Abstract
Human isolates of Streptococcus pyogenes, a Gram-positive bacterium, are characterized by significant genetic and phenotypic variation. The rgg locus, also known as ropB, is a global transcriptional regulator of genes associated with metabolism, stress responses, and virulence in S. pyogenes strain NZ131 (serotype M49). To assess the breadth of the Rgg regulon, the rgg gene was inactivated in three additional strains representing serotypes M1 (strains SF370 and MGAS5005) and M49 (strain CS101). Changes in gene expression were identified in the post-exponential phase of growth using Affymetrix NimbleExpress Arrays. The results identified an Rgg core-regulon consisting of speB and adjacent hypothetical protein gene, spy2040, and a variable and strain-specific sub-regulon, ranging in size from a single gene (spy1793) in strain MGAS5005 to 43 genes in strain SF370. Thus, both interserotypic and intraserotypic variation is characteristic of the Rgg regulon in S. pyogenes.
Keywords: Streptococcus pyogenes, Rgg, transcriptional regulation
INTRODUCTION
Streptococcus pyogenes (group A streptococcus) is a Gram-positive pathogen that exhibits significant phenotypic diversity, which is likely to contribute to the wide variety of clinical outcomes associated with human infection. The clinical severity of infection ranges from asymptomatic colonization to severe invasive diseases, such as streptococcal toxic shock syndrome and necrotizing fasciitis. Moreover, post-infection sequelae including rheumatic fever, glomerulonephritis, and neurological disorders contribute substantially to the morbidity and mortality associated with the pathogen (Cunningham, 2002).
Clinical isolates of S. pyogenes are differentiated into more than 100 emm-types based on variability of the 5′-region of the emm gene (http://www.cdc.gov/ncidod/biotech/strep/M-ProteinGene_typing.htm), which encodes the LPXTG-anchored adhesin and antiphagocytic M protein (Fischetti, 1989). More generally, clinical isolates can be differentiated into two classes (Class I and Class II) based on reactivity with antibodies directed against the C repeat region of the M protein and the organization and composition of genes proximal to emm in the chromosome. For example, class I, but not class II, strains possess the gene encoding the streptococcal inhibitor of complement (SIC), which contributes to virulence in murine models (Lukomski et al., 2000). Class II, but not class I, strains possess the gene encoding serum opacity factor (SOF), which also contributes to virulence and promotes adherence (Timmer et al., 2006). Class I strains are associated with invasive diseases and acute rheumatic fever, while class II strains are associated with pyoderma and acute glomerulonephritis (Cunningham, 2000). Although differences in the host response are clearly important, specific strains of S. pyogenes are more likely to cause certain diseases than others, which indicates that strain-variable genetic elements contribute to the disease process. The complete genome sequences of more than 12 strains of S. pyogenes have been determined. These include isolates from invasive disease episodes (Ferretti et al., 2001; Beres et al., 2006), rheumatic fever (Smoot et al., 2002; Holden et al., 2007), and puerperal sepsis (Green et al., 2005). The genome content of the strains is heterogeneous, mostly due to various bacteriophages (Banks et al., 2002) and integrated conjugative elements (Beres & Musser, 2007). These strain-variable elements contribute to the so-called pan-genome (Tettelin et al., 2005), or metagenome (Beres & Musser, 2007), which is estimated to consist of approximately 2,500 genes (Lefébure & Stanhope, 2007).
Rgg, also known as RopB, (Chaussee et al., 1999; Lyon et al., 1998) is one member of a family of transcriptional regulators (TIGR01716) encoded in the genomes of some species of low G+C Gram-positive bacteria. Inactivation of rgg in the class II strain NZ131 (serotype M49) is associated with changes in the transcript levels of 706 genes compared to the parental wild-type strain (Dmitriev et al., 2006). Many of these genes encode known or putative virulence factors, such as the hyaluronic acid capsule, C5a peptidase, streptokinase, streptolysins S and O, and mitogenic factor (Chaussee et al., 2001; Dmitriev et al., 2006). Corresponding phenotypic differences identified in the mutant strain include: (i) tolerance to penicillin-mediated killing and thermal and oxidative stressors (Chaussee et al., 2004; Chaussee et al., 2006); (ii) catabolism of arginine during the exponential phase of growth in the presence of glucose (Chaussee et al., 2003); (iii) increased production of DNase, NADase, and SLO; (iv) an inability to grow in chemically defined media (CDM) containing sucrose, fructose or mannose as the primary carbon source; and (v) decreased frequency of prophage NZ131.1 induction (Dmitriev et al., 2006). Thus, Rgg is an important global regulator of genes associated with metabolism, stress response, and virulence in strain NZ131.
The purpose of this study was to assess the breadth of the Rgg regulon in additional isolates of S. pyogenes including two M1 serotypes representing class I strains and an additional M49 serotype representing class II strains. DNA microarrays identified an Rgg core-regulon and a variable and strain-specific sub-regulon, which suggests that diversity among regulatory circuits contributes to the phenotypic diversity of S. pyogenes.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions
The wild-type S. pyogenes strains MGAS5005, SF370, and CS101 have been described (Ferretti et al., 2001; Sumby et al., 2005; Haanes et al., 1992). S. pyogenes was grown at 37 °C in a 5% CO2 atmosphere without agitation in either Todd-Hewitt broth (Becton Dickinson, Sparks, MD) containing 0.2% (wt/vol) yeast extract or CDM (Dmitriev et al., 2006). Escherichia coli strain DH5α was purchased from Gibco-BRL (Gaithersburg, MD), and the suicide cloning vector pVA891-2 was kindly provided by H. Malke (Malke et al., 1994).
