Summary
Surface expression of human leukocyte antigen (HLA) class I antigens on melanoma lines was evaluated by locus-specific monoclonal antibodies (mAbs) with three different techniques: Fluorescence-activated cell sorting (FACS), immunohistochemistry with cytospin preparation (ICP), and complement-mediated cytotoxicity (CMC). Eleven HLA class I-expressing cell lines developed from metastases were used. Specific expression of HLA loci was examined under routine culture conditions and after 48-h incubation in interferon-γ (IFN-γ: 500 U/ml). Loss of allelic expression was seen in one line (586-MEL): Products of genes coding for HLA-A29 and -B44, in strong linkage disequilibrium, were not detectable. HLA-A antigens were consistently detected by all methodologies and minimally affected by pretreatment with IFN-γ. HLA-B antigens were detectable in 8 of 11 lines by ICP and 3 of 11 lines by CMC. By FACS the supratypic specificity HLA-Bw6 was expressed at low levels in most lines (mean fluorescence 47.2 ± 13.4 and rose to 259.8 ± 45.9 after incubation with IFN-γ; P < 0.001). HLA-Cw antigen detection by CMC correlated with HLA-B (p < 0.01), suggesting that down-regulation and sensitivity to IFN-γ are shared by the two loci. This low expression of the HLA-B antigens may play a role in the evasion of the host immune response and its up-regulation may be useful in allowing tumor antigen recognition.
Keywords: Melanoma, Human leukocyte antigen class I, Immunotherapy
There is strong experimental evidence that major histocompatibility complex (MHC) class I antigen expression is important in tumor immunology and immunotherapy. In animal studies the expression of MHC class I antigens appears essential for tumor rejection (1,2). In vitro studies with either human or mouse tumor-infiltrating lymphocytes (TILs) as effectors have shown that target cell lysis is MHC class I restricted (3–5). Moreover, adoptive transfer of TILs to patients with advanced cancer has resulted in tumor regression (6), suggesting that TIL recognition of tumor antigen in the context of MHC class I may be responsible for the anti-tumor antigen response.
Evasion of host immune recognition by tumors may account for different responses to TIL therapy and may involve one or more of several mechanisms, including altered expression of cell surface molecules. Solid tumors may vary in degree of expression of human leukocyte antigen (HLA) class I (7–11). Complete loss of expression of HLA class I molecules on the cell surface of some melanomas (12,13) as well as other tumors (14) has been associated with deficient transcription (12) or translation (13) of mutated β2-microglobulin genes. Other mechanisms could cause altered expression of MHC class I molecules on the cell surface; these may include abnormalities of transcription/translation of class I genes or defects in the antigen-processing machinery (15–17). A reduced expression of HLA-B locus antigens compared with HLA-A has been described in cultured cell lines established from small cell lung cancer and laryngeal carcinoma (18) as in some melanoma cell lines (19), while alterations of HLA-A and -C have been described by others in colon (9) and ovarian carcinomas (20) and sarcomas (20). The locus-specific down-regulation of the HLA-B antigens has been associated with increased expression of the c-myc oncogene (19), but this finding was not confirmed by others (18). Differential inducibility of HLA-A3 and -B7 alleles to interferon-α (IFN-α) and IFN-γ has been reported and correlated to two nucleotide differences in the IFN-responsive sequence located in the promoter region of the two loci (21).
While different mechanisms responsible for the alteration of tumor cell surface expression of HLA class I antigens have been described, the actual incidence of abnormal expression of HLA molecules has not been definitely established. Studies on tissue specimens have yielded inconsistent results because of the differences in the methodologies used (7). Immunohistochemical techniques, most commonly employed, depend on several factors including the specificity and sensitivity of the antibodies used, the different site of origin of the lesions tested, and the different criteria used to define a lesion as “positive” (7,22–26). Thus, the pattern of surface expression of HLA class I molecules on cancer cells is confusing, with some studies showing that class I molecules are almost always present in cancer cells and other studies supporting a more heterogeneous distribution (7,27–29).
This study was designed to assess in detail the pattern of locus-specific HLA class I molecule surface expression in W6/32-positive melanoma cell lines by comparing three commonly used methodologies to better understand the conflicting data reported in the literature.
MATERIALS AND METHODS
Tumor Cell Lines
The cell lines 397-MEL, 526-MEL, 537-MEL, 586-MEL, 624-MEL, 697-MEL, 888-MEL, 938-MEL, 1011-MEL, 1088-MEL, and 1102-MEL were established from metastatic melanoma lesions and were randomly selected among those known to express MHC class I antigens as determined by binding of the monoclonal antibody (mAb) W6/32. Melanoma lines were maintained in monolayer culture in complete medium (CM) consisting of RPMI 1640 (Biofluids, Rockville, MD, U.S.A.) supplemented with 0.1 mM nonessential amino acids (Biofluids), 1.0 mM sodium pyruvate (Biofluids), 5 × 10−5 M 2-mercaptoethanol (ME) (Aldrich Chemical Co., Milwaukee, WI, U.S.A.), 0.03% glutamine, 100 U/ml penicillin (both from NIH Media Unit), 0.5 g/ml amphotericin B (Flow Laboratories, McLean, VA, U.S.A.), and 10% heat-inactivated fetal calf serum (Biofluids) as previously described (30). To elicit expression of HLA class I antigens, cell lines were also incubated for 48 h in CM containing recombinant IFN-γ at the dose of 500 U/ml (Biogen, Cambridge, MA, U.S.A.). This concentration was five times higher than the concentration necessary to elicit optimal expression (>95% detection) by fluorescence-activated cell sorting (FACS) of HLA class II and HLA-B antigens in each population tested (with the exception of HLA-Bw4 specificity in 586-MEL). Minimal incremental effect on HLA antigen expression was noted above this concentration.
Epstein-Barr Virus B-Cell Lines, Cultured T-Lymphocytes, and Other Cell Lines
B lymphoblastoid cells (501-EBV, 537-EBV, 583-EBV, 586-EBV, 888-EBV) derived from patient peripheral blood were transformed with exogenous Epstein-Barr virus (EBV) (31). Tumor-infiltrating lymphocytes (1043-TIL, 1128-TIL, 1143-TIL) were grown and maintained as previously described (32). EBV lines and TILs, which constitutionally express HLA class I antigens, were used as HLA-matched positive controls for mAbs in each analysis [FACS and immunohistochemistry with cytospin preparation (ICP)]. The fibroblast strains of 1154-FIB and Malme-3 (33) were used to assess expression, respectively, of HLA-Bw4 and HLA-Bw6 supratypic specificity in nonneoplastic, nonlymphoid cells.
