7165. doi: 10.1093/nar/17.18.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wu XQ, Gross HJ. The length and the secondary structure of the D-stem of human selenocysteine tRNA are the major identity determinants for serine phosphorylation. EMBO J. 1994;13:241–248. doi: 10.1002/j.1460-2075.1994.tb06254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sherrer RL, Ho J, Söll D. Divergence of selenocysteine tRNA recognition by archaeal and eukaryotic O-phosphoseryl-tRNASec kinase. Nucleic Acids Res. 2008;36:1871–1880. doi: 10.1093/nar/gkn036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Baron C, Westhof E, Böck A, Giege R. Solution structure of selenocysteine-inserting tRNASec from Escherichia coli Comparison with canonical tRNASer. J Mol Biol. 1993;231:274–292. doi: 10.1006/jmbi.1993.1282. [DOI] [PubMed] [Google Scholar]
- 122.Hubert N, Sturchler C, Westhof E, Carbon P, Krol A. The 9/4 secondary structure of eukaryotic selenocysteine tRNA: more pieces of evidence. RNA. 1998;4:1029–1033. doi: 10.1017/s1355838298980888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ioudovitch A, Steinberg SV. Structural compensation in an archaeal selenocysteine transfer RNA. J Mol Biol. 1999;290:365–371. doi: 10.1006/jmbi.1999.2901. [DOI] [PubMed] [Google Scholar]
- 124.Baron C, Sturchler C, Wu XQ, Gross HJ, Krol A, Böck A. Eukaryotic selenocysteine inserting tRNA species support selenoprotein synthesis in Escherichia coli. Nucleic Acids Res. 1994;22:2228–2233. doi: 10.1093/nar/22.12.2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Araiso Y, Palioura S, Ishitani R, Sherrer RL, O’Donoghue P, Yuan J, Oshikane H, et al. Structural insights into RNA-dependent eukaryal and archaeal selenocysteine formaiton. Nucleic Acids Res. 2007;36:1187–1199. doi: 10.1093/nar/gkm1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ganichkin OM, Xu XM, Carlson BA, Mix H, Hatfield DL, Gladyshev VN, Wahl MC. Structure and catalytic mechanism of eukaryotic selenocysteine synthase. J Biol Chem. 2007;283:5849–5865. doi: 10.1074/jbc.M709342200. [DOI] [PubMed] [Google Scholar]
- 127.Zhang Y, Romero H, Salinas G, Gladyshev VN. Dynamic evolution of selenocysteine utilization in bacteria: a balance between selenoprotein loss and evolution of selenocysteine from redox active cysteine residues. Genome Biol. 2006;7:R94. doi: 10.1186/gb-2006-7-10-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Stanzel M, Schön A, Sprinzl M. Discrimination against misacylated tRNA by chloroplast elongation factor Tu. Eur J Biochem. 1994;219:435–439. doi: 10.1111/j.1432-1033.1994.tb19956.x. [DOI] [PubMed] [Google Scholar]
- 129.Rother M, Wilting R, Commans S, Böck A. Identification and characterisation of the selenocysteine-specific translation factor SelB from the archaeon Methanococcus jannaschii. J Mol Biol. 2000;299:351–358. doi: 10.1006/jmbi.2000.3756. [DOI] [PubMed] [Google Scholar]
- 130.Roy H, Becker HD, Mazauric MH, Kern D. Structural elements defining elongation factor Tu mediated suppression of codon ambiguity. Nucleic Acids Res. 2007;35:3420–3430. doi: 10.1093/nar/gkm211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Min B, Kitabatake M, Polycarpo C, Pelaschier J, Raczniak G, Ruan B, Kobayashi H, et al. Protein synthesis in Escherichia coli with mischarged tRNA. J Bacteriol. 2003;185:3524–3526. doi: 10.1128/JB.185.12.3524-3526.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sheppard K, Yuan J, Hohn MJ, Jester B, Devine KM, Soll D. From one amino acid to another: tRNA-dependent amino acid biosynthesis. Nucleic Acids Res. 2008;36:1813–1825. doi: 10.1093/nar/gkn015. [DOI] [PMC free article] [PubMed] [Google Scholar]