DNA techniques
Plasmid DNA was isolated from E. coli using Plasmid Midi Kit (QIAGEN, Valencia, CA). From agarose gels, DNA was isolated using the QIAquick PCR Purification Kit (QIAGEN). PCR products were purified using DNA Clean & Concentrator (Orange, Calif.). Most of other routine molecular techniques were done as previously described (Sambrook et al., 1989).
Insertional inactivation of rgg
The rgg gene was insertional inactivated in each strain as previously described (Chaussee et al., 1999). Briefly, the entire rgg gene was amplified using the primers RGG+1 (5′-CTG GAG CTG TTG AGA TAA ACT AC-3′) and RGG-4 (5′-GGC TAT TGA CCT TAT GCA CC-3′), and digested with EcoRI and HindIII, which had corresponding restriction sites within rgg. The resulting 592 bp fragment was cloned into the vector pVA891-2. Following E. coli transformation, the recombinant plasmid was isolated and used to transform S. pyogenes. Transformants were selected on agar plates containing 2.5 μg mL−1 of erythromycin. Insertional inactivation was confirmed in each strain by nucleotide sequencing and Southern blotting, as previously described (Chaussee et al., 1999). The heterologous DNA inserted into the rgg locus was identical in all three rgg mutant strains and identical to the previously described NZ131 rgg mutant (Chaussee et al., 1999).
Pulsed field gel electrophoresis (PFGE) analysis
Chromosomal DNA for PFGE analysis was isolated and digested with SmaI as described (Elliott et al., 1998). Fragments were separated in 1% agarose gel in 0.5 × TBE buffer (GeneLine apparatus, Beckman Instruments, Calif.) using the following conditions: 5 min, 170 V, 5 s pulse time; 16 h, 200 V, 40 s pulse time; and 8 h, 200 V, 8 s pulse time, and visualized with ethidium bromide, 0.5 μg mL−1.
DNA microarray analysis
RNA was isolated from 40 ml cultures of S. pyogenes in the post-exponential phase of growth (Fig. 1) with an RNeasy Mini Kit (QIAGEN). Affymetrix NimbleExpress Arrays designed based on the S. pyogenes strain SF370 genome sequence (Ferretti et al., 2001) were purchased from Affymetrix (Santa Clara, CA). In addition to the SF370 genome sequence, 660 known streptococcal bacteriophage genes were included in the array platform. Together, the arrays consisted of 3,203 qualifiers representing 2,354 predicted S. pyogenes ORFs, 804 intergenic region probes, and 45 control oligos used for spike-ins. Microarray hybridization and analysis of the data was done as recently published (Dmitriev et al., 2006). The average signal intensity value of each gene was transformed to a log2 (log base 2) value. The change between two experimental conditions (n-fold) was calculated by taking the ratio of the signal intensity (difference of the log2 value) between experimental conditions. Present and absent calls were assessed and statistically significant genes (T-test; P value ≤ 0.05) were considered to be differently expressed. All of the microarray data are available through the Gene Expression Omnibus data repository via accession numbers GSE 7335 and 7341.
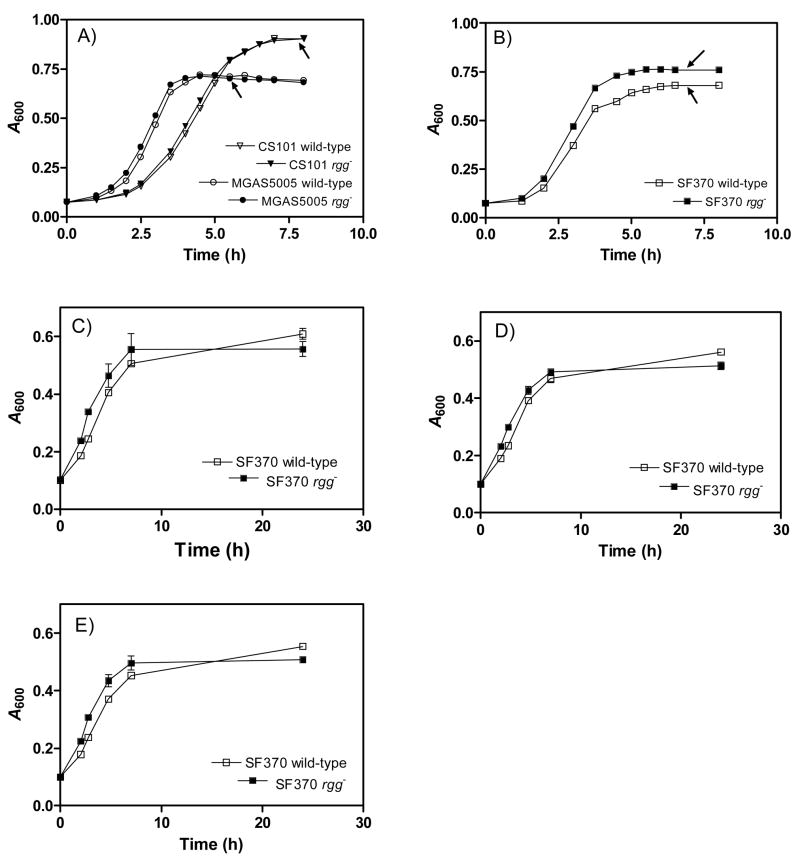
Fig. 1.
