Abstract
The genome of Pneumocystis, which causes life-threatening pneumonia in immunosuppressed patients, contains a multi-copy gene family that encodes the major surface glycoprotein (Msg). Pneumocystis can vary the expressed Msg, presumably as a mechanism to avoid host immune responses. Analysis of 24 msg gene sequences obtained from a single human Pneumocystis isolate demonstrated that the sequences segregate into two branches. Based on a number of analyses, recombination among msg genes appears to be an important mechanism for generating msg diversity. Intra-branch recombination occurred more frequently than inter-branch recombination. Restriction fragment length polymorphism analysis demonstrated substantial variation in the repertoire of the msg gene family among isolates of human Pneumocystis, which was not observed in laboratory isolates of rat or mouse Pneumocystis; this may be the result of examining outbred vs. captive populations. Increased diversity in the Msg repertoire, generated in part by recombination, increases the potential for antigenic variation in this abundant surface protein.
Keywords: AIDS, Pneumocystis, antigenic variation, repertoire, immune evasion, major surface glycoprotein
Introduction
Pneumocystis causes life-threatening pneumonia in immunosuppressed hosts. The most abundant surface protein of Pneumocystis is the major surface glycoprotein (Msg), which is encoded by a multi-copy gene family with an estimated 50 to 100 copies (of ~3,000 kb each) per genome that are clustered in tandem arrays near the telomeres of each chromosome [1]. These genes encode an incomplete protein that lacks an N-terminal peptide and are not expressed unless they are translocated downstream of a unique subtelomeric expression site which encodes the upstream conserved sequence (UCS) [2-7]. Only the variant present at the expression site is translated in a given organism. However, the ~50-100 variant msg genes provide a large potential for antigenic variation [8-12]. Variation of the expressed Msg presumably facilitates evasion of immune responses in hosts. Since Pneumocystis are haploid [13], a single organism can express only a single Msg variant. However, multiple variants can be expressed in a single infected lung from an immunosuppressed host [14, 15]. A similar msg gene organization has been identified in human, rat, mouse, and ferret Pneumocystis. [4, 6, 7, 12]. Recombination may play a role in generating multiple msg variants [3, 16, 17].
Antigenic variability in other species such as African trypanosomes and Borrelia species is associated with evasion of host immune, primarily antibody, responses. The potential for antigenic variation in these organisms is increased not only by having multiple unique copies of genes encoding their surface proteins per organism genome, but also by variation in the repertoire of these multi-copy (i.e. different isolates have unique sets of these genes), which likely further contributes to successful immune evasion [18-20]. To better characterize the family of genes that compose the msg repertoire in human Pneumocystis (P. jirovecii), we undertook to sequence individual msg variants in a patient with Pneumocystis pneumonia (PcP), and determine their relationships to each other, specifically focusing on possible recombination between msgs. To see if Pneumocystis have variable repertoires of the msg gene family, similar to other organisms, we used restriction fragment length polymorphism analysis to examine the msg repertoire in human, rat, and mouse Pneumocystis.
Methods
Pneumocystis DNA preparation
P. jirovecii-infected autopsy lung from 6 patients with PcP (5 with HIV infection) were used for DNA extraction. For P. murina, infected lungs from scid mice were used. P. carinii lungs were obtained from immunosuppressed rats maintained in two facilities (Biocon, Rockville, MD and Indiana University, Indianapolis, IN) and partially purified by Ficoll-Hypaque density gradient centrifugations [21]. Genomic DNA was isolated either by using QIAamp DNA mini kit (Qiagen) or by proteinase K treatment [22]. Guidelines of the National Institutes of Health with regard to human and animal experimentation were followed in the conduct of these studies.
PCR Amplification
Pneumocystis DNA was amplified with TaqPlus Long (Stratagene), using primers from conserved regions (based on alignment of available msg sequences) designed to amplify the entire ~3.3 kb of the msg variable region (Supplemental Table and Supplemental Figure 1). PCR conditions for P. carinii (primers GK521 and GK527) and P. murina (primers GK257 and GK261) were: 2 min at 94° C, followed by 35 cycles of 30 s at 94° C, 30 s at 56° C, 4 min at 72° C and a final extension of 10 min at 72° C. PCR conditions for P. jirovecii (primers GK126 and GK452) were: 2 minutes at 95°C, followed by 10 cycles of 30 s at 94°C, 1 min at 68°C with 1°C decremental steps in each cycle, and 4 min at 72° C, then 35 cycles of 30 s at 94°C, 30 s at 58°C, and 4 min at 72°C.
Sequencing of msg variants
Genomic DNA from a single infected patient was analyzed by nested PCR using HotstarTaq (Qiagen) following limiting dilution [23]. The first round was as described above, but with a 15 min initial denaturation. The second round conditions (primers GK508 and GK506) were: 15 min at 95 °C; 35 cycles of 30s at 94°C, 30s at 58°C, 4 min at 72 °C; and a final extension of 10 min at 72 ° C. For the limiting dilution, DNA was serially diluted (3- to 10-fold per dilution) in preliminary studies and 10 replicate nested PCR reactions were performed at each dilution. The dilution at which only ~3 PCR reactions yielded a product as determined by agarose gel electrophoresis was utilized for subsequent subcloning [23]. Amplification products were subcloned into TOPO TA cloning PCR 2.1 (Invitrogen) and clones showing distinct sequences (>1% variability) following initial sequencing of the 3’ and 5’ ends were completely sequenced. To examine the likelihood of PCR-mediated recombination between 2 msgs on a single DNA fragment, we used the identical procedure to amplify and sequence a genomic clone with 2 full-length msgs in tandem repeats (GenBank #AF038556). We found no recombinants among 52 fully sequenced clones that were generated during 6 PCR reactions.