Flow Cytometric Analysis (FACS)
To examine MHC protein expression, cultured tumor cell lines or appropriate controls were harvested with 0.05% trypsin and 0.02% versene, washed twice in ice-cold FACS buffer (Ca2+, Mg2+, phenol red–free Hanks’ Balanced Salt Solution with 5% heat-inactivated fetal calf serum and 0.2% sodium azide), and incubated for 45 min at 4°C with 20 μl of the appropriate mAb. Staining was performed with 20 μl of the appropriate fluorescein [fluorescein isothiocyanate (FITC)]–conjugated secondary antibody for 45 min at 4°C. Fluorescence was analyzed by a FACScan 440 (Becton Dickinson, San Jose, CA, U.S.A.). Nonviable cells were gated with propidium iodide. FACS data are presented as mean channel number (mcn) of fluorescence.
Immunocytochemistry on Cytospin Preparations (ICP)
Cell cultures were spun at 500 rpm for 5 min in a Shandon Cytospin II (Shandon Southern Instruments, Sewickley, PA, U.S.A.) and allowed to air dry for 30 min. Cytospin slides were fixed in acetone, quenched for 5 min in 3% H2O2 solution, and, after blocking of nonspecific binding in a 0.05 M Tris-buffered saline (TBS) containing 3% horse serum, were incubated for 2 h at room temperature with the appropriate HLA specific antibody. After washing in TBS, secondary antibody incubation was carried for 45 min followed by 30-min incubation with avidin-biotin (ABC) reagent (Vector Elite Standard ABC Kit Pk-6100) and 5-min incubation in diaminobenzidine (Sigma). Reading was done in a blinded fashion by a cytopathologist and scored from 0 to 3+ according to a subjective scale with 0 being equal to the negative control using nonspecific myeloma-associated protein for background staining and 3+ being staining of 88-EBV with W6/32.
Complemented-Mediated Cytotoxicity
Complement-mediated cytotoxicity (CMC) was done using the Amos modified method (34) with alterations in complement concentration, time of incubation, and number of tumor cells per well found to be appropriate after previous titration experiments for the low sensitivity of melanoma lines by complement. These changes were not necessary for the mAb trays in which the same standard conditions used for typing of peripheral blood lymphocytes were found to be optimal. Briefly, after trypsinization, viability was checked using trypan blue (Gibco Laboratories, Grand Island, NY, U.S.A.) exclusion method and the cell concentration was adjusted to 2.0 × 106 cells/ml.
mAb Typing Trays
One microliter of cell suspension was added to each well and the tray (One Lambda, Canoga Park, CA, U.S.A.) was incubated for 1 h at room temperature. This is an all-inclusive step since complement is preloaded in the trays. After incubation 5 μl of trypan blue in 2% ethylenediaminetetraacetate was added to each well for 15 min; then excess dye was removed and 20 μl of 1% fetal calf serum in RPMI 1640 was added to each well. mAb was a useful control for possible false-positive results due to the presence of contaminating antibodies in the standard sera that could react with other HLA specificities.
Typing with Sera from Multiparous Donors
One microliter of cell suspension was added to each typing well and incubated for 40 min at room temperature. Then 5 μl of rabbit complement (C-six Diagnostics, Mequon, WI, U.S.A.) was added to each well. Complement was preabsorbed with human packed red blood cells and used at a dilution of 1:2.25 found to be optimal for typing of tumor cell lines after previous titration studies done using various types and different numbers of tumor lines (data not shown). After 60 min the trays were processed as the mAb trays described before. Results were recorded according to the standard NIH reading scale for microcytotoxicity assays (8 = 81–100% cell death, 6 = 50–80% cell death, 4 = 21–50% cell death, 2 = 11–20% cell death, and 1 = 0–10% cell death). An HLA allele was considered present if at least two of three mAbs or three of four sera from multiparous donors scored ≥6.
mAbs for FACS and ICP
For FACS and ICP the following primary mAbs were used: W6/32 (Sera Labs, Westbury, NY, U.S.A.; mouse IgG2a) reacting with monomorphic determinants on the HLA-A,-B,-C molecules (35) (1:50 dilution for both FACS and ICP); IVA12 (100 g/ml; IgG1, ATCC HB 145) reacting against HLA class II antigens (DR, DQ, and possibly DP) (36); MA2.2 (mouse IgG2a) reactive against the HLA-A2 and -A29 antigen (Incstar Co., Stillwater, MN, U.S.A.) (37) used at a dilution of 1:30 for FACS analysis and undiluted for ICP. The following mouse IgM mAbs were purchased from One Lambda: anti-Al,36,-B63 (71/HA-1), anti-A25 (136/ HA-1), anti-26,34 (101/HS-l), anti-A29 (266/HS-l), anti-30,31 (273/HA-1), and anti-A9,32 (238/HA-1); these mAbs were used undiluted. Supernatants from hybridomas were used undiluted and included GAP A-3 mAb (mouse IgG2ak) specific for HLA-A3 (38), mAb A11.1M (mouse IgG3) recognizing HLA-A11 and 24 (39), and mAb SFR8-B6 (rat IgG2b) directed against the HLA-Bw6 superspecificity (40). The HLA-Bw4 superspecificity was tested using 820310 (Biotest Diagnostics, Fairfield, NJ, U.S.A.) undiluted for FACS and ICP. As a negative control for ICP, a purified mouse myeloma protein, IgGl-(MOPC), was used at a 1:50 dilution (Organon Teknika Corp., Durham, NC, U.S.A.). Secondary antibodies for FACS included fluorescein (FITC)-conjugated goat anti-mouse IgGl,2a,2b,3 (Becton Dickinson) at a dilution of 1:8, FITC-conjugated goat anti-mouse IgM (μ-chain specific) (Jackson Immune Research, West Grove, PA, U.S.A.) at a dilution of 1:20, and FITC-conjugated mouse anti-rat IgG2b,3b, F(ab′)2 fragment specific (Jackson Immune Research), at a dilution of 1:30. For ICP secondary antibodies included biotinylated goat anti-mouse IgG (H + L) at 1:1,000 dilution (Kirkegaard & Perry Laboratories, Gaithersburg, MD, U.S.A.), biotinylated goat anti-mouse IgM (μ-chain specific) (Vector Laboratories, Burlingame, CA, U.S.A.) at 1:50 dilution, and biotinylated rabbit, mouse-absorbed, anti-rat IgG (H + L chain) (Vector Laboratories) at 1:50 dilution. Titration of antibodies was determined by strength of staining of HLA-matched T-lymphocytes or EBV lines.
Preliminary HLA Typing of Patient Lymphocytes
To assess the original HLA type of the patients from whom the melanoma cell lines were originated, lymphocytes from tumor donors were used for HLA typing. This was done in a standard fashion by the NIH HLA Laboratory using the modified Amos microcytotoxicity test (34).
Statistical Analysis
Paired sample Student t test was used for parametric comparison of HLA class I expression in the presence or absence of stimulation with IFN-γ using as a parameter the mean channel number calculated by FACS. All other nonparametric comparisons were done using the Fisher exact test (CMC) or the Wilcoxon rank test (ICP). Data are presented as means ± SEM.