Growth of the wild-type strains and corresponding rgg mutants in Todd-Hewitt broth (A, B) and CDM containing 2% glucose (C), fructose (D) or sucrose (E) as the primary carbon and energy source. Arrows designate points of RNA isolation for microarray analysis. The results are shown as the means and standard errors of the means from two independent experiments.
Determination of SpeB, NADase, DNase and SLO cytolytic activities
SpeB, NADase, DNase and SLO cytolytic activities were determined as previously described (Chaussee et al., 1999; Bricker et al., 2002; Dmitriev, 2006).
Quantitative reverse transcription (RT)-PCR
Oligonucleotide primers and TaqMan probes used in the study were previously published (Dmitriev et al., 2006). In addition, primers and fluorescent probes for the phosphoribosylglycinamide formyltransferase purN gene (5′-CTT GGC CTA TGA GAG GCG TAT T-3′, 5′-CCG TGG GCA CCT GGA A-3′, and 5′-TCA ATA TTC ACC CAG CCT ACC TGC CTG AA-3′) and the streptolysin S sagA gene (5′-TTG CTC CTG GAG GCT GCT-3′, 5′-CTT CCG CTA CCA CCT TGA GAA T-3′, and 5′-ACC ACT TCC AGT AGC AAT TGA GAA GCA ACA AG-3′) were designed with Primer Express 2.0 software (ABI Prism, PE Biosystems, Framingham, Mass.) and purchased from Sigma-Genosys (The Woodlands, TX). Amplification and detection were done with the ABI Prism 7700 Sequence Detection System (PE Applied Biosystems) using TaqMan One-Step RT-PCR Master Mix reagents (Roche, Indianapolis, In.), as previously described (Chaussee et al., 2003).
Sequence analysis
The rgg genes from S. pyogenes strains MGAS5005, SF370, and CS101 were sequenced using ABI Prism 377 Perkin-Elmer Sequencer and Big Dye Terminator Kit (Applied Biosystems) with the primers RGG+1 and RGG-4. The data were deposited into the GenBank database under accession numbers DQ009036 and DQ176644. The deduced Rgg proteins of all three strains were identical to that of the NZ131 strain. The 498 bp regions upstream the rgg genes sequenced with the primers 5′-CGG CAA ATA CTG GGT TAG CAA GA-3′ and 5′-GGA TGC CTA ATG AAT TCA ACG GTT T-3′ were also identical in all the strains.
RESULTS
Expression of virulence factors in the rgg mutant strains
To assess the Rgg regulon in S. pyogenes, three widely studied strains, representing both class I (MGAS5005 and SF370, serotypes M1) and class II (CS101, serotype M49) organisms, were selected for study. Inactivation of rgg in strains CS101, MGAS5005, and SF370 as described in the Materials and Methods, abrogated SpeB expression (Fig. 2), similar to previously described mutants created in strains NZ131 and HSC5 (Chaussee et al., 1999; Lyon et al., 1998). Southern blotting showed that the heterologous DNA inserted into only the rgg locus (data not shown). These results indicate that Rgg-dependent activation of speB expression is conserved among both class I and II clinical isolates of S. pyogenes.
Fig. 2.
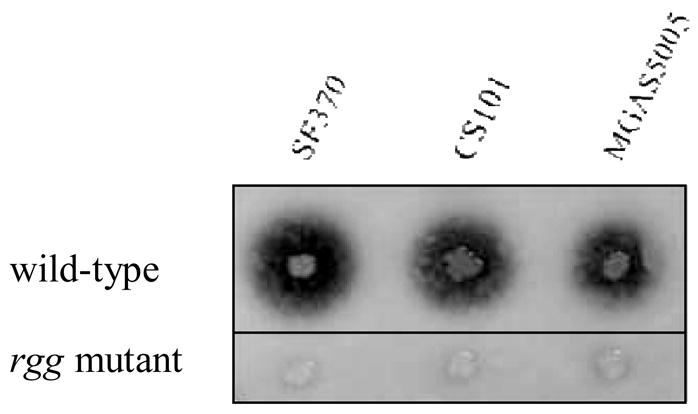
Abrogation of SpeB expression in the strains SF370, CS101, and MGAS5005 following rgg inactivation.
The SLO, NADase, and DNase activities, which were elevated in the NZ131 rgg mutant strain (Dmitriev et al., 2006), were similarly measured in strains MGAS5005, SF370, and CS101 and the corresponding rgg mutant derivatives. Despite variation in SLO, NADase, and DNase activities among the wild-type strains, no difference between wild-type and corresponding rgg mutant strains was observed (data not shown).
Growth of wild-type and the rgg mutant strains in Todd-Hewitt broth and CDM
Inactivation of rgg in strain NZ131 is associated with altered growth in Todd-Hewitt broth and an inability to use non-glucose carbohydrates as the primary carbon and energy sources when grown with CDM (Chaussee et al., 2003; Dmitriev et al., 2006). To determine if similar changes occurred in strains CS101, MGAS5005, and SF370, the growth of wild-type strains and mutant derivatives was examined. Growth of wild-type CS101 and MGAS5005 strains was identical to the corresponding mutant strains in both Todd-Hewitt broth (Fig. 1) and CDM containing 2% glucose, sucrose or fructose (data not shown). In contrast, the SF370 rgg mutant strain had a slightly shorter lag period and different growth yield compared to the parental strain when grown with Todd-Hewitt broth (Fig. 1) or CDM containing various carbohydrates as the primary carbon and energy source (Fig. 1). Thus, while there was a minor effect on growth in strain SF370 following rgg inactivation, mutant strains retained the ability to use sucrose, glucose and fructose as the primary carbon and energy source during growth with CDM, in contrast to results previously obtained with strain NZ131 (Dmitriev et al., 2006).
