Summary
dHAND and eHAND are bHLH transcription factors expressed during embryogenesis and are required for the proper development of cardiac and extraembryonic tissues. HAND genes, like the myogenic bHLH genes, are classified as class B bHLH genes, which are expressed in a tissue-restricted pattern and function by forming heterodimers with class A bHLH proteins. Myogenic bHLH genes are shown not to form homodimers efficiently suggesting that their activity is dependent on their E-protein partners. To identify HAND Interacting Proteins (HIPs) that regulate the activity of the HAND genes, we screened an E9.5–10.5 mouse embryonic yeast 2-hybrid library with eHAND. Several HIPs held high sequence identity to eHAND, indicating that eHAND could form and function as a homodimer. Based on the high degree of amino acid identity between eHAND and dHAND, it is possible that dHAND could also form homodimers and heterodimers with eHAND. We show using yeast and mammalian 2-hybrid as well as biochemical pull-down assays that eHAND and dHAND are capable of forming both HAND homo- and heterodimers in vivo. To investigate whether HAND genes form heterodimers with other biologically relevant bHLH proteins, we tested and show HAND heterodimerization with the recently identified Hairy-related transcription factors, HRT1-3. This finding is exciting, as both HRT and HAND genes are coexpressed in the developing heart and limb and both have been implicated in establishing tissue boundaries and pattern formation. Moreover, competition gel shift analysis demonstrates that dHAND and eHAND can negatively regulate the DNA binding of MyoD/E12 heterodimers in a manner similar to MISTI and Id proteins suggesting a possible transcriptional inhibitory role for HAND genes. Taken together, these results show that dHAND and eHAND can form homo- and heterodimer combinations with multiple bHLH partners and this broad dimerization profile reflects the mechanisms by which HAND genes regulate transcription.
Introduction
Members of the basic Helix-Loop-Helix (bHLH) superfamily of transcription factors are expressed in a wide range of tissues during development and play a major role in cell specification and differentiation (1,2). bHLH proteins bind DNA as either homo or heterodimers, such that the juxtaposition of the basic region of each factor creates a combined DNA binding domain that recognizes 6 bp sequences termed an Ebox (3). Members of the bHLH superfamily have been categorized into 2 main groups. The Class A bHLH genes, which include the gene products of the E2A gene, E12 and E47, and HEB, are defined by their ubiquitous expression in all tissues and their ability to form homodimers as well as heterodimers with a large range of other bHLH proteins (4,5). In contrast, the class B bHLH genes exhibit tissue restricted expression and in the example the myogenic bHLH genes, MyoD, myogenin, Myf5, and MRF4, do not form homodimers efficiently (7). Thus requiring the formation of heterodimers with Class A bHLH genes (1,6). This observation established the paradigm that class B bHLH proteins form heterodimers with class A bHLH factors to bind DNA and regulate transcription.
This paradigm has prompted numerous laboratories to use Class A bHLH proteins as baits to identify novel Class B genes in yeast 2-hybrid screens (8). The HAND genes, dHAND (HAND2, Thing2, and Hed) and eHAND (HAND1, Thing1, and Hxt) are class B bHLH transcription factors that were cloned using Class A proteins as bait (9–12). Both HAND genes exhibit tissue restricted expression patterns that are partially overlapping during development (9–12). dHAND is expressed within the heart, neural crest, lateral mesoderm, deciduum, and limb bud of mouse embryos (12). While eHAND expression overlaps with dHAND’s in the lateral mesoderm, neural crest, and outflow tract of the heart, eHAND is expressed uniquely in extraembryonic mesoderm (9–11,13). In the developing ventricles, the HAND genes exhibit a sided expression pattern where dHAND is expressed predominantly within the developing right ventricle and eHAND is expressed predominantly within the left ventricle (9–11,13, 14). The result of high levels of dHAND expression in the right ventricle, high levels of eHAND expression in the left ventricle and overlapping expression of d- and eHAND in the outflow tract and at the boundary of both ventricles, is a gradient of HAND transcription factors within the developing heart. In addition, the expression of the HAND genes within the developing heart has been shown to be essential for proper cardiac development as mice harboring null mutations of both dHAND and eHAND show severe developmental defects in both cardiac and extraembryonic mesoderm (13–15). Taken together, these results demonstrate that the HAND genes are critical for proper embryonic development but the mechanism by which these genes function has yet to be determined.