RESULTS
Effect of IFN-γ on Surface Expression of HLA Antigens
To ascertain the effect of IFN-γ on HLA antigen expression, we tested by FACS analysis 11 melanoma cell lines before and after incubation with IFN-γ at the dose of 500 U/ml for 48 h (Table 1). Prior to treatment all cell lines expressed HLA class I and none expressed HLA class II antigens. Up-regulation of HLA class I by IFN-γ as determined by binding of W6/32 mAb, was noted in 4 of 11 lines (526-MEL, 586-MEL, 1011-MEL, and 1102-MEL), while it was not noted in the remaining 7 cell lines; the average mean channel number change after treatment with IFN-γ was not statistically significant (men = 217.2 ± 55.7 vs. 264.3 ± 63.7; p > 0.05). This dose of IFN-γ moderately increased expression of HLA-A antigens (men = 116.1 ± 24.7 vs. 202.2 ± 42.6; p > 0.05). The HLA-B locus was very sensitive to the effect of IFN-γ and all cell lines showed consistent up-regulation of this locus (men = 26.5 ± 8.7 vs. 284.9 ± 53.0; p < 0.01). IFN-γ was also extremely effective in eliciting expression of HLA class II in 10 of the 11 lines (men = 5.5 ± 0.5 to 369.8 ± 78.7; p < 0.01); on several occasions 888-MEL failed to express HLA class II antigens after treatment with IFN-γ, while it was still able to up-regulate the HLA-B locus. Discussion of the defect of HLA class II expression in this line is beyond the purpose of this study. In a separate set of experiments, HLA class I expression as detected by mAb W6/32 was analyzed comparing FACS analysis and ICP (Table 2). The average expression of HLA class I with or without pretreatment with IFN-γ was again not significantly different (139.0 ± 33.4 vs. 138.6 ± 32.7; p > 0.05). HLA class I antigens were detected by both methodologies independently from treatment with IFN-γ.
TABLE 1.
Effect of interferon-γ (IFN-γ) on expression of human leukocyte antigen (HLA) class 1 and II antigens
| n | IFN-γ(−) | IFN-γ(+) | |
|---|---|---|---|
| W6/32 | 11 | 217.2 ± 55.7 | 264.3 ± 63.7 |
| IVA12 | 10a | 5.5 ± 0.5 | 369.8 ± 78.7b |
| HLA-A | 11 | 116.1 ± 24.7 | 201.2 ± 42.6 |
| HLA-B | 11 | 26.5 ± 8.7 | 284.9 ± 53.0b |
Expression of HLA class I antigens as detected by fluorescence cell sorter analysis. Mean channel number is presented. IFN-γ(−), detectability of HLA loci in baseline conditions; IFN-γ(+), detectability of HLA loci after pretreatment with IFN-γ (500 U/ml × 48 h).
Include analysis of 10 of 11 lines because 888-MEL expression of HLA class II is unresponsive to IFN-γ at this and higher doses. HLA-A and HLA-B refer to the mean of pooled data from all cell lines: Data for HLA-A were obtained using allele-specific monoclonal antibodies (mAbs); data for HLA-B were collected using supratypic specific mAbs (see Materials and Methods).
Only significant p values are shown:
p < 0.01. p values refer to paired analysis for each cell line in the presence or absence of IFN-γ stimulation.
TABLE 2.
Melanoma cell line human leukocyte antigen (HLA) type by three methodologies
| FACS data mean channel number
|
CMC
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Control
|
Specific HLA mAb
|
ICP
|
Anti-human seraa |
mAbb |
|||||||
| Cell line | Patient HLA type | Pre IFN | Post IFN | Pre IFN | Post IFN | Pre IFN | Post IFN | Pre IFN | Post IFN | Pre IFN | Post IFN |
| 624-MEL | MHC-I (W6/32) | 6 | 7 | 206 | 161 | 3+ | 3+ | NR | NR | NR | NR |
| A2 | 6 | 7 | 180 | 120 | 2+ | 2+ | + | + | + | + | |
| A3 | 6 | 7 | 109 | 148 | 1+ | 1+ | + | + | + | + | |
| Bw6(B7) | 8 | 8 | 21 | 335 | 0 | 1+ | −(−) | +(+) | +(−) | +(+) | |
| Bw6 (B14) | −(−) | +(+) | +(+) | +(+) | |||||||
| 526-MEL | MHC-I (W6/32) | 5 | 5 | 84 | 108 | 2+ | 3+ | NR | NR | NR | NR |
| A2 | 5 | 5 | 95 | 150 | 2+ | 2+ | + | + | + | + | |
| A3 | 5 | 5 | 63 | 97 | 1+ | 1+ | + | + | + | + | |
| Bw6 (B50) | 6 | 7 | 14 | 267 | 1+ | 2+ | −(−) | +(−) | −(−) | +(−) | |
| Bw6 (B62) | −(−) | +(+) | −(−) | +(+) | |||||||
| 1102-MEL | MHC-I (W6/32) | 5 | 4 | 94 | 179 | 3+ | 3+ | NR | NR | NR | NR |
| A2 | 5 | 4 | 47 | 160 | 2+ | 3+ | + | + | + | + | |
| A24 | 5 | 4 | 68 | 200 | ND | ND | + | + | + | + | |
| Bw6 (B55) | 5 | 4 | 144 | 521 | 2+ | 3+ | +(+) | +(+) | +(+) | +(+) | |
| Bw6 (B62) | +(+) | +(+) | +(+) | +(+) | |||||||
| 697-MEL | MHC-I (W6/32) | 5 | 4 | 236 | 202 | 2+ | 3+ | NR | NR | NR | NR |
| A2 | 5 | 5 | 206 | 156 | 1+ | 1+ | + | + | + | + | |
| A11 | 5 | 5 | 98 | 102 | 1+ | 1+ | + | + | + | + | |
| Bw6(B7) | 6 | 6 | 13 | 290 | 1+ | 2+ | −(−) | +(+) | −(−) | +(+) | |
| Bw6 (B15) | −(−) | +(+) | −(−) | +(+) | |||||||
| 938-MEL | MHC-I (W6/32) | 6 | 4 | 214 | 108 | 4+ | 3+ | NR | NR | NR | NR |
| A1 | 5 | 5 | 21 | 27 | 1+ | 1+ | + | + | + | + | |
| A24 | 5 | 5 | 70 | 103 | 1+ | 2+ | + | + | + | + | |
| Bw6 (B7) | 6 | 6 | 93 | 495 | 1+ | 3+ | +(+) | +(+) | +(+) | +(+) | |
| Bw6 (B8) | +(−) | +(+) | +(−) | +(+) | |||||||
| 1088-MEL | MHC-I (W6/32) | 8 | 7 | 373 | 411 | 2+ | 2+ | NR | NR | NR | NR |
| A1 | 7 | 6 | 26 | 21 | 0 | 0 | + | + | + | + | |
| A2 | 8 | 6 | 98 | 127 | 1+ | 1+ | + | + | + | + | |
| Bw4 (B44) | 7 | 8 | 14 | 64 | 1+ | 0 | −(−) | +(−) | −(−) | +(+) | |
| Bw6 (B8) | 8 | 6 | 64 | 257 | 0 | 2+ | −(−) | +(+) | −(−) | +(+) | |
| 888-MEL | MHC-I (W6/32) | 5 | 5 | 47 | 47 | 3+ | 3+ | NR | NR | NR | NR |
| A1 | 6 | 6 | 18 | 13 | 0 | 0 | + | + | + | + | |
| A24 | 5 | 5 | 81 | 46 | 0 | 1+ | + | + | + | + | |
| Bw4 (B52) | 5 | 5 | 8 | 36 | 0 | 1+ | −(−) | −(−) | −(−) | −(−) | |
| Bw6 (B55) | 5 | 5 | 45 | 106 | 0 | 2+ | −(−) | +(+) | −(−) | +(+) | |
| 397-MEL | MHC-I (W6/32) | 5 | 4 | 118 | 100 | 2+ | 1+ | NR | NR | NR | NR |
| A1 | 7 | 4 | 30 | 23 | 1+ | 1+ | + | + | + | + | |
| A25 | 7 | 4 | 8 | 5 | 2+ | 2+ | + | + | + | + | |
| Bw6 (B8) | 7 | 4 | 20 | 157 | 1+ | 1+ | −(−) | +(+) | −(−) | +(−) | |
| Bw6 (B62) | −(−) | +(+) | −(−) | +(+) | |||||||