DNA microarray analysis of the wild-type and rgg mutant strains
To identify Rgg-regulated genes, the transcriptomes of CS101, MGAS5005, and SF370 wild-type and rgg mutant strains were analyzed. Our initial experiments revealed few differences in the transcriptomes of MGAS5005 rgg mutant and CS101 rgg mutant compared to corresponding wild-type strains during exponential phase (data not shown). These data are in agreement with earlier observations in strain NZ131, in which rgg inactivation alters a larger number of genes during the post-exponential phase of growth compared to the exponential phase (Dmitriev et al., 2006). Therefore, to identify the largest number of Rgg-regulated genes in MGAS5005, CS101, and SF370 strains, RNA was isolated from post-exponential phase cultures and gene transcripts were measured with Affymetrix NimbleExpress Arrays. Inactivation of rgg in strains MGAS5005, CS101, and SF370 was associated with a 2-fold or more (P ≤ 0.05) change in the expression of 3, 13, and 45 loci, respectively (Table 1). In strain MGAS5005, 1 and 2 gene transcripts were more abundant and less abundant, respectively, in the mutant strain compared to the wild-type strain. In strain CS101, 10 and 3 gene transcripts were more and less abundant, respectively, in the mutant strain compared to the wild-type strain. Finally, in strain SF370, 28 and 17 gene transcripts were more abundant and less abundant, respectively, in the mutant strain compared to the wild-type strain (Table 1). The known or putative functions of the majority of Rgg-regulated genes in all of the strains were associated with replication, transcription and translation, and metabolism. Quantitative RT-PCR was used to validate the microarray data (Table 2).
Table 1.
Transcriptome changes (2-fold or greater; P values ≤ 0.05) associated with rgg nactivation during post-exponential growth
| Fold changes (rgg/wt)b |
|||||
|---|---|---|---|---|---|
| SPy no.a | Gene | Description | SF370 | CS101 | MGAS 5005 |
|
More abundant genes in rgg mutants
| |||||
| 25 | phosphoribosylformylglycinamidine synthase II | 6 | -c | - | |
| 26 | purF | phosphoribosylpyrophosphate amidotransferase | 5 | - | - |
| 27 | purM | phosphoribosylformylglycinamide cyclo-ligase | 6 | - | - |
| 28 | purN | phosphoribosylglycinamide formyltransferase | 7 | - | - |
| 32 | purD | phosphoribosylamine-glycine ligase | 3 | - | - |
| 34 | purK | phosphoribosylaminoimidazole carboxylase II | 4 | - | - |
| 35 | abiR | abortive infection phage resistance protein | 2 | - | - |
| 36 | purB | adenylosuccinate lyase | 2 | - | - |
| 37*d | hypothetical protein | 3 | - | - | |
| 39 | protein-tyrosine phosphatase | 2 | - | - | |
| 663 | hypothetical protein | 2 | - | - | |
| 670 | hypothetical protein | 2 | - | - | |
| 673 | hypothetical protein | 2 | - | - | |
| 728 | hypothetical protein | - | 2 | - | |
| 732* | hypothetical protein | 3 | - | - | |
| 733* | hypothetical protein | 2 | - | - | |
| 738* | sagA | streptolysin S associated protein | 2 | - | - |
| 739* | sagB | streptolysin S associated ORF | 4 | - | - |
| 742* | hypothetical protein | 4 | - | - | |
| 744* | ABC transporter | 3 | - | - | |
| 746* | ABC-2 type transport system permease protein | 3 | - | - | |
| 1037 | ybbR | hypothetical protein | - | 3 | - |
| 1125* | GTP pyrophosphokinase | - | 3 | - | |
| 1126* | hypothetical protein | - | 3 | - | |
| 1154 | hypothetical protein | - | 2 | - | |
| 1243 | pstC | phosphate ABC transporter (permease protein) | - | 3 | - |
| 1267 | pcrA | ATP-dependent DNA helicase | 3 | - | - |
| 1326 | hypothetical protein | - | 2 | - | |
| 1600 | hyaluronidase | 2 | - | - | |
| 1718 | esterase | 2 | - | - | |
| 1793 | ABC transporter | - | - | 2 | |
| 1815* | scrA | sucrose-specific PTS permease, enzyme II | 2 | - | - |
| 1858 | pepXP | X-Pro dipeptidyl-peptidase IV | - | 3 | - |
| 1884 | similar to several eukaryotic hypersensitive- | 2 | - | - | |
| induced response proteins | |||||
| 1916 | lacG | phospho-beta-D-galactosidase | 2 | - | - |
| 2032 | ATP-binding cassette transporter-like protein | - | 3 | - | |
| 2107 | oxidoreductase | 2 | - | - | |
| 2148 | mutS | DNA mismatch repair protein | 2 | - | - |
|
| |||||
| Less abundant genes in rgg mutants | |||||
|
| |||||
| 246 | rnpA | ribonuclease P protein component | −3 | - | - |
| 600 | hypothetical protein | −4 | - | - | |
| 609 | ftsW | cell division protein | −2 | - | - |
| 627 | regR | transcriptional regulator (LacI family) | −2 | 2 | - |
| 1016 | hypothetical protein | −3 | - | - | |
| 1017 | hypothetical protein | −3 | - | - | |
| 1140 | tdk2 | thymidine kinase | −3 | - | - |
| 1141 | prfA | peptide chain release factor 1 | −2 | - | - |
| 1201 | ylxM | DNA-binding protein | −3 | - | - |
| 1202 | gntR | transcription regulator, GntR family | −4 | - | - |
| 1210 | hypothetical protein | −3 | - | - | |
| 1233 | coaA | pantothenate kinase | −2 | - | - |
| 1632 | gmk | guanylate kinase | −2 | - | - |
| 1741 | hypothetical protein | −3 | - | - | |
| 2037* | prsA | peptidyl prolyl isomerase | - | −11 | - |
| 2039* | speB | pyrogenic exotoxin B | −37 | −71 | −35 |
| 2040* | hypothetical protein | −146 | −335 | −69 | |
| 2207 | trsA | tryptophanyl-tRNA synthetase | −3 | - | - |
SPy numbers designate open reading frames based on the SF370 S. pyogenes genome annotation (Ferretti et al., 2001).