Restriction fragment length polymorphism and Southern blot analysis
PCR products amplified as above were separated on an agarose gel, excised, purified using Qiaquick gel extraction kit (Qiagen), digested with the indicated restriction enzymes, analyzed on 1% agarose gels in 1X TBE buffer and visualized by SYBR green staining (Molecular Probes). Preliminary studies demonstrated that the RFLP pattern was stable during repeat PCR reactions and not dependent on DNA concentration. DNA blotted onto Nytran membranes (Schleicher&Schuell) was probed with oligonucleotides labeled using DIG Oligonucleotide Tailing Kit (Roche, Indianapolis, IN) or DIG-labeled DNA probes (PCR DIG Probe Synthesis Kit, Roche); Southern blot analysis was performed as described [24]. Blots were stripped at 37°C in buffer containing 0.2M NaOH and 0.1% SDS before rehybridization.
Statistical analysis
msg sequences were aligned using Clustal W (Megalign module of Lasergene, DNASTAR Inc., Madison, WI) using a gap penalty of 15 and gap length penalty of 6.66 [25]. Neighbor-joining trees of gap-stripped msg sequences were constructed using PAUP* 4.0 (Sinauer Associates, Inc., Sunderland, MA), with a rat P. carinii msg sequence as the outgroup. Bootstrap values were calculated using 1000 resampling replicates. Average pairwise differences between groups and population structure tests were performed on gap-stripped sequences using the method of Hudson [24] (see also http://wwwabi.snv.jussieu.fr/~achaz/hudsontest.html for online version of this analysis). Based on permutation, this test gives the probability that two a priori defined groups have more similarity within groups than between groups. Homogeneity was defined as an absence of structure. Simplot and Bootscanning analyses of the alignments were performed to identify regions where recombination occurred. SimPlot compares similarity of a short window (200 nucleotides) of an individual sequence to the corresponding window for the entire complement of sequences in the alignment. The degree of relatedness is calculated as the window is moved in steps (200 nucleotides) across the alignment. Marked changes in similarity indicate the presence of recombination [26]. Bootscanning analyzes phylogenetic relationships of sequences, and calculates bootstrap values of sequences that cluster phylogenetically in a sliding window (200 nucleotides) to a reference set of potential parental sequences [27]. The bootstrap values are plotted as a function of the window position along the sequence. Sharp changes in bootstrap values indicate strong support for recombination.
Results
Analysis of msg genes from a single patient isolate
To examine the relationship among msg genes of human Pneumocystis, we sequenced 24 unique msg variant genes from a patient infected with a single Pneumocystis isolate (based on typing using ITS1 as well as tandem repeats in the UCS [28, 29]). To minimize artifacts resulting from recombination among msg genes during PCR, we utilized limiting dilution followed by PCR. msgs are arranged in tandem repeats on individual chromosomes, which required us to subclone the PCR product prior to sequencing; this can potentially introduce point mutations at a frequency of ~1 in 1000 bp (based on studies with a plasmid containing an msg). The effect of such mutations was minimized by using consensus sequences from clones that were >99% identical, as well as additional sequencing using genomic DNA and primers specific for individual msgs. All clones contained a complete open reading frame for Msg. Sequences have been deposited in GenBank (accession codes EF371022-26, EF371028-33, EF371035-36, EF371038, EF371040-42, EF371045, EF371050-53, and EF371055-56).
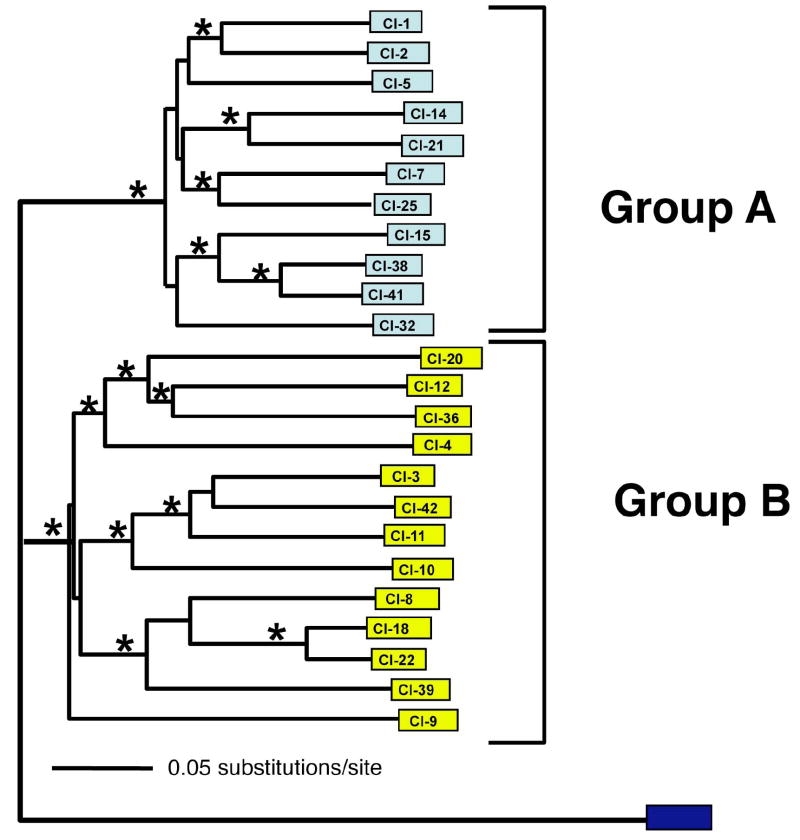
Phylogenetic analysis revealed that msg sequences segregated into subgroups (Figure 1). Robust bootstrap values supported a tree structure consisting of two principal branches, designated A and B, which contained 11 and 13 sequences, respectively. Detailed analysis of msg sequence heterogeneity revealed substantial nucleotide diversity; 1981/3445 sites within msg were polymorphic. The overall average pairwise differences of msg sequences was 0.252. Sequences in group A have a significantly lower diversity compared with those in group B (average pairwise difference = 0.200 and 0.295, respectively; p<0.0001). The average pairwise differences between groups was 0.364, suggesting that differences between groups are greater than those within either individual group.
Figure 1.