| 537-MEL | MHC-I (W6/32) | 5 | 5 | 63 | 59 | 2+ | 2+ | NR | NR | NR | NR |
| A1 | 5 | 5 | 16 | 13 | 1+ | 1+ | + | + | + | + | |
| A26 | 5 | 5 | 8 | 143 | 2+ | 2+ | + | + | + | + | |
| Bw4 (B44) | 5 | 5 | 10 | 33 | 1+ | 2* | +(−) | +(−) | +(−) | +(+) | |
| Bw6 (B70) | 5 | 5 | 71 | 122 | 1+ | 3+ | +(+) | +(+) | +(+) | +(+) | |
| 586-MEL | MHC-I (W6/32) | 4 | 5 | 69 | 89 | 2+ | 2+ | NR | NR | NR | NR |
| A29 | 4 | 5 | 4 | 5 | 1+ | 1+ | — | — | — | — | |
| A31 | 4 | 5 | 96 | 111 | 3+ | 3+ | + | + | + | + | |
| Bw4 (B44) | 4 | 5 | 4 | 5 | 1+ | 0 | −(−) | −(−) | −(−) | −(−) | |
| Bw6 (B8) | 4 | 5 | 15 | 107 | 1+ | 2+ | −(−) | +(+) | −(−) | +(+) | |
| 1011-MEL | MHC-I (W6/32) | 5 | 5 | 25 | 61 | 2+ | 2+ | NR | NR | NR | NR |
| A11 | 5 | 5 | 26 | 32 | 1+ | 1+ | + | + | + | + | |
| A32 | 8 | 8 | 16 | 163 | 0 | 0 | + | + | + | + | |
| Bw6 (B7) | 5 | 5 | 19 | 200 | 1+ | 2+ | −(−) | +(+) | −(−) | +(+) | |
| Bw6 (B40) | −(−) | +(+) | −(−) | +(+) | |||||||
Expression of HLA class I loci in 11 HLA class I-positive cell lines as detected by three methodologies: fluorescence-activated cell sorting (FACS), immunocytochemistry with cytospin preparation (ICP), and complement-mediated cytotoxicity (CMC). FACS data are expressed as mean channel number. ICP data are expressed as a subjective scale from 0 to 3+ as noted in a blinded fashion by the cytopathologist. CMC data are presented as positive or negative (see Materials and Methods). For the HLA-B locus, results are expressed as Bw4 and/or Bw6 supratypic specificity and for the actual HLA-B allele expected for that patient (in parentheses). FACS and ICP data from cell lines with the single HLA-Bw6 supratypic specificity are given only once in the first row for that line.
Detection of HLA loci by CMC using sera from multiparous donors.
Detection of HLA loci by CMC using mouse monoclonal antibodies (mAbs).
This specificity was tested at a later stage when an mAb against HLA-A25 became available.
Expression of HLA-A Allospecificity
The HLA-A locus was more consistently detected than the HLA-B locus in the melanoma lines studied (Table 2). In two cell lines one HLA-A allele was not detected (586-MEL and 537-MEL). In the case of 586-MEL, the HLA-A allele was not detected by FACS and CMC even after treatment with IFN-γ. IFN-γ caused HLA-A antigen up-regulation of >50% in three cell lines (1102-MEL, 537-MEL, and 1011-MEL). In 536-MEL, HLA-A26 was not detectable before treatment with IFN-γ and became detectable after treatment with this cytokine. The average mean channel number for HLA-A antigen detection was moderately affected by incubation with IFN-γ (74.3 ± 14.4 vs. 102.9 ± 16.4; p < 0.05). The same was true for the most common alleles (HLA-A2: 125.4 ± 32.7 vs. 142.6 ± 9.0, n = 5; HLA-A1: 22.2 ± 2.9 vs. 19.4 ± 3.1, n = 5). The lower mean channel number reported for the HLA-A 1 compared with the HLA-A2 allospecificity does not reflect differential expression, but it is due to weaker binding of the anti-HLA-Al mAb as determined by titration on EBV cell lines (data not shown). ICP demonstrated the presence of the HLA-A locus in 17 of 21 alleles tested. In the cases in which the HLA-A locus was detected without pretreatment with IFN-γ, no up-regulation was noted after IFN-γ. CMC using either sera from multiparous donors or mAbs always detected the HLA-A locus except for HLA-A29 in 586-MEL. IFN-γ was not necessary for the detection of HLA-A locus by CMC.
Expression of HLA-B Locus
There was only one true example of total loss of expression of HLA-B locus (586-MEL, HLA-B44), which occurred in the same cell line in which loss of allelic expression of HLA-A29 was noted (see Table 2). In this case no up-regulation was noted after IFN-γ. In the absence of incubation with IFN-γ, the HLA-B locus generally was expressed at low levels (Bw4: men = 9.0 ± 2.4, n = 4; Bw6: men = 47.2 ± 13.4, n = 11). After IFNγ the B locus was expressed at significantly higher levels (Bw4: men = 34.5 ± 13.9, p < 0.05; Bw6: men = 259.8 ± 45.9, p < 0.01). When both HLA-B alleles belonged to the HLA-Bw6 supratypic specificity, the mean channel number was significantly higher than in lines Bw4/ Bw6 “heterozygous” (323.6 ± 56.7, n = 7, and 148.0 ± 42.2, respectively; n = 4, p < 0.05), suggesting that both HLA-B loci were equally up-regulated by IFN-γ pretreatment. Three different behaviors of HLA-B locus are exemplified in Fig. 1. In lymphoid cell lines such as TILs, the mean channel number for HLA-B locus of EBV B-cell lines and fibroblasts was always above the average mean channel number for melanoma cell lines (Fig. 2). ICP detected the HLA-Bw6 supratypic specificity in 8 of 11 cases. After IFN-γ pretreatment, this specificity was always detected and in 10 of 11 cases up-regulated (Fig. 3). The subjective quantitation of up-regulation did not correlate with data from FACS analysis. Overall, ICP was less sensitive than FACS and less accurate than CMC. CMC with sera from multiparous donors showed a very poor detection of HLA-B locus: HLA-Bw6 expression was not detected in 8 of 11 lines tested and HLA-Bw4 was not detected in 3 of 4 cell lines. CMC using mAbs detected the HLA-Bw6 specificity in 4 of 11 lines tested. Detection by CMC strongly correlates with expression of HLA-B by FACS. With CMC it was possible to evaluate the behavior of the two separate HLA-B alleles since sera or mAbs specific for the different subspecificities were available for this technique. In most cases HLA-B alleles were symmetrically up-regulated by pretreatment with IFN-γ (Table 2). The use of mAb trays avoided the false positivity due to cross reactivity against HLA class II antigens present in human sera. This was particularly useful when testing cell lines treated with IFN-γ because of their high HLA class II antigen expression.