Change in transcript level of rgg mutant compared to that of the wild-type.
rgg inactivation doesn’t affect transcript level.
Asterisks indicate genes similarly regulated in NZ131 strain (Dmitriev et al., 2006).
Table 2.
Correlation between results obtained with DNA microarrays and quantitative TaqMan RT-PCR
| SPy no.a | Designation b | Strain | Fold changes (rgg/wt) c |
|
|---|---|---|---|---|
| RT-PCR (Log10) | Microarrays (Log10) | |||
| 0028 | purN (phosphoribosylglycinamide formyltransferase) | SF370 | 1.72 | 0.87 |
| 0165 | spn (NAD-glycohydrolase) | CS101 | 0.33 | 0.46 |
| 0165 | spn (NAD-glycohydrolase) | MGAS5005 | 0.21 | 0.24 |
| 0738 | sagA (streptolysin S) | SF370 | 0.76 | 0.32 |
| 0738 | sagA (streptolysin S) | MGAS5005 | −0.06 | −0.16 |
| 1547 | arcA (arginine deiminase) | SF370 | 0.35 | 0.12 |
| 2039 | speB (pyrogenic exotoxin B) | CS101 | −3.5 | −1.8 |
| 2039 | speB (pyrogenic exotoxin B) | SF370 | −2.28 | −1.6 |
| 2039 | speB (pyrogenic exotoxin B) | MGAS5005 | −1.7 | −1.5 |
| 2043 | mf (mitogenic factor) | CS101 | −0.02 | −0.16 |
| 2043 | mf (mitogenic factor) | SF370 | 0.45 | 0.14 |
SPy numbers designate open reading frames based on the SF370 S. pyogenes genome annotation (Ferretti et al., 2001).
Gene designations are in italics, and the corresponding protein function is in parentheses.
Change in transcript level of rgg mutant compared to that of the wild-type.
Surprisingly, the only genes similarly affected by rgg inactivation in all three strains were speB and spy2040, which are co-transcribed (Neely et al., 2003; Ma et al., 2006). In addition, only the expression of purine metabolism genes (spy25-spy28) in strain SF370 and the peptidyl prolyl isomerase gene (prsA) in strain CS101 were altered by more than 5-fold following rgg inactivation (Table 1). The results indicate that Rgg influences transcription in strains MGAS5005, SF370, and CS101 during the post-exponential phase of growth; however, the magnitude and extent of transcriptional changes is not nearly as great as those previously identified in strain NZ131 (Dmitriev et al., 2006).
Inactivation of rgg alters transcription of regulatory genes in strains SF370 and CS101, but not in strain MGAS5005
Inactivation of rgg in strain NZ131 altered the transcript levels of 20 genes encoding transcriptional regulatory proteins (Dmitriev et al., 2006). Such perturbation of other regulatory networks presumably contributes to the genomewide changes associated with rgg inactivation in this strain (Chaussee et al., 2002; Dmitriev et al., 2006). In contrast, rgg inactivation altered the abundance of three regulatory gene transcripts (sagA, regR and gntR) in strain SF370, and only one (regR) in CS101. No change in regulatory gene expression was detected in the rgg mutant of strain MGAS5005 (Table 1). Thus, the number of differences in the expression of regulatory genes in rgg mutant strains correlated with the number of differences in structural gene expression (Table 1). The results indicate that many genes in the Rgg regulon are controlled by strain-specific secondary mechanisms that remain to be elucidated.