Population structure of P. jirovecii msg sequences present in a single infected host. Sequences were aligned and neighbor joining phylogenetic trees constructed as described in the methods using a rat msg sequence (black box; MSG100, GenBank # D31909) to root the tree. Bootstrap analysis (*=bootstrap values≥ 99) identified two large distinct branches (Groups A and B), each of which consisting of several smaller groups of distinct sequences.
In order to investigate whether the segregation of groups A and B were statistically significant, we employed an adaptation of the population structure of Hudson [30, 31]. This nonparametric analysis yields a quantitative measure of the probability of homogeneity (absence of structure), the likelihood that two sets of sequences are derived from a single population. By this analysis, the probability that these two sets of sequences do not show any structure (i.e. are homogeneous) is less than 10-9. This structure was present only when sequences were grouped according to the phylogenetic branches A and B; analysis of groups that were obtained by purely random assignment of msg sequences revealed no evidence of structure. These data suggest that groups A and B identified by phylogenetic analysis are organized as two segregated subpopulations.
Analysis for recombination among msg genes
Because msgs are a family of related but unique genes, we undertook to examine if recombination was playing a role in generating msg diversity. We initially performed a separate phylogenetic analysis with each of 3 fragments of the gap-stripped msg sequence: nucleotides 1-932, nucleotides 933-1862, and nucleotides 1863-2813. These msg fragments also segregated into two distinct groups (Supplemental Figure 2). Some, but not all, branch associations with 100% bootstrap support in the full-length sequence were maintained in analysis of the smaller fragments. However, several branches were reassorted in the gene fragment analysis compared to the gene-wide analysis, with 100% bootstrap support (e.g. see sequences Cl-1, Cl-7, Cl-14, and Cl-25). Branch switching with strong bootstrap support as a function of nucleotide position strongly suggests the presence of recombination. (We use recombination to refer to both reciprocal and non-reciprocal genetic exchange, the latter being properly called conversion).
Because msg alignments revealed numerous insertions and/or deletions (indels), we undertook specific analysis of the indels. This demonstrated that a number of msg indels segregated primarily according to phylogenetic grouping. As shown in figure 2, an insertion of 3 nucleotides after position 1339 were found almost exclusively in sequences in principal branch A of the phylogenetic tree, but this insertion was also identified in one of the Group B sequences. Similarly a deletion of 6 nucleotides was uniformly present after nucleotide 2908 in Group A and in 3/13 sequences of group B; an insertion of 3 nucleotides after nucleotide 2925 was uniformly present in group B and in one sequence in group A. The finding of an indel uniformly present in one branch and appearing in the other branch strongly supports the presence of recombination among these msgs.
Figure 2.
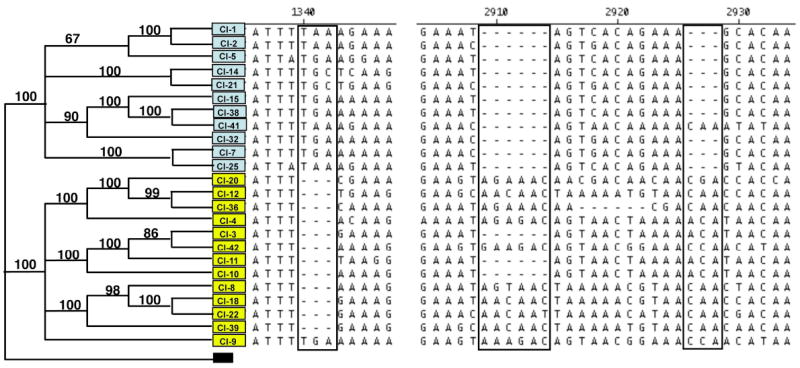
Analysis of indels reveals evidence of recombination. Sequences were arranged according to the phylogenetic analysis shown in Figure 1 with groups A and B color coded and with bootstrap values for branches indicated (left). Sequences in the nucleotide region ~1340 and ~2920 with obvious indels (boxed) showed strong but not exclusive segregation according to groups. An insertion at position 1340 in group A was absent in 12/13 of group B sequences, but the same 3 nucleotide insertion was present in one group B sequence (Cl-9). Similar segregation of indels at position 2910 and 2926 revealed strong evidence of recombination between groups A and B.
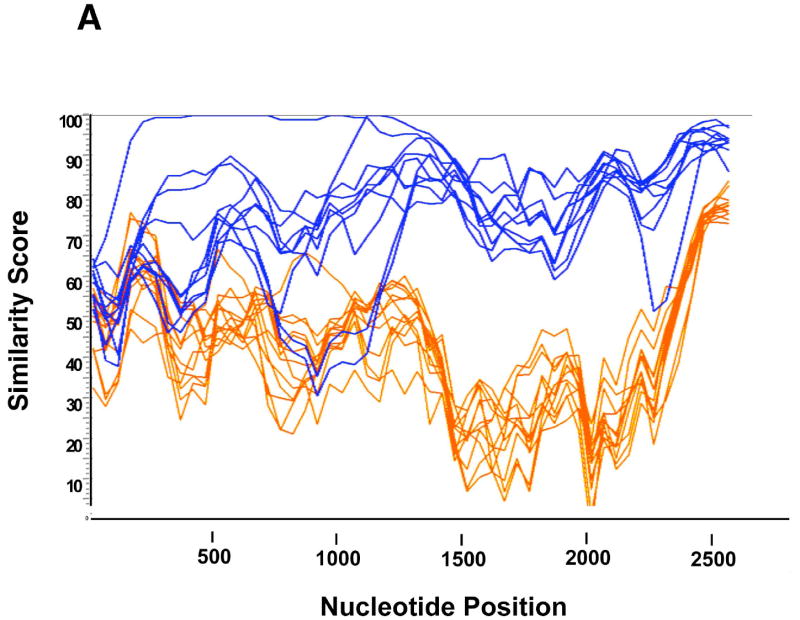
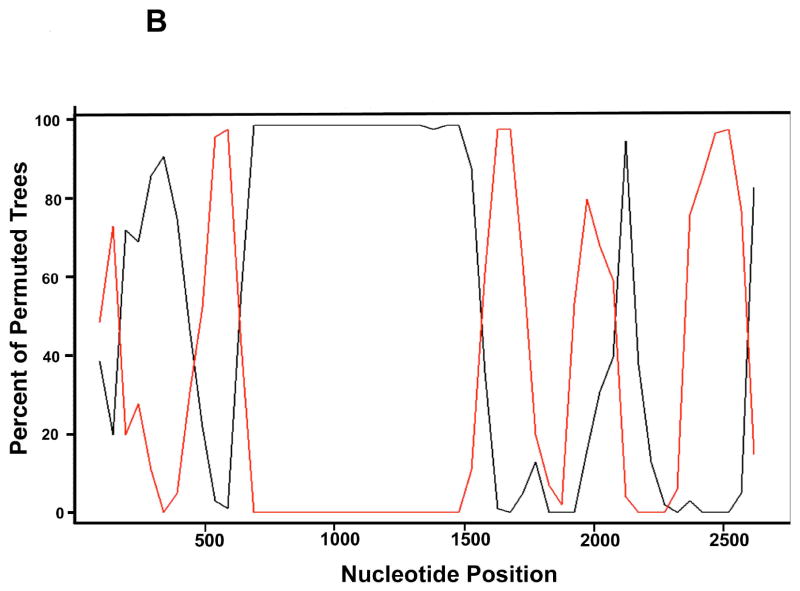