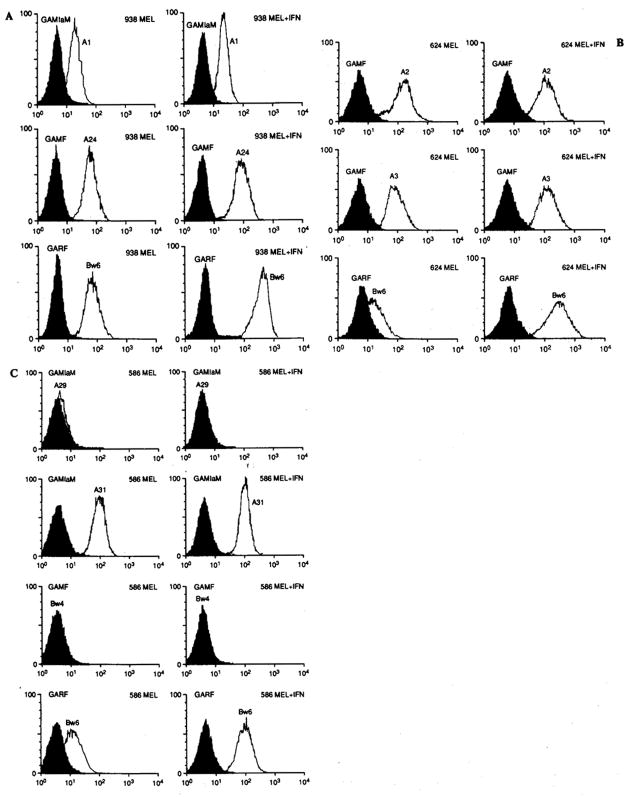
FIG. 1.
Three examples of different patterns of expression of human leukocyte antigen (HLA) class I in three distinct melanoma cell lines as detected by fluorescence-activated cell sorting analysis. A: Line 938-MEL expressing both HLA-A and -B antigens in baseline conditions. After 48-h incubation in interferon-γ (IFN-γ; 500 U/ml), only the HLA-B specificities are up-regulated. B: Line 624-MEL (most common pattern) expressing HLA-A but only minimally HLA-B loci in baseline conditions. Also in this case treatment with IFN-γ up-regulates the expression of HLA-B. C: Line 586-MEL. Total loss of HLA-A and -B allelic expression not detectable even after IFN-γ. GAMF, fluorescein-conjugated goat anti-mouse IgG; GAMIgM, fluorescein-conjugated goat anti-mouse IgM; GARF, fluorescein-conjugated goat anti-rat IgG.
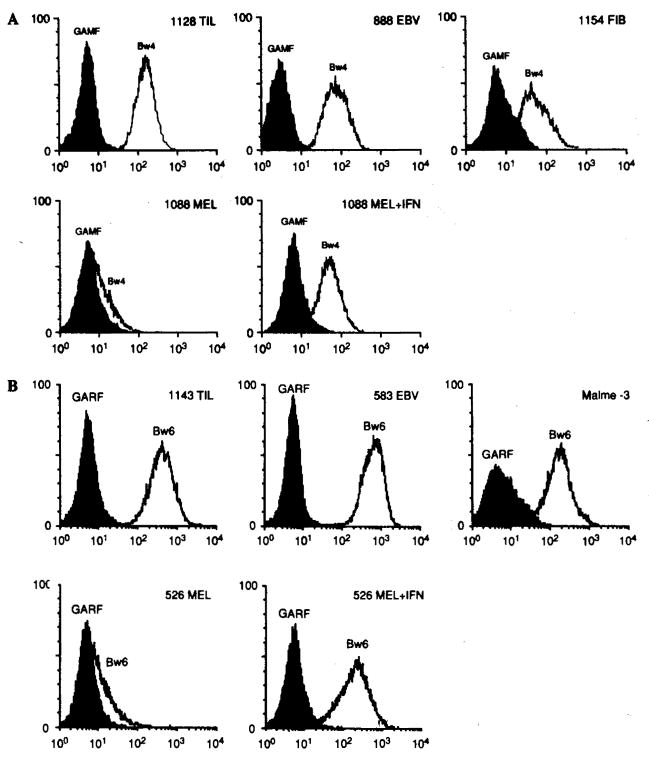
FIG. 2.
A: Example of differential expression of human leukocyte antigen (HLA)-Bw4 supratypic specificity by different cell types including a tumor-infiltrating lymphocyte (TIL) line (1128 TIL), an Epstein-Barr virus (EBV) B-cell line (888-EBV), a fibroblast line (1154-FIB), and a melanoma line (1088 MEL) with and without pretreatment with interferon-γ (IFN-γ). B: Example of differential expression of HLA-Bw6 supratypic specificity by different cell types including a TIL line (1143 TIL), an EBV B-cell line (583-EBV), a fibroblast line (Malme-3), and a melanoma line (526-MEL) with and without pretreatment with IFN-γ. For other abbreviations see the legend to Fig. 1.
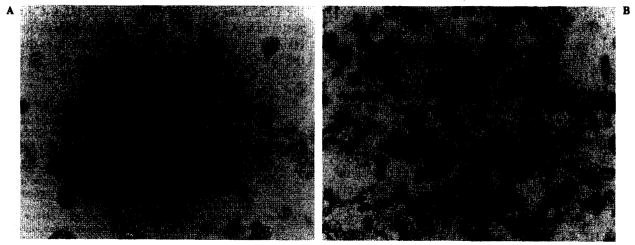
FIG. 3.
Immunohistochemistry with cytospin preparation of line 537-MEL stained with anti-HLA-Bw6 antibody in baseline culture conditions (A) and after 48-h incubation with interferon-γ (IFN-γ) (B). Obviously increased expression of human leukocyte antigen (HLA)-B locus is detected after IFN-γ.
Expression of HLA-C Locus Antigens by CMC
HLA-C locus antigen expression was evaluated by CMC only since no anti-C locus-specific mAbs were available. A correlation was noted between detection of HLA-B and HLA-C loci by CMC (Table 3). The HLA-C locus was detected in only 4 of 11 lines, 3 of which had detectable HLA-B. After IFN-γ HLA-C was detectable in 10 of 11 lines and their expression was significantly correlated with that of HLA-B (p < 0.05).