DISCUSSION
Rgg core- and sub-regulons
Strain CS101 (serotype M49; class II) was selected to facilitate the identification of intraserotypic variation in the Rgg regulon by comparing the results to those previously obtained with the strain NZ131 (serotype M49; class II) (Dmitriev et al., 2006). In addition, strains MGAS5005 and SF370 (serotypes M1; class I) were selected to identify potential differences within serotype M1 and to identify potential interserotypic and class differences in the regulon by comparing the results to those obtained with M49 strains. The genome sequences of MGAS5005, SF370, and NZ131, but not CS101, have been determined (Ferretti et al., 2001; McShan et al., 2006; Sumby et al., 2005). The M1 strains have similar PFGE patterns of chromosomal DNA, which were different from those of M49 strains (data not shown). Furthermore, MGAS5005 and SF370 belong to the same Multi Locus Sequence Type ST28 (http://www.mlst.net), and their Rgg regulons were expected to be similar. Surprisingly, the Rgg regulon varied significantly not only among the serotypes, but even among strains of the same serotype and Multi Locus Sequence Type. Only speB and adjacent spy2040 (encoding a 56 amino acid hypothetical protein) gene transcripts were less abundant in all of the rgg mutant strains (Table 1), indicating that these co-transcribed genes (Neely et al., 2003; Ma et al., 2006; Dmitriev et al.; 2006) comprise the Rgg core-regulon. Rgg binds to DNA in the promoter region of speB (Neely et al., 2003), indicating that the core-regulon is directly regulated by Rgg. Other genes, which are influenced by Rgg, varied from a single gene (spy1793) in strain MGAS5005 to 43 genes in the strain SF370. These genes can be considered as part of the Rgg sub-regulon, which is strain-variable.
Rgg interacts with other regulons
The strains used in this study were selected, in part, because they encode an identical Rgg polypeptide, have identical rgg promoter regions, and have similar levels of rgg expression, as determined with TaqMan quantitative RT-PCR (data not shown). Thus, the strain-associated differences in the regulon are not the result of Rgg sequence variation or rgg transcript levels, as previously observed for other streptococcal regulatory proteins (Vickerman et al., 2003; Vahling & McIver, 2005; Loughman & Caparon, 2007).
An association between the number of Rgg-regulated transcription factors and the total number of Rgg-regulated structural genes suggests that many changes in the sub-regulon are due to indirect effects associated with the perturbation of other regulatory circuits. For example, inactivation of rgg in strain NZ131 alters the expression of several regulatory genes (Dmitriev et al., 2006), each of which is present in the genomes of strains SF370 and MGAS5005; however, only sagA, regR, and gntR expression was altered following inactivation of rgg in strain SF370. All chromosomally encoded regulatory proteins present in strain SF370 are also encoded by strain 5005, suggesting that the differences in the sub-regulon between the strains is not due to compositional variation in the set of chromosomally encoded regulatory genes. Of course allelic variation or differential expression of regulatory proteins, including novel regulators such as LacD.1 (Loughman & Caparon, 2006), might contribute to strain-associated differences in the Rgg sub-regulon. In addition, S. pyogenes strains are polylysogenic and may differ in the number and types of bacteriophages present in the chromosome. For example, three bacteriophages (370.1, 370.2, and 370.4) are present in the SF370 strain but absent in the MGAS5005 strain. Similarly, two bacteriophages (5005.1 and 5005.3) are present in strain MGAS5005 but absent in the SF370 strain. Each prophage encodes several regulatory proteins involved in structural gene expression and lysogeny. Given that Rgg shares similarity with bacteriophage-encoded regulators, it is possible that the strain-associated differences in bacteriophage-encoded regulatory proteins influences the Rgg sub-regulon; although, further experiments are clearly necessary to test this hypothesis.
Regulon variation in human bacterial pathogens
The diverse clinical manifestations associated with S. pyogenes are due, in part, to variation in the gene content of strains. In addition, strain- or isolate-specific variation in gene expression described in human pathogens significantly contributes to phenotypic diversity and significantly impacts host-pathogen interactions (Kwinn et al., 2007). In Streptococcus pneumoniae, inactivation of a response regulator gene (rr09) altered the transcript levels of 102 and 80 genes in strains D39 and TIGR4, respectively; however, the expression of only 7 of these genes were similarly affected in the two strains (Hendriksen et al., 2007). Similar diversity has been described in the BvgAS regulon of Bordetella bronchiseptica and Bordetella pertussis (Cummings et al., 2006). In S. pyogenes, the global transcriptional regulator Mga promotes the expression of a number virulence factors, including M protein (emm) and the C5a peptidase (scpA) (Hondorp & McIver, 2007). Mga influences the expression of 204, 201, and 37 genes in strains of serotypes serotypes M4, M1, M6, respectively. Notably, only emm, scpA, and spy2036 were similarly affected by mga inactivation in all three strains (Ribardo & McIver, 2006). The variation in Rgg regulon, in conjunction with variation in the Mga regulon, indicates that significant diversity exists among virulence-associated regulatory circuits of S. pyogenes. Additional information related to the molecular basis for such variation is thus necessary to understand the regulation of virulence factor expression in S. pyogenes.
In summary, we identified Rgg-regulated genes in strains representing class I and II organisms (M1 and M49 serotypes). The results show both inter- and intraserotypic variation in the Rgg regulon, which was consistent with results of biochemical, microbiological and quantitative real-time PCR assays. Such plasticity in regulatory circuits may provide pathogens with a means to adapt rapidly to changes in host-pathogen interactions.
Acknowledgments
We thank A. Erkine, K. Weaver, A. Manna, and A. Ballal for critical review of the manuscript and technical assistance, and B. Buttaro for strain CS101. This work was supported by NIAID/NIH grant RO1 AIO52147 to M.S.C., NIH grant 2 P20 RR016479, and Russian President Grant MD-374.2007.4 to A.V.D.