To further investigate potential recombination events, we used two sliding window approaches to measure region-specific similarity (SimPlot), and to quantitate bootstrap values for phylogenetic trees (bootscanning). SimPlot analysis identified a divergence in sequence similarity, with a segregation of sequences after ~1000-1200 nucleotides, that corresponded exactly to branches A and B in the phylogenetic analysis (Figure 3A). A recombination event can explain this sharp divergence. Bootscanning analysis of tree structure was used to identify potential areas of recombination more precisely. Phylogenetic trees constructed using a sliding window (200 nucleotides) of sequences from candidate recombinants and potential parental sequences are compared by bootstrap analyses; switching in bootstrap support from one parental sequence to another is consistent with recombination. We analyzed sets of sequences that had strong bootstrap support in full length msg sequence analysis, but which switched support in partial sequence analyses. In the bootscanning analysis (Figure 3B) of Cl-14 and Cl-21, and of Cl-14 and CI-25, clear evidence of two recombination events were detected (at positions ~700 and ~1520). Exhaustive bootscanning comparisons identified numerous additional areas where switching (presumably secondary to recombination) occurred in sequences within and between groups A and B (data not shown).
Figure 3.
A. Simplot analysis of msg sequences. Sequences were subjected to Simplot analysis comparing all other msg sequences to sequence Cl-5 of group A. Each line represents a single clone. All group A sequences (blue lines) are relatively closely related to Cl-5 (similarity to Cl-5 score 70-100), while group B sequences (orange lines) diverged sharply between nucleotides ~1200-2300 (similarity to Cl-5 score < 60), suggesting recombination at the divergance site. Similarity scores of group B sequences increased near the end of the msg sequence, suggesting additional recombination events.
B. Bootscanning detects evidence of frequent recombination. msg sequences were subjected to bootscanning analysis using Cl-14 as the query sequence and Cl -7, Cl -21, and Cl -25 as comparator sequences. For simplicity, only the comparisons between Cl-14 and Cl-21 (in black), and that between Cl-14 and Cl-25 (in red) are shown. The X axis depicts the nucleotide position within the msg sequence, and the Y axis, the percent of trees grouping with the query sequence. The number of permuted trees grouping Cl -21 with Cl -14 using a sliding window of 200 nucleotides showed sharp switches in frequency, with demarcations suggesting a recombination event approximately at nucleotides 700 and 1520. In contrast the number of Cl-14 permuted trees grouping with Cl-25 show reciprocal switch in homology at approximately the same locations. Additional potential switches in homology were seen in both comparisons, indicating multiple recombination events.
Analysis of msg gene variation in different Pneumocystis isolates
Given the evidence for recombination events among different msgs, we determined whether the msg repertoires (the 50-100 msg genes per genome) are identical among P. jirovecii isolates from different patients. Primers based on highly conserved regions (among the known msg variants) at the beginning of the coding region and downstream of the stop codon (Supplemental Figure 1) were used to amplify the whole msg repertoire of all individual genomes from an isolate. The resultant PCR product of ~3.3 kb was subjected to restriction fragment length polymorphism (RFLP) analysis. If the msg repertoire is highly conserved, the RFLP pattern among different isolates should be very similar or identical. Among 6 isolates from 6 patients, RFLP patterns were strikingly different when digested with Mbo I, Hind III or Dra I (Figure 4A). Southern blot analysis using an oligonucleotide designed from a conserved area near the 3’-end of msg, which was selected based on an alignment of known P. jirovecii msg sequences, is shown in Figure 4B. The 6 isolates showed distinct patterns, especially when digested with Hind III or Dra I. To further examine this, the blot was reprobed with an oligonucleotide specific for msg32, a previously characterized msg variant [22]. Strong hybridization was seen only for isolate #1 (Figure 4C), which shows that this specific variant is not present in all P. jirovecii, again supporting the high diversity of repertoire in this species. To verify that PCR amplification was not introducing an artifact into the RFLP analysis, restriction digestion using genomic DNA from 4 samples followed by hybridization was performed. Again a highly variable pattern was seen (Figure 4D) among the isolates. These observations demonstrate that the msg repertoires of P. jirovecii are highly variable.
Figure 4.