In our efforts to determine the functional role the HAND genes play in development, we employed a yeast 2-hybrid screen with full-length eHAND as bait using an E9.5–10.5 embryonic library. Several HIPs isolated from this screen were homologous to eHAND, suggesting that eHAND could form homodimers. Given that the amino acid identity between the bHLH regions of d- and eHAND is greater than 90%, we hypothesized that dHAND could form homodimers and that dHAND and eHAND could form heterodimers. To address these questions, we employed yeast and mammalian 2-hybrid analysis as well as biochemical pull-down techniques to show that eHAND and dHAND are capable of forming homodimer and heterodimers with each other. To determine if d- and eHAND were capable of forming heterodimers with other class B bHLH factors, we tested heterodimerization of the HAND genes with the newly described Hairy-related transcription factors (HRT) HRT1-3. HRTs are coexpressed with HAND genes within the developing heart, limb buds and other mesodermally derived tissues and thus are biologically relevant HAND partners (16–18). Our data show that indeed HAND genes can form heterodimers with the HRT genes, which is an intriguing finding as both HRT and HAND genes have implicated roles in establishing tissue boundaries and tissue patterning. Moreover, competitive gel-shift assays using an Ebox probe demonstrate that both dHAND and eHAND can negatively regulate the DNA binding of MyoD/E12 heterodimers and thus can regulate the transcription of other bHLH genes independent of direct DNA binding. When considering the slightly overlapping sided expression of the HAND genes in cardiac development, a complex combinatorial array of interactions between d- and eHAND E-proteins, HRT’s, and any unknown bHLH factors are possible. The dimerization properties of the HAND genes suggests complex regulatory functions which allow extremely fine transcriptional control of downstream target genes. This multitude of dimeric complexes may facilitate gene transcription via DNA-binding or repress transcription by preventing the dimerization and DNA binding of other bHLH factors.
Experimental Procedures
Plasmids
The plasmids pAS eHAND and VP16 eHAND were generated using a 650bp PCR product of eHAND beginning at the initiating Methionine and ending at the termination codon 5′ primer 5′GACGGCGAATTCATGAACCTCGTGGGCAGCTAC3′ and 3′ primer 5′GACGGCCCGGGTCACTGCAAATCGAGGTCGCG3′. PCR conditions were 94°C 30 sec, 55°C 45sec, and 72°C 1 minute for 30 cycles. This PCR fragment was subcloned into pCR2.1 (Invitrogen) and excised as an EcoRI fragment for cloning into pAS and VP16-3 vectors. pACT PAN1 is a HAND interacting protein (HIP) that contains the bHLH region of PAN1 and was isolated from a yeast 2-hybrid screen of an E9.5–10.5 embryo library using pAS eHAND as bait (19). WW3NEDD4 and 5.68VP16 were gifts from Dr. Guy James, UTHSCSA. WW3NEDD4 is the third WW domain of NEDD4 cloned into pGBD (20). 5.68VP16 is a non-specific prey that was cloned from a yeast 2-hybrid screen using the WW domains 1 and 2 of MAGI-I (21). pAS dHAND-bHLH was generated by restricting pSG424 dHAND-bHLH (a gift from Dr. Brian Black, UCSF) with EcoRI and subcloning the dHAND-bHLH insert into the EcoRI site of pAS. pACT HEIRI, pCITE E12, and pSVE47VP16 were generous gifts from Dr. Brian Black, UCSF.