References
- Banks DJ, Beres SB, Musser JM. The fundamental contribution of phages to GAS evolution, genome diversification and strain emergence. Trends Microbiol. 2002;10:515–521. doi: 10.1016/s0966-842x(02)02461-7. [DOI] [PubMed] [Google Scholar]
- Beres SB, Musser JM. Contribution of exogenous genetic elements to the group a streptococcus metagenome. PLoS ONE. 2007;2(8):e800. doi: 10.1371/journal.pone.0000800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beres SB, Richter EW, Nagiec MJ, Sumby P, Porcella SF, DeLeo FR, Musser JM. Molecular genetic anatomy of inter- and intraserotype variation in the human bacterial pathogen group A Streptococcus. Proc Natl Acad Sci USA. 2006;103:7059–7064. doi: 10.1073/pnas.0510279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricker AL, Cywes C, Ashbaugh CD, Wessels MR. NAD+-glycohydrolase acts as an intracellular toxin to enhance the extracellular survival of group A streptococci. Mol Microbiol. 2002;44:257–269. doi: 10.1046/j.1365-2958.2002.02876.x. [DOI] [PubMed] [Google Scholar]
- Chaussee MS, Ajdic D, Ferretti JJ. The rgg gene of Streptococcus pyogenes NZ131 positively influences extracellular SPE B production. Infect Immun. 1999;67:1715–1722. doi: 10.1128/iai.67.4.1715-1722.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaussee MA, Callegari EA, Chaussee MS. Rgg regulates growth phase-dependent expression of proteins associated with secondary metabolism and stress in Streptococcus pyogenes. J Bacteriol. 2004;186:7091–7099. doi: 10.1128/JB.186.21.7091-7099.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaussee MS, Watson RO, Smoot JC, Musser JM. Identification of Rgg-regulated exoproteins of Streptococcus pyogenes. Infect Immun. 2001;69:822–831. doi: 10.1128/IAI.69.2.822-831.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaussee MS, Somerville GA, Reitzer L, Musser JM. Rgg coordinates virulence factor synthesis and metabolism in Streptococcus pyogenes. J Bacteriol. 2003;185:6016–6024. doi: 10.1128/JB.185.20.6016-6024.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaussee MA, McDowell EJ, Rieck LD, Callegari EA, Chaussee MS. Proteomic analysis of a penicillin-tolerant rgg mutant strain of Streptococcus pyogenes. J Antimicrob Chemother. 2006;58:752–759. doi: 10.1093/jac/dkl319. [DOI] [PubMed] [Google Scholar]
- Chaussee MS, Sylva GL, Sturdevant DE, Smoot LM, Graham MR, Watson RO, Musser JM. Rgg influences the expression of multiple regulatory loci to coregulate virulence factor expression in Streptococcus pyogenes. Infect Immun. 2002;70:762–770. doi: 10.1128/iai.70.2.762-770.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings CA, Bootsma HJ, Relman DA, Miller JF. Species- and strain-specific control of a complex, flexible regulon by Bordetella BvgAS. J Bacteriol. 2006;188:1775–1785. doi: 10.1128/JB.188.5.1775-1785.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham MW. Pathogenesis of group A streptococcal infections. Clin Microbiol Rev. 2000;13:470–511. doi: 10.1128/cmr.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitriev AV, McDowell EJ, Kappeler KV, Chaussee MA, Rieck LD, Chaussee MS. The Rgg regulator of Streptococcus pyogenes influences the utilization of nonglucose carbohydrates, prophage induction, and expression of the NAD-glycohydrolase virulence operon. J Bacteriol. 2006;188:7230–7241. doi: 10.1128/JB.00877-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott JA, Farmer KD, Facklam RR. Sudden increase in isolation of group B streptococci, serotype V, is not due to emergence of a new pulsed-field gel electrophoresis type. J Clin Microbiol. 1998;36:2115–2116. doi: 10.1128/jcm.36.7.2115-2116.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti JJ, McShan WM, Ajdic D, et al. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc Natl Acad Sci USA. 2001;98:4658–4663. doi: 10.1073/pnas.071559398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischetti VA. Streptococcal M protein: molecular design and biological behavior. Clin Microbiol Rev. 1989;2:285–314. doi: 10.1128/cmr.2.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham MR, Smoot LM, Migliaccio CA, Virtaneva K, Sturdevant DE, Porcella SF, Federle MJ, Adams GJ, Scott JR, Musser JM. Virulence control in group A streptococcus by a two-component gene regulatory system: global expression profiling and in vivo infection modeling. Proc Natl Acad Sci USA. 2002;99:13855–13860. doi: 10.1073/pnas.202353699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green NM, Zhang S, Porcella SF, Nagiec MJ, Barbian KD, Beres SB, LeFebvre RB, Musser JM. Genome sequence of a serotype M28 strain of group A Streptococcus: potential new insights into puerperal sepsis and bacterial disease specificity. J Infect Dis. 2005;192:760–770. doi: 10.1086/430618. [DOI] [PubMed] [Google Scholar]