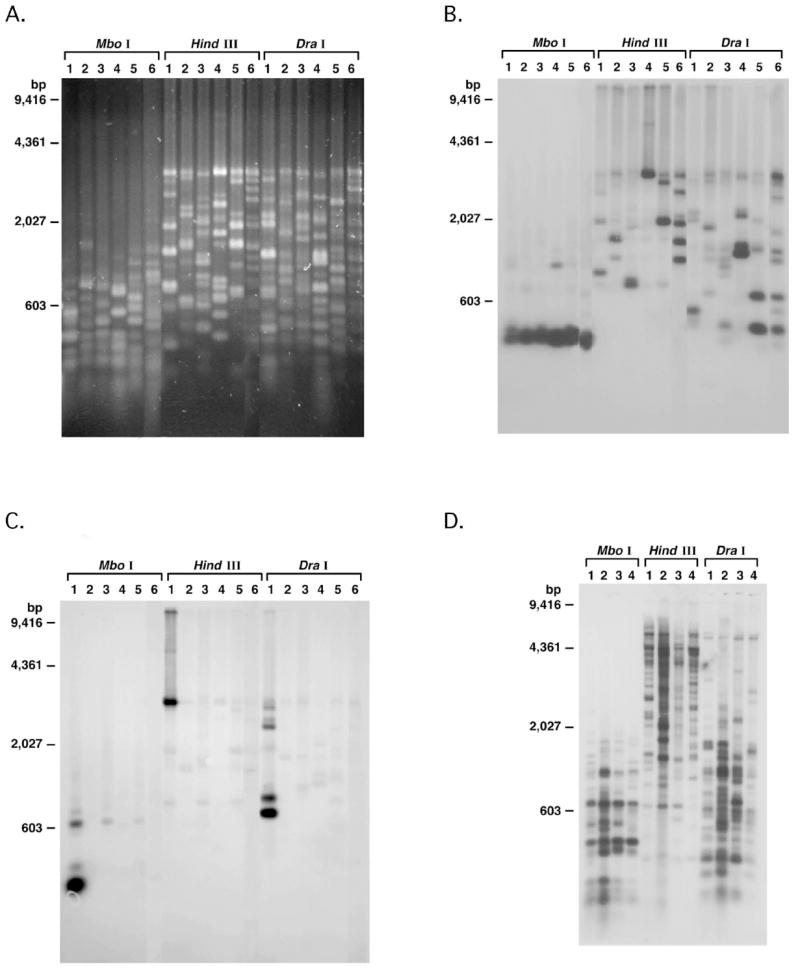
A: RFLP analysis of msg of P. jirovecii. msg was amplified from DNA prepared from the pulmonary samples of P. jirovecii infected patients (1-6: pulmonary samples collected between 1985 and 1999). By ITS1 and UCS typing, the 6 isolates were respectively E and 3 repeats; E and G and 3, 4, and 6 repeats; E and 6 repeats; E and 4 repeats; E and 3 and 4 repeats; E and 3 repeats. The PCR products were digested with Mbo I, Hind III, or Dra I restriction enzymes and analyzed using 1% agarose gels in 1X TBE buffer and then stained with SYBR green.
B-C. Southern blot analysis of msg of P. jirovecii. The DNA fragments from the gel (RFLP analysis, Fig. 4A) were blotted onto a membrane and hybridized successively with oligonucleotides GK195 (B) and GK576 (C). The blot was stripped between hybridizations. GK195 is from a highly conserved region near the 3’ end of msg and GK576 is specific for a single msg clone (msg 32).
D. Southern blot analysis of DNA from P. jirovecii-infected pulmonary samples. Genomic DNA extracted from the pulmonary samples of P. jirovecii infected patients (1-4) was digested with Mbo I, Hind III, or Dra I restriction enzymes. The blot was hybridized with a probe spanning nucleotides 1576-2974 of HuMSG14.
Given that different species of Pneumocystis infect rats and mice, and that these Pneumocystis also have similar multicopy msg gene families and systems for expressing msg, we performed RFLP analysis to examine the diversity of the msg repertoires in rat and mouse Pneumocystis [6, 10]. Using DNA samples from the lungs of 6 P. murina-infected scid mice housed in a single cage, an identical RFLP pattern was seen when the PCR products were digested with Hind III or Dra I (data not shown). Using lung samples collected at our facility from 1999-2004 (Figure 5A), RFLP patterns were again identical in all 6 mice. Southern blot analysis using an oligonucleotide designed from a conserved region of P. murina msg showed an identical pattern of hybridization for all isolates in each study (results not shown). Because all the mice were from a single colony, we conducted a similar analysis of rat P. carinii that were obtained from two different facilities over a period of years. RFLP patterns are very similar in all rats when digested with Hind III or Dra I (Figure 5B). Southern blot analysis using an oligonucleotide from the conserved region of P. carinii msg showed a nearly identical pattern of hybridization in all 7 rats (results not shown). Southern blot analysis of genomic DNA further confirmed that the RFLP pattern was highly conserved among isolates (Figure 5C). Thus, rat and mouse Pneumocystis from animals that are bred in captivity did not demonstrate the same degree of variability in msg repertoires as P. jirovecii.
Figure 5.