The plasmids pBIND, pBIND ID, pACT, pACT MyoD and G5 Luciferase were obtained from Promega’s CheckMate mammalian 2-hybrid system. pBIND eHAND and pACT eHAND were constructed using the same EcoRI insert from pAS eHAND but the insert was digested out of VP16 eHAND as a BamHI/NotI fragment. pACT dHAND was constructed by digesting the dHAND cDNA with BssHII, filling in the site with Klenow, ligating on a BamHI linker, digesting with BamHI and XbaI for subcloning. pGEX eHAND was generated using the EcoRI fragment from VP16 eHAND. pGEX dHAND was generated by taking an NcoI/XbaI dHAND fragment from pCITE dHAND. pGEX 5.68 was a generous gift from Dr. Guy James, UTHSCSA. PCRII Topo HRT 1, HRT2, and HRT3 were generous gifts from Dr. Eric Olson, UTSWMC Dallas. These clones were digested with EcoRV and BamHI, the inserts were cloned into PCITE4B, and the resulting constructs were used to generate in vitro labeled proteins.
Yeast interaction studies
The yeast strain PJ69-4 A (22) was transformed with the various baits (pAS eHAND, pAS dHAND-bHLH, or WW3NEDD4) and prey plasmids (VP16-3, eHANDVP16-3, 5.68VP16-3, pACT Heir I, or pACT PAN1) as previously described using a lithium acetate procedure (22). After transformation, the yeast were plated on selective media that lacked tryptophan (-Trp) and leucine (-Leu) and media that lacked tryptophan, leucine, histidine (-His) and adenine (-Ade) and were grown at 30°C as described (22). Yeast grown on the -Trp/-Leu plates was then inoculated into liquid cultures in -Trp/-Leu media, grown at 30°C, and protein lysates from these yeast were prepared and used in liquid β-galactosidase analysis as previously described (22).
Biochemical pull-down assays
The bacteria BL21 (Novagen) were transfected with the constructs pGEX eHAND, pGEX dHAND or pGEX 5.68 and then were induced to produce GST fusion proteins (23). The proteins were isolated from bacteria using standard methods and bound to glutathione-linked agarose beads (23). 35S labeled in vitro transcribed and translated proteins for the bHLH genes, eHAND, dHAND, HRT1, HRT2, HRT3, and E12, were made using TNT rabbit reticulocyte lysate system (Promega), and were incubated with the bound beads for18hr at 4°C in binding buffer (0.3M sucrose, 1mM EDTA, 10mM TRIS pH 8.0, 0.15MNaCl, 1% TritonX 100, 1ug/ml leupeptin, 2ug/ml aprotinin, and 1mM PMSF) (24). The samples were then washed 4 times in wash buffer (binding buffer with 0.6M NaCl). The beads were boiled in loading buffer and run through a 10% SDS PAGE gel. Gels were then dried and exposed to phosphorimager screens or X-ray film.
Mammalian 2-Hybrid analysis
Equal amounts (2μg) of pG5LUC, pBIND bait plasmid (pBIND, pBIND eHAND, or pBIND Id), and prey plasmid (pACT, pACT eHAND, pACT dHAND, pACT MyoD or pSVE47VP16), were transfected into COS cells seeded onto 3.5 cm dishes using TransIT PanPak (Mirus Corp.) lipofectin using their recommended protocol. Cells were harvested 48hr post transfection, lysates were prepared, and luciferase assays were performed using the Dual-Luciferase reporter assay system (Promega) or Bioluminescent reporter gene assay system (Tropix Inc.) following recommended protocols. All samples were read on a Dynex MLX microtitre plate Luminometer.