TABLE 3.
Detection of human leukocyte antigen (HLA) class IB and Cw antigens by complement-mediated cytotoxicity (CMC)a
| HLA-B
|
HLA-C
|
||||||
|---|---|---|---|---|---|---|---|
| IFN-γ (−)c | IFN-γ (+) | IFN-γ (−) | IFN-γ (+) | Linkageb IFN-γ | |||
| 624-MEL | B7 | − | + | Cw7 | − | + | +++ |
| B14 | − | + | − | + | |||
| 526-MEL | B50 | − | − | − | + | NA | |
| B62 | − | + | Cw3 | − | − | ++++ | |
| 1102-MEL | B55 | + | + | Cw6 | + | + | ++++ |
| B62 | + | + | Cw3 | + | + | ++++ | |
| 697-MEL | B7 | − | + | Cw6 | − | + | ++++ |
| B15 | − | + | Cw3 | − | + | NA | |
| 938-MEL | B7 | + | + | Cw6 | + | + | ++++ |
| B8 | − | + | Cw7 | + | + | ++++ | |
| 1088-MEL | B8 | − | + | − | |||
| B44 | − | − | Cw5 | − | + | +++ | |
| 888-MEL | B52 | − | − | Cw1 | − | + | NA |
| B55 | − | + | Cw7 | + | + | NA | |
| 397-MEL | B8 | − | + | Cw7 | − | + | ++++ |
| B62 | − | + | Cw6 | − | + | NA | |
| 537-MEL | B44 | − | − | Cw5 | + | + | +++ |
| B70 | + | + | Cw1 | + | + | NA | |
| 586-MEL | B8 | − | + | Cw7 | − | + | ++++ |
| B44 | − | − | − | ||||
| 1011-MEL | B7 | − | + | Cw7 | − | + | +++ |
| B40 | − | + | Cw3 | − | + | ++++ | |
Detection of HLA class IB and Cw antigens in 11 HLA class I-positive cell lines by CMC. Positive are considered CMC scores of ≥6 (see text). The HLA-B and -C loci were paired according to their linkage disequilibrium scores as reported by Grange et al. (41).
Correlation of HLA-B vs. HLA-C locus expression in baseline conditions and after pretreatment with interferon-γ (IFN-γ): p < 0.05.
Linkage score: level of linkage disequilibrium reported to exist between HLA-B and -C shown in the same row. NA, information not available.
IFN-γ (−), detectability of HLA loci in baseline conditions; IFN-γ(+), detectability of HLA loci after pretreatment with IFN-γ (500 U/ml × 48 h).
DISCUSSION
It has been postulated that alterations in the expression of MHC molecules on the surface of cancer cells could be part of a chain of events leading to escape from immune recognition. Loss of (β2-microglobulin leading to complete loss of MHC class I antigen expression has been described for some melanoma (12,13) and other tumor cell (14) lines. We recently noted four cases of true loss of β2-microglobulin in melanoma lines (manuscript in preparation). All of these occurred in association with rapid tumor growth in patients failing to respond to interleukin-2-based immunotherapy.
In this study we focused our attention on HLA class I antigens-expressing melanoma cell lines as detected by mAb W6/32. This was done with the purpose of assessing (a) whether other patterns of HLA class I antigen modulation were to be found in cell lines and (b) the degree of sensitivity and accuracy of the methodologies commonly used for this purpose. Total loss of allelic expression (defined as inability, even after IFN-γ, to detect the allele by at least two methodologies using different antibodies such FACS and CMC) was a relatively rare event, affecting only 1 of 11 lines tested (586-MEL). In 586-MEL the loss of an HLA-A allele (A29) was associated with the loss of an HLA-B allele (B44) and DR7 (data not shown), all of them part of the original patient’s haplotype (A29,31, B8,44, and DR1,7) (5). The simultaneous loss of expression of these alleles suggests loss of a large genomic unit within the same chromosome because of the strong linkage disequilibrium of this ancestral haplotype HLA-A29, -B44, and -DR7 (41,42). Allele-specific polymerase chain reaction amplification of HLA class I and class II genes in this tumor line as well as the autologous EBV line might provide confirmation of loss of genomic DNA (43). The frequency of loss of allelic expression is lower than that reported for tumors of other histologies (20) and lower than the frequency reported by Kageshita et al. (44) on frozen sections of metastatic melanoma. These authors reported allelic-specific loss of HLA-A2 antigens in 44% of 9 metastatic HLA class I-positive lesions; we noted such loss in 1 of 11 cell lines by FACS analysis (9%). In both studies the tumors were HLA class I positive as determined by monomorphic determinant mAbs. While the lower frequency that we noted in cell lines may partially be due to a higher sensitivity of FACS analysis, it is interesting to note that a similar pattern of allelic-specific loss has been documented in vivo.
A more common phenomenon was detected, the functional significance of which is unknown, i.e., the “fading” of HLA-B locus antigen expression in most melanoma lines. All 11 lines were generated from patients carrying the HLA-Bw6 supratypic specificity. Of these lines, only three showed levels of expression that could be detected by methodologies commonly used for tissue typing, i.e., CMC. Two other lines had moderate expression of the HLA-B locus, while the other six had minimal expression by FACS analysis. We plan to examine other melanocytic cells (including normal melanocytes) to ascertain whether this phenomenon is melanoma specific or a characteristic of melanocytes per se. Other nonneoplastic cell lines tested included TILs, EBV B-cell lines, and fibroblasts and showed a severalfold higher degree of expression of HLA-B locus (in the absence of pretreatment with IFN-γ). This low expression of HLA-B antigens was unlikely due to the enzymatic digestion necessary to remove the tumor cell lines from the cell culture flasks since fibroblasts, which were treated in identical fashion, had easily detectable amounts of HLA-B locus antigens on their surface (Fig. 2).
Parallel to the HLA-B locus was the expression of the HLA-Cw locus by CMC. HLA-C is usually expressed at the cell surface at ~10% the level of expression of HLA-A or -B (45); not surprisingly, therefore, this locus was difficult to detect in standard conditions but was easily detectable after stimulation with IFN-γ. Further, it appeared that a strict correlation was present in detectability of the two loci by CMC, suggesting that regulation of their expression is closely related. As for the HLA-B locus, the significance of this phenomenon and its possible relevance in cancer immunology are unknown. The HLA-B and -C loci were very sensitive to up-regulation by IFN-γ, while the HLA-A locus was less reproducibly affected. It is impossible to as certain with these data whether the HLA-B locus down-regulation is a culture artifact. Ideally it would be best to assess HLA-B locus expression on cryopreserved tissue sections of tumors using immunohistochemistry, although this technique may not be as sensitive and specific as FACS analysis. This study was specifically aimed at the analysis of cell lines that are often used as stimulators for the growth and expansion of tumor-specific lymphocytes or as targets in cytotoxicity or cytokine release assays using MHC-restricted effectors. Knowledge of HLA expression in tumor cell lines is therefore extremely relevant to basic tumor immunology studies independently from their in vivo correlation.