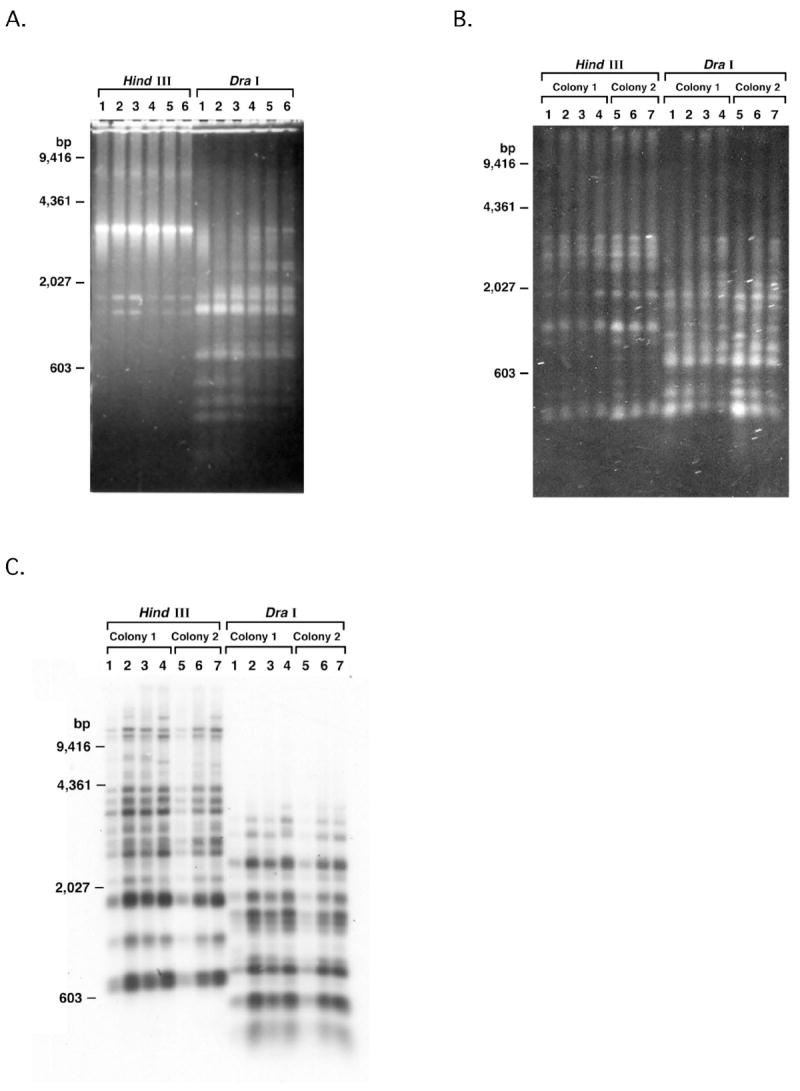
A: RFLP analysis of msg of P. murina. msg was amplified from DNA prepared from the lungs of infected scid mice collected over a 5 year period (1-6: lungs collected from 1999 to 2004, respectively). The PCR products were digested with the restriction enzymes Hind III or Dra I, analyzed using 1% agarose gels in 1X TBE buffer and stained with SYBR green.
B: RFLP analysis of msg of P. carinii. msg was amplified from the DNA preparations from the lungs of P. carinii infected rats. Lanes 1-4: rats from one facility; lungs were collected from 1998 to 2001; lanes 5-7: rats from a second facility; the lungs were collected from 1993 to 1995. The PCR products were digested with Hind III or Dra I restriction endonucleases, analyzed on 1% agarose gels in 1X TBE buffer and stained with SYBR green.
C. Southern blot analysis of DNA from P. carinii-infected lung samples. Genomic DNA was digested with Hind III or Dra I restriction enzymes. The blot was hybridized with a probe generated by amplifying genomic DNA from P. carinii with primers GK524 and GK526.
Discussion
We have demonstrated by RFLP analysis that there is substantial diversity of msg genes in P. jirovecii among different isolates: no 2 isolates had the same repertoire of 50-100 genes. Based upon sequence analysis of 24 unique msg genes from a single isolate, recombination among msg variants plays a major role in generating msg diversity. Since all msgs appear to be located in clusters near telomeres [3, 15, 32], recombination upstream of the clusters or within the msgs can further increase msg diversity [17]. Recombination has been previously hypothesized to play an important role in generating such haplotype diversity [3, 16,17].
The identification of 2 distinct branches of msg genes in P. jirovecii obtained from a single infected patient was surprising. It is possible that the patient was infected with 2 unique strains of Pneumocystis, although ITS1 as well as UCS typing suggested infection with a single strain. Alternatively, the 2 branches may represent msg genes in a single isolate that were inherited from 2 parental strains via sexual reproduction, and have not yet had a chance to genetically intermingle extensively. This is consistent with the identification of msgs from both branches in individual PCR reactions, suggesting that they are on one DNA fragment. It is also possible that for either biological or other reasons the 2 sets of genes are limited to recombination primarily within a branch rather than across a branch. We do not believe that the 2 branches represent unique families of genes similar to those that we and others have previously identified in P. carinii (e.g. msg and msr) [17, 33], since the upstream primer region for amplification of P. jirovecii msgs includes a highly conserved sequence homologous to the conserved recombination junction element (CRJE) in P. carinii. A primary distinguishing characteristic in P. carinii between msgs and variants is the presence of this CRJE in the former but not the latter.
Recombination between genetically distinct organisms, between genetically identical organisms, or between different msgs within an individual organism could all increase the diversity of the msg repertoire. The conservation of the RFLP pattern in rat and mouse Pneumocystis compared to the diverse patterns seen in human Pneumocystis is striking, but may simply have resulted from examining a captive vs. outbred population; alternatively, it is possible that P. jirovecii has developed a mechanism for increasing msg diversity not present in the other Pneumocystis species. RFLP analysis of Pneumocystis obtained from wild animals rather than colony-bred animals would definitively address this issue.
While the function of Msg may be to facilitate adherence to host cells or proteins [34, 35], Msg antigenic variation likely confers an immunologic advantage to the organism in its interaction with the host. Similar antigenic variability in other species such as African trypanosomes and Borrelia species is associated with evasion of host immune, primarily antibody, responses; repertoire variation in these organisms has been demonstrated and likely contributes to successful immune evasion [18-20]. Given that cell-mediated immune responses, especially CD4+ T lymphocyte responses, appear to be the most critical to controlling Pneumocystis infection [36-38], and that antibody responses to Msg are easily detected in humans [39, 40], the primary function of Msg diversity may be to evade cell-mediated responses. Consistent with this hypothesis, we have not been able to detect in vitro proliferative responses to a recombinant Msg isoform when using human peripheral blood mononuclear cells, despite easily detectable antibodies to the same antigen (unpublished observations); this may reflect a low probability that individuals had been infected with P. jirovecii expressing the cloned msg isoform. While Pneumocystis infection in healthy hosts does not appear to result in a chronic waxing and waning infection [41], as is seen in other species with antigenic variation, such as trypanosomes [42, 43], msg diversity may facilitate re-infection of healthy hosts by delaying development of effective cellular immune responses.
The substantial variability in the msg repertoire demonstrated by RFLP analysis potentially provides a robust method for typing P. jirovecii. Current typing methods rely primarily on examining variations in one or more SNPs within a single or limited number of loci [44], whereas RFLP analysis of amplified msg genes allows interrogation of potentially 50 to 100 genes. Such analysis may help determine the relationship among isolates from putative outbreaks of Pneumocystis pneumonia [45].