Gel Shifts
The various bHLH proteins were in vitro transcribed and translated using Promega’s TNT system as described in the manufacture’s protocol. Oligonucleotides corresponding to the Ebox from the muscle creatine kinase (MCK) enhancer Ebox 5′ CCCCCCCAACACCTGCTGCCTGAGC 3′ reported to bind myogenic bHLH/E12 complexes were 5′ end labeled and annealed with their cold complement. 40 fMol of the labeled oligo were incubated with 10ul of unprogrammed reticulocyte lysate or a total of 10ul of programmed reticulocyte lysate with the indicated bHLH factor with or without 100 fold cold oligo or mutant oligo 5′ GATTCCATTGGTGCTTGATTCCAGAG 3′ as described (25) and then run through a 5% polyacrylamide gel, dried, and exposed via phosphorimager.
Results
To identity HAND interacting proteins (HIPs) that are potentially involved in the regulation of HAND bioactivity, we screened an E9.5–10.5 embryo library using full-length eHAND as bait. Several of these HIPs exhibited high levels of LACZ activation and were sequenced. Surprisingly, one of these HIPs held a 98% sequence identity to mouse eHAND between amino acid 44 and 217 which includes the entire bHLH domain (data not shown). Southern blot analysis of PCR products generated from the 80 HIPs identified in this screen showed an additional 4 clones to be mouse eHAND (data not shown). The concept that eHAND could form an efficient homodimer had never occurred to us as its expression pattern defined the gene within class B of the bHLH superfamily, which are generally thought not to form homodimers efficiently. To investigate this finding in more detail, we constructed a full-length eHANDVP16 fusion to determine if full-length eHAND could form homodimers in yeast. Concurrently, we looked at eHAND’s ability to form heterodimers with the HLH protein Heir I, the mouse E-protein PAN1, and the bHLH domain of dHAND. As expected from our initial findings, eHAND failed to rescue yeast plated on quadruple dropout media when transfected with our empty VP16-3 containing prey construct or when transfected with irrelevant prey clone 5.68 (Figure 1A). β-galactosidase activity confirms that no significant interaction occurs between eHAND and VP16 or with clone 5.68 (Figure 1B). We also observed no significant interaction by growth or β-galactosidase analysis of eHAND with the HLH protein HEIR1. As expected, eHAND and PAN1 rescues growth of yeast plated on quadruple dropout media and shows 25-fold increase in activation of β-galactosidase activity (Figure 1B).
Figure 1.
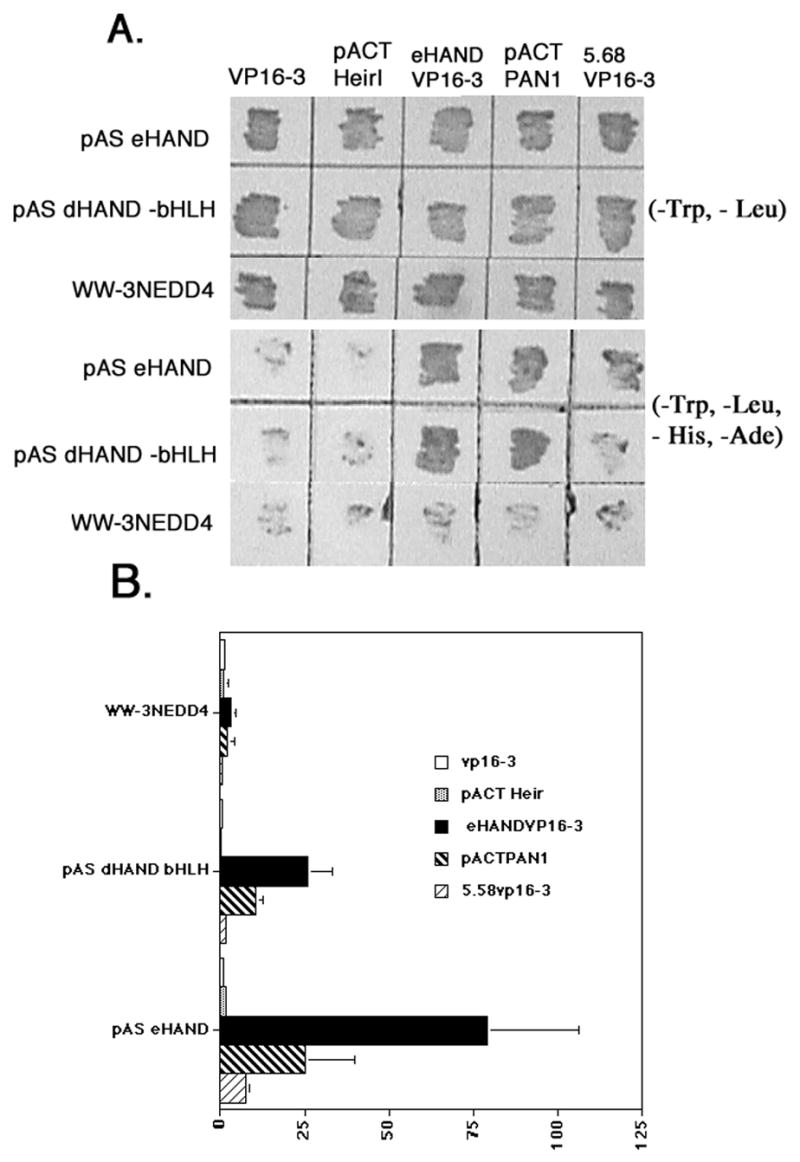