In summary, we have identified two ways in which melanoma cell lines could partially lose their ability to express HLA antigens at the cell surface. We observed loss of alleles (HLA-A and -B) in one melanoma and a more common “fading” of the HLA-B locus that could be restored by IFN-γ treatment in a locus- rather than allelic-specific fashion. A combination of methodologies proved useful for two reasons: It allowed verification of results and at the same time allowed evaluation of HLA-C locus patterns by CMC and detection of low expression of HLA-B by FACS, avoiding false negativities due to the lower sensitivity of the other methods.
References
- 1.Weber JS, Rosenberg SA. Effects of murine tumor class I major histocompatibility complex expression on antitumor activity of tumor-infiltrating lymphocytes. J Nail Cancer Inst. 1990;82:755–61. doi: 10.1093/jnci/82.9.755. [DOI] [PubMed] [Google Scholar]
- 2.Restifo NP, Esquivel F, Asher AL, et al. Defective presentation of endogenous antigens by a murine sarcoma: implications for the failure of an anti-tumor immune response. J Immunol. 1991;147:1453–9. [PMC free article] [PubMed] [Google Scholar]
- 3.Wolfel T, Klehmann E, Muller C, Schutt KH, Meyer Zum Buschenfelde KH, Knuth A. Lysis of human melanoma cells by autologous cytolytic T cell clones. J Exp Med. 1989;170:797–810. doi: 10.1084/jem.170.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crowley NJ, Darrow TL, Quinn-Allen MA, Seigler HF. MHC-restricted recognition of autologous melanoma by tumor-specific cytotoxic T cells. Evidence for restriction by a dominant HLA-A allele. J Immunol. 1991;146:1692–9. [PubMed] [Google Scholar]
- 5.Horn SS, Topalian SL, Simonis T, Mancini M, Rosenberg SA. Common expression of melanoma tumor associated antigens recognized by human tumor infiltrating lymphocytes: analysis by human lymphocyte antigen restriction. J Immunother. 1991;10:153–64. [PubMed] [Google Scholar]
- 6.Rosenberg SA, Packard BS, Aebersold PM, et al. Use of tumor infiltrating lymphocytes and interleukin-2 in the immunotherapy of patients with metastatic melanoma. N Engl J Med. 1998;319:1676–80. doi: 10.1056/NEJM198812223192527. [DOI] [PubMed] [Google Scholar]
- 7.Ruiter DJ, Mattijssen V, Broecker E-B, Ferrone S. MHC antigens in human melanomas. Semin Cancer Biol. 1991;2:35–45. [PubMed] [Google Scholar]
- 8.Natali PG, Nicotra MR, Bigotti A, et al. Selective changes in expression of HLA class I polymorphic determinants in human solid tumors. Proc Natl Acad Sci USA. 1989;86:6719–23. doi: 10.1073/pnas.86.17.6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Momburg F, Ziegler A, Harpprecht J, Moller P, Moldenhauer G, Hammerling GJ. Selective loss of HLA-A or HLA-B antigen expression in colon carcinoma. J Immunol. 1989;149:352–8. [PubMed] [Google Scholar]
- 10.Kaklamanis L, Gatter KC, Hill AB, et al. Loss of HLA class-I alleles, heavy chains and beta 2-microglobulin in colorectal cancer. Int J Cancer. 1992;51:379–85. doi: 10.1002/ijc.2910510308. [DOI] [PubMed] [Google Scholar]
- 11.Brocker E-B, Suter L, Bruggen J, Ruiter DJ, Macher E, Sorg C. Phenotypic dynamics of tumor progression in human malignant melanoma. Int J Cancer. 1985;36:29–35. doi: 10.1002/ijc.2910360106. [DOI] [PubMed] [Google Scholar]
- 12.D’Urso C, Wang Z, Cao Y, Takate R, Zeff R, Ferrone S. Lack of HLA class I antigen expression by cultured melanoma cell FO-I due to a defect in beta 2-MU gene expression. J Clin Invest. 1991;87:284–92. doi: 10.1172/JCI114984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z, Cao Y, Albino AP, Zeff R, Houghton A, Ferrone S. Lack of HLA class I expression by melanoma cells SK-MEL-33 caused by a reading frameshift in beta 2-microglobulin messenger RNA. J Clin Invest. 1993;91:684–92. doi: 10.1172/JCI116249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gattoni-Celli S, Kirsch K, Timpane R, Isselbacher KJ. 2-Microglobulin gene is mutated in a human colon cancer cell line (HCT) deficient in the expression of HLA class I antigens on the cell surface. Cancer Res. 1992;52:1201–4. [PubMed] [Google Scholar]
- 15.Restifo NP, Esquivel F, Kawakami Y, et al. Identification of human cancers deficient in antigen processing. J Exp Med. 1993;177:265–72. doi: 10.1084/jem.177.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blanchet O, Bourge JF, Zinszner H, Tatari Z, Degos L, Paul P. DNA binding of regulatory factors interacting with MHC-class-I gene enhancer correlates with MHC-class-I transcriptional level in class-I-defective cell lines. Int J Cancer. 1991;6(suppl):138–45. doi: 10.1002/ijc.2910470725. [DOI] [PubMed] [Google Scholar]
- 17.Restifo NP, Kawakami Y, Marincola FM, et al. Molecular mechanisms used by tumors to escape from immune recognition: immunogenetherapy and the cell biology of MHC class I. J Immunother. 1993;14:182–90. doi: 10.1097/00002371-199310000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruiz-Cabello F, Perez-Ayala M, Gomez O, et al. Molecular analysis of MHC-class I expression in human tumor cell lines. Int J Cancer. 1991;6(suppl):123–30. doi: 10.1002/ijc.2910470723. [DOI] [PubMed] [Google Scholar]
- 19.Versteeg R, Kruse-Wolters M, Plomp AC, et al. Suppression of class I human histocompatibility leucocyte antigen by the c-myc is locus specific. J Exp Med. 1989;170:621–35. doi: 10.1084/jem.170.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang P, Vanky F, Li S-L, Vegh Zs, Persson U, Klein E. Expression of MHC-class I antigens in human carcinomas and sarcomas analyzed by isoelectric focusing. Int J Cancer. 1991;6(suppl):106–16. doi: 10.1002/ijc.2910470721. [DOI] [PubMed] [Google Scholar]
- 21.Hakem R, Le Bouteiller P, Jezo-Bremond A, Harper K, Campese D, Lemonnier FA. Differential regulation of HLA-A3 and HLA-B7 MHC class I genes by IFN is due to two nucleotide differences in their IFN response sequences. J Immunol. 1991;147:2384–90. [PubMed] [Google Scholar]