Supplementary Material
Acknowledgments
We would like to thank Rene Costello and Howard Mostowski for their assistance with the animal studies.
This research was supported by the Intramural Research Program of the NIH Clinical Center and the National Cancer Institute.
Footnotes
The authors have no conflicting financial interests.
References
- 1.Stringer JR, Keely SP. Genetics of surface antigen expression in Pneumocystis carinii. Infect Immun. 2001;69:627–39. doi: 10.1128/IAI.69.2.627-639.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wada M, Sunkin SM, Stringer JR, Nakamura Y. Antigenic variation by positional control of major surface glycoprotein gene expression in Pneumocystis carinii. J Infect Dis. 1995;171:1563–8. doi: 10.1093/infdis/171.6.1563. [DOI] [PubMed] [Google Scholar]
- 3.Wada M, Nakamura Y. Unique telomeric expression site of major-surface-glycoprotein genes of Pneumocystis carinii. DNA Res. 1996;3:55–64. doi: 10.1093/dnares/3.2.55. [DOI] [PubMed] [Google Scholar]
- 4.Edman JC, Hatton TW, Nam M, et al. A single expression site with a conserved leader sequence regulates variation of expression of the Pneumocystis carinii family of major surface glycoprotein genes. DNA Cell Biol. 1996;15:989–99. doi: 10.1089/dna.1996.15.989. [DOI] [PubMed] [Google Scholar]
- 5.Sunkin SM, Stringer JR. Residence at the expression site is necessary and sufficient for the transcription of surface antigen genes of Pneumocystis carinii. Mol Microbiol. 1997;25:147–60. doi: 10.1046/j.1365-2958.1997.4461806.x. [DOI] [PubMed] [Google Scholar]
- 6.Haidaris CG, Medzihradsky OF, Gigliotti F, Simpson-Haidaris PJ. Molecular characterization of mouse Pneumocystis carinii surface glycoprotein A. DNA Res. 1998;5:77–85. doi: 10.1093/dnares/5.2.77. [DOI] [PubMed] [Google Scholar]
- 7.Kutty G, Ma L, Kovacs JA. Characterization of the expression site of the major surface glycoprotein of human-derived Pneumocystis carinii. Mol Microbiol. 2001;42:183–93. doi: 10.1046/j.1365-2958.2001.02620.x. [DOI] [PubMed] [Google Scholar]
- 8.Radding JA, Armstrong MY, Ullu E, Richards FF. Identification and isolation of a major cell surface glycoprotein of Pneumocystis carinii. Infect Immun. 1989;57:2149–57. doi: 10.1128/iai.57.7.2149-2157.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haidaris PJ, Wright TW, Gigliotti F, Haidaris CG. Expression and characterization of a cDNA clone encoding an immunodominant surface glycoprotein of Pneumocystis carinii. J Infect Dis. 1992;166:1113–23. doi: 10.1093/infdis/166.5.1113. [DOI] [PubMed] [Google Scholar]
- 10.Kovacs JA, Powell F, Edman JC, et al. Multiple genes encode the major surface glycoprotein of Pneumocystis carinii. J Biol Chem. 1993;268:6034–40. [PubMed] [Google Scholar]
- 11.Garbe TR, Stringer JR. Molecular characterization of clustered variants of genes encoding major surface antigens of human Pneumocystis carinii. Infect Immun. 1994;62:3092–101. doi: 10.1128/iai.62.8.3092-3101.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wright TW, Bissoondial TY, Haidaris CG, Gigliotti F, Haidaris PJ. Isoform diversity and tandem duplication of the glycoprotein A gene in ferret Pneumocystis carinii. DNA Res. 1995;2:77–88. doi: 10.1093/dnares/2.2.77. [DOI] [PubMed] [Google Scholar]
- 13.Wyder MA, Rasch EM, Kaneshiro ES. Quantitation of absolute Pneumocystis carinii nuclear DNA content. Trophic and cystic forms isolated from infected rat lungs are haploid organisms. J Eukaryot Microbiol. 1998;45:233–9. doi: 10.1111/j.1550-7408.1998.tb04531.x. [DOI] [PubMed] [Google Scholar]
- 14.Angus CW, Tu A, Vogel P, Qin M, Kovacs JA. Expression of variants of the major surface glycoprotein of Pneumocystis carinii. J Exp Med. 1996;183:1229–34. doi: 10.1084/jem.183.3.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sunkin SM, Stringer JR. Translocation of surface antigen genes to a unique telomeric expression site in Pneumocystis carinii. Mol Microbiol. 1996;19:283–95. doi: 10.1046/j.1365-2958.1996.375905.x. [DOI] [PubMed] [Google Scholar]
- 16.Stringer JR. Antigenic variation in pneumocystis. J Eukaryot Microbiol. 2007;54:8–13. doi: 10.1111/j.1550-7408.2006.00225.x. [DOI] [PubMed] [Google Scholar]
- 17.Keely SP, Renauld H, Wakefield AE, et al. Gene arrays at Pneumocystis carinii telomeres. Genetics. 2005;170:1589–600. doi: 10.1534/genetics.105.040733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barbour AG, Restrepo BI. Antigenic variation in vector-borne pathogens. Emerg Infect Dis. 2000;6:449–57. doi: 10.3201/eid0605.000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borst P. Molecular genetics of antigenic variation. Parasitol Today. 1991;7:29–33. doi: 10.1016/S0167-5699(05)80009-X. [DOI] [PubMed] [Google Scholar]
- 20.Rudenko G. The polymorphic telomeres of the African Trypanosome Trypanosoma brucei. Biochem Soc Trans. 2000;28:536–40. doi: 10.1042/bst0280536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kovacs JA, Halpern JL, Swan JC, Moss J, Parrillo JE, Masur H. Identification of antigens and antibodies specific for Pneumocystis carinii. J Immunol. 1988;140:2023–31. [PubMed] [Google Scholar]