- Haanes EJ, Heath DG, Cleary PP. Architecture of the vir regulons of group A streptococci parallels opacity factor phenotype and M protein class. J Bacteriol. 1992;174:4967–4976. doi: 10.1128/jb.174.15.4967-4976.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriksen WT, Silva N, Bootsma HJ, Blue CE, Paterson GK, Kerr AR, de Jong A, Kuipers OP, Hermans PW, Mitchell TJ. Regulation of gene expression in Streptococcus pneumoniae by response regulator 09 is strain dependent. J Bacteriol. 2007;189:1382–1389. doi: 10.1128/JB.01144-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden MTG, Scott A, Cherevach I, et al. Complete genome of acute rheumatic fever-associated serotype M5 Streptococcus pyogenes strain Manfredo. J Bacteriol. 2007;189:1473–1477. doi: 10.1128/JB.01227-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondorp ER, McIver KS. The Mga virulence regulon: infection where the grass is greener. Mol Microbiol. 2007;66:1056–1065. doi: 10.1111/j.1365-2958.2007.06006.x. [DOI] [PubMed] [Google Scholar]
- Kwinn LA, Khosravi A, Aziz RK, Timmer AM, Doran KS, Kotb M, Nizet V. Genetic characterization and virulence role of the RALP3/LSA locus upstream of the streptolysin S operon in invasive M1T1 group A streptococci. J Bacteriol. 2007;189:1322–1329. doi: 10.1128/JB.01256-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefébure T, Stanhope MJ. Evolution of the core and pan-genome of Streptococcus: positive selection, recombination, and genome composition. Genome Biol. 2007;8(5):R71. doi: 10.1186/gb-2007-8-5-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughman JA, Caparon MG. Contribution of invariant residues to the function of Rgg family transcription regulators. J Bacteriol. 2007;189:650–655. doi: 10.1128/JB.01437-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughman JA, Caparon MG. A novel adaptation of aldolase regulates virulence in Streptococcus pyogenes. EMBO J. 2006;25:5412–5422. doi: 10.1038/sj.emboj.7601393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukomski C, Hoe NP, Abdi I, Rurangirwa J, Kordari P, Liu M, Dou SJ, Adams GG, Musser JM. Nonpolar inactivation of the hypervariable streptococcal inhibitor of complement gene (sic) in serotype M1 Streptococcus pyogenes significantly decreases mouse mucosal colonization. Infect Immun. 2000;68:535–542. doi: 10.1128/iai.68.2.535-542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon WR, Gibson CM, Caparon MG. A role for trigger factor and an rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 1998;17:6263–6275. doi: 10.1093/emboj/17.21.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Bryant AE, Salmi DB, Hayes-Schroer SM, McIndoo E, Aldape MJ, Stevens DL. Identification and characterization of bicistronic speB and prsA gene expression in the group A streptococcus. J Bacteriol. 2006;188:7626–7634. doi: 10.1128/JB.01059-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malke H, Mechold U, Gase K, Gerlach D. Inactivation of the streptokinase gene prevents Streptococcus equisimilis H46A from acquiring cell-associated plasmin activity in the presence of plasminogen. FEMS Microbiol Lett. 1994;116:107–112. doi: 10.1111/j.1574-6968.1994.tb06683.x. [DOI] [PubMed] [Google Scholar]
- Neely MN, Lyon WR, Runft DL, Caparon M. Role of RopB in growth phase expression of the SpeB cysteine protease of Streptococcus pyogenes. J Bacteriol. 2003;185:5166–5174. doi: 10.1128/JB.185.17.5166-5174.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribardo DA, McIver KS. Defining the Mga regulon: comparative transcriptome analysis reveals both direct and indirect regulation by Mga in the group A streptococcus. Mol Microbiol. 2006;62:491–508. doi: 10.1111/j.1365-2958.2006.05381.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- McShan WM, Savic DJ, Karasawa T, Lin SP, Roe B, Ferretti JJ. The Streptococcus genome era. International Congress Series. 2006;1289:171–174. [Google Scholar]
- Sitkiewicz I, Musser JM. Expression microarray and mouse virulence analysis of four conserved two-component gene regulatory systems in group A Streptococcus. Infect Immun. 2006;74:1339–1351. doi: 10.1128/IAI.74.2.1339-1351.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot JC, Barbian KD, Van Gompel JJ, et al. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc Natl Acad Sci USA. 2002;99:4668–4673. doi: 10.1073/pnas.062526099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumby P, Porcella SF, Madrigal AG, et al. Evolutionary origin and emergence of a highly successful clone of serotype M1 group A Streptococcus involved multiple horizontal gene transfer events. J Infect Dis. 2005;192:771–782. doi: 10.1086/432514. [DOI] [PubMed] [Google Scholar]
- Tettelin H, Masignani V, Cieslewicz MJ, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome. Proc Natl Acad Sci USA. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmer AM, Kristian SA, Datta V, Jeng A, Gillen CM, Walker MJ, Beall B, Nizet V. Serum opacity factor promotes group A streptococcal epithelial cell invasion and virulence. Mol Microbiol. 2006;62:15–25. doi: 10.1111/j.1365-2958.2006.05337.x. [DOI] [PubMed] [Google Scholar]
- Vahling CM, McIver KS. Identification of residues responsible for the defective virulence gene regulator Mga produced by a natural mutant of Streptococcus pyogenes. J Bacteriol. 2005;187:5955–5966. doi: 10.1128/JB.187.17.5955-5966.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickerman MM, Wang M, Baker LJ. An amino acid change near the carboxyl terminus of the Streptococcus gordonii regulatory protein Rgg affects its abilities to bind DNA and influence expression of the glucosyltransferase gene gtfG. Microbiol. 2003;149:399–406. doi: 10.1099/mic.0.25983-0. [DOI] [PubMed] [Google Scholar]