- 22.Holzmann B, Broker EB, Lehmann JM, et al. Tumor progression in human malignant melanoma: five stages defined by their antigenic phenotypes. Int J Cancer. 1987;39:466–71. doi: 10.1002/ijc.2910390410. [DOI] [PubMed] [Google Scholar]
- 23.D’Alessandro G, Zardawi I, Grace J, McCarthy WH, Hersey P. Immunohistological evaluation of MHC class I and II antigen expression on nevi and melanoma: relation to biology of melanoma. Pathology. 1987;19:339–46. doi: 10.3109/00313028709103880. [DOI] [PubMed] [Google Scholar]
- 24.Takata M, Hirone T, Matsamura H. β2-microglobulin expression in normal melanocytes, nevocellular nevi and malignant melanoma. J Invest Dermatol. 1989;92:243S–7S. doi: 10.1111/1523-1747.ep13075770. [DOI] [PubMed] [Google Scholar]
- 25.Cordon-Cardo C, Fuks Z, Drobnjak M, Moreno C, Eisenbach L, Fetdman M. Expression of HLA-A,B,C, antigens on primary and metastatic tumor cell populations of human carcinomas. Cancer Res. 1991;51:6372–80. [PubMed] [Google Scholar]
- 26.Mechtersheimer G, Staudter M, Majdic O, Dórken B, Moldenhauer G, Möller P. Expression of HLA-A,B,C, beta 2-microglobulin (beta 2-m), HLA-DR, -DP, -DQ and of HLA-D-associated invariant chain (Ii) in soft tissue tumors. Int J Cancer. 1990;46:813–23. doi: 10.1002/ijc.2910460512. [DOI] [PubMed] [Google Scholar]
- 27.Elliot BE, Carlow DA, Rodricks A, Wade A. Perspective on the role of MHC antigens in normal and malignant cell development. Adv Cancer Res. 1989;53:181–9. doi: 10.1016/s0065-230x(08)60282-1. [DOI] [PubMed] [Google Scholar]
- 28.Pellegrino MA, Ferrone S, Reisfeld RA, Irie RF, Golub SH. Expression of histocompatibility (HLA) antigens on tumor cells and normal cells from patients with melanoma. Cancer. 1977;40:36–41. doi: 10.1002/1097-0142(197707)40:1<36::aid-cncr2820400108>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 29.Pollack MS, Heagney SD, Livingston PO, Fogh J. HLA-A, -B, -C alloantigen expression on forty-six cultured tumor cell lines. JNatl Cancer Inst. 1981;66:1003–12. doi: 10.1093/jnci/66.6.1003. [DOI] [PubMed] [Google Scholar]
- 30.Topalian SL, Kasid A, Rosenberg SA. Immunoselection of human melanoma resistant to specific lysis by autologous tumor-infiltrating lymphocytes. J Immunol. 1990;144:4487–95. [PubMed] [Google Scholar]
- 31.Moss DJ, Misko IS, Burrows SR, Burman K, McCarthy R, Sculley TB. Cytotoxic T-cell clones discriminate between A-and B-type Epstein-Barr virus transformants. Nature. 1988;331:719. doi: 10.1038/331719a0. [DOI] [PubMed] [Google Scholar]
- 32.Topalian SL, Muul LM, Solomon D, Rosenberg SA. Expansion of human tumor-infiltrating lymphocytes for use in immunotherapy trials. J Immunol Methods. 1987;102:127–41. doi: 10.1016/s0022-1759(87)80018-2. [DOI] [PubMed] [Google Scholar]
- 33.Fogh J, Trempe G. New human tumor cell lines. In: Fogh J, editor. Human tumor cells in vitro. New York: Plenum Press; 1975. pp. 115–59. [Google Scholar]
- 34.Hopkins KA. ASHI Laboratory Manual. 2. American Society for Histocompatibility and Immunogenetics; Lenexa, KS: 1990. Basic microlymphocytotoxicity test; pp. 195–201. [Google Scholar]
- 35.Brodsky FM, Parham P. Monomorphic anti-HLA-A,B,C monoclonal antibodies detecting molecular subunits and combinatorial determinants. J Immunol. 1982;128:129–35. [PubMed] [Google Scholar]
- 36.Shaw S, Ziegler A, DeMars R. Specificity of monoclonal antibodies directed against human and murine class II histocompatibility antigens as analyzed by binding to HLA-deletion mutant cell lines. Hum Immunol. 1985;12:191–211. doi: 10.1016/0198-8859(85)90336-2. [DOI] [PubMed] [Google Scholar]
- 37.Williams AF, Porter RR, Ada GL, editors. Contemporary topics in molecular immunology. New York: Plenum Press; 1977. p. 83. [DOI] [PubMed] [Google Scholar]
- 38.Berger AE, Davis JE, Cresswell P. Monoclonal antibody to HLA-A3. Hybridoma. 1982;1:87–90. doi: 10.1089/hyb.1.1982.1.87. [DOI] [PubMed] [Google Scholar]
- 39.Foung KH, Taidi B, Ness D, Grumet FC. A monoclonal antibody against HLA-A11 and A24. Hum Immunol. 1986;15:316–9. doi: 10.1016/0198-8859(86)90006-6. [DOI] [PubMed] [Google Scholar]
- 40.Radka SF, Kostyu DD, Amos DB. A monoclonal antibody directed against the HLA-Bw6 epitope. J Immunol. 1982;128:2804–6. [PubMed] [Google Scholar]
- 41.Grange D, Tongio MM, Hauptmann G, et al. Linkage between HLA-A, C, B, Bf and DR alleles: haplotype study of healthy families by factorial correspondence analysis. In: Albert ED, Baur MP, Mayr WR, editors. Histocompatibility testing. Berlin: Springer-Verlag; 1984. pp. 326–9. [Google Scholar]
- 42.Dawkins RL, Degli-Esposti MP, Abraham LJ, Zhang W, Christiansen FT. Conservation versus polymorphism of the MHC in relation to transplantation, immune responses and autoimmune disease. In: Klein J, Klein D, editors. Molecular evolution of the major histocompatibility complex. Berlin: Springer-Verlag; 1991. pp. 391–402. [Google Scholar]
- 43.Browning MI, Krausa P, Rowan A, Bicknell DC, Bodmer JG, Bodmer WF. Tissue typing in HLA-A locus from genomic DNA by sequence specific PCR: comparison of HLA genotype and surface expression on colorectal tumor cell lines. Proc Natl Acad Sci USA. 1993;90:2842–5. doi: 10.1073/pnas.90.7.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kageshita T, Wang Z, Calorini L, et al. Selective loss of human leukocyte class I allospecificities and staining of melanoma cells by monoclonal antibodies recognizing monomorphic determinants of class I human leukocyte antigens. Cancer Res. 1993;53:3349–54. [PubMed] [Google Scholar]
- 45.Zemmour J, Parham P. Distinctive polymorphism at the HLA-C locus: implications for the expression of HLA-C. J Exp Med. 1992;176:937–50. doi: 10.1084/jem.176.4.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Der Bruggen P, Traversari C, Chomez P, et al. A gene encoding an antigen recognized by cytolytic T lymphocytes on human melanoma cells. Science. 1991;254:1643–7. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]