- 22.Mei Q, Turner RE, Sorial V, Klivington D, Angus CW, Kovacs JA. Characterization of major surface glycoprotein genes of human Pneumocystis carinii and high-level expression of a conserved region. Infect Immun. 1998;66:4268–73. doi: 10.1128/iai.66.9.4268-4273.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palmer S, Kearney M, Maldarelli F, et al. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J Clin Microbiol. 2005;43:406–13. doi: 10.1128/JCM.43.1.406-413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma L, Kutty G, Jia Q, Kovacs JA. Characterization of variants of the gene encoding the p55 antigen in Pneumocystis from rats and mice. J Med Microbiol. 2003;52:955–60. doi: 10.1099/jmm.0.05131-0. [DOI] [PubMed] [Google Scholar]
- 25.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lole KS, Bollinger RC, Paranjape RS, et al. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152–60. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salminen MO, Carr JK, Burke DS, McCutchan FE. Identification of breakpoints in intergenotypic recombinants of HIV type 1 by bootscanning. AIDS Res Hum Retroviruses. 1995;11:1423–5. doi: 10.1089/aid.1995.11.1423. [DOI] [PubMed] [Google Scholar]
- 28.Ma L, Kutty G, Jia Q, et al. Analysis of variation in tandem repeats in the intron of the major surface glycoprotein expression site of the human form of Pneumocystis carinii. J Infect Dis. 2002;186:1647–54. doi: 10.1086/345721. [DOI] [PubMed] [Google Scholar]
- 29.Lu JJ, Bartlett MS, Shaw MM, et al. Typing of Pneumocystis carinii strains that infect humans based on nucleotide sequence variations of internal transcribed spacers of rRNA genes. J Clin Microbiol. 1994;32:2904–12. doi: 10.1128/jcm.32.12.2904-2912.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hudson RR, Kaplan NL. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics. 1985;111:147–64. doi: 10.1093/genetics/111.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Achaz G, Palmer S, Kearney M, et al. A robust measure of HIV-1 population turnover within chronically infected individuals. Mol Biol Evol. 2004;21:1902–12. doi: 10.1093/molbev/msh196. [DOI] [PubMed] [Google Scholar]
- 32.Stringer JR, Cushion MT. The genome of Pneumocystis carinii. FEMS Immunol Med Microbiol. 1998;22:15–26. doi: 10.1111/j.1574-695X.1998.tb01183.x. [DOI] [PubMed] [Google Scholar]
- 33.Huang SN, Angus CW, Turner RE, Sorial V, Kovacs JA. Identification and characterization of novel variant major surface glycoprotein gene families in rat Pneumocystis carinii. J Infect Dis. 1999;179:192–200. doi: 10.1086/314558. [DOI] [PubMed] [Google Scholar]
- 34.Wisniowski P, Pasula R, Martin WJ., 2nd Isolation of Pneumocystis carinii gp120 by fibronectin affinity: evidence for manganese dependence. Am J Respir Cell Mol Biol. 1994;11:262–9. doi: 10.1165/ajrcmb.11.3.8086164. [DOI] [PubMed] [Google Scholar]
- 35.Pottratz ST, Paulsrud J, Smith JS, Martin WJ., 2nd Pneumocystis carinii attachment to cultured lung cells by pneumocystis gp 120, a fibronectin binding protein. J Clin Invest. 1991;88:403–7. doi: 10.1172/JCI115318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phair J, Munoz A, Detels R, Kaslow R, Rinaldo C, Saah A. The risk of Pneumocystis carinii pneumonia among men infected with human immunodeficiency virus type 1. Multicenter AIDS Cohort Study Group. N Engl J Med. 1990;322:161–5. doi: 10.1056/NEJM199001183220304. [DOI] [PubMed] [Google Scholar]
- 37.Shellito J, Suzara VV, Blumenfeld W, Beck JM, Steger HJ, Ermak TH. A new model of Pneumocystis carinii infection in mice selectively depleted of helper T lymphocytes. J Clin Invest. 1990;85:1686–93. doi: 10.1172/JCI114621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McFadden DC, Powles MA, Smith JG, Flattery AM, Bartizal K, Schmatz DM. Use of anti-CD4+ hybridoma cells to induce Pneumocystis carinii in mice. Infect Immun. 1994;62:4887–92. doi: 10.1128/iai.62.11.4887-4892.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bishop LR, Kovacs JA. Quantitation of anti-Pneumocystis jiroveci antibodies in healthy persons and immunocompromised patients. J Infect Dis. 2003;187:1844–8. doi: 10.1086/375354. [DOI] [PubMed] [Google Scholar]
- 40.Daly KR, Koch J, Levin L, Walzer PD. Enzyme-linked immunosorbent assay and serologic responses to Pneumocystis jiroveci. Emerg Infect Dis. 2004;10:848–54. doi: 10.3201/eid1005.030497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vestereng VH, Bishop LR, Hernandez B, Kutty G, Larsen HH, Kovacs JA. Quantitative real-time polymerase chain-reaction assay allows characterization of Pneumocystis infection in immunocompetent mice. J Infect Dis. 2004;189:1540–4. doi: 10.1086/382486. [DOI] [PubMed] [Google Scholar]
- 42.Donelson JE. Antigenic variation and the African trypanosome genome. Acta Trop. 2003;85:391–404. doi: 10.1016/s0001-706x(02)00237-1. [DOI] [PubMed] [Google Scholar]
- 43.Horn D. The molecular control of antigenic variation in Trypanosoma brucei. Curr Mol Med. 2004;4:563–76. doi: 10.2174/1566524043360078. [DOI] [PubMed] [Google Scholar]
- 44.Beard CB. Strain typing methods and molecular epidemiology of Pneumocystis pneumonia. Emerg Infect Dis. 2004;10:1729–35. doi: 10.3201/eid1010.030981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manoloff ES, Francioli P, Taffe P, Van Melle G, Bille J, Hauser PM. Risk for Pneumocystis carinii transmission among patients with pneumonia: a molecular epidemiology study. Emerg Infect Dis. 2003;9:132–4. doi: 10.3201/eid0901.020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.