Dimerization properties of HAND genes using yeast 2-hybrid analysis. A) Growth assay shows patched yeast transformations containing the various bait plasmids listed on right pAS eHAND, pAS dHAND-bHLH, and WW-3NEDD4. The various preys listed at top VP16-3, pACT HEIRI (ID1), eHANDVP16-3, pACT PAN1 (E12) and 5.68VP16-3 onto plates lacking Trp and Leu (-TL) and plates lacking Trp, Leu, His and Ade (-THLA). Only protein pairs that directly interact will rescue yeast growth on -THLA media. B) Liquid β galactosidase assays showing strength of the protein-protein interactions. At least 4 independent transformations were used for each interaction pair. Data expressed as fold activation with pAS eHAND/VP16-3 activity equal to 1.0. Error bars denote standard error.
The non-specific control bait containing the WW 3 domain of NEDD4 (WW3NEDD4) did not rescue significantly yeast plated on quadruple drop out media when cotransformed with eHANDVP16 or any of our other preys and only accounted for a slight increase in β-galactosidase activity over control when cotransformed with eHAND (Figure 1). Full-length eHAND transfected with itself, rescues yeast growth and produced a more than 70-fold activation of β-galactosidase activity over control (Figure 1). As expected, we observe yeast rescue when eHAND is cotransfected with the bHLH of dHAND. The HAND proteins share a high amino acid identity within their bHLH domains and growth rescue and a 25 -fold increase in β-galactosidase activity for the HAND heterodimer are similar to results obtained with PAN1 (Figure 1). Moreover, as only the bHLH of dHAND was used in this assay, it indicates that the interaction observed between d- and eHAND and e- and eHAND is likely to be through the bHLH domain. Although these results are suggestive that the HAND genes are capable of forming homo- and heterodimers with each other, it is not clear whether these interactions occur in a non-yeast system. To address this question, we used a biochemical pull-down approach to determine if the dimerization properties of d- and eHAND observed in yeast would occur in an in vitro system.
To demonstrate the ability of the HAND genes to form homo and heterodimers in a non-yeast system, we employed a pull-down assay using a bacterially expressed GST fusion proteins of dHAND and eHAND that were immobilized onto glutathione-linked agarose beads and performed co-incubations with 35S labeled in vitro transcribed and translated proteins. Co-incubations were then washed and all protein retained on the beads was separated by SDS-PAGE and detected by autoradiography or phosphorimager exposure (Figure 2). As a control, the irrelevant bait 5.68 was constructed as a GST fusion to insure that interactions observed are specific. Completely consistent with the yeast 2-hybrid results, both dHAND and eHAND are retained on the GSTeHAND beads indicating that the eHAND can form homo and dHAND/eHAND heterodimers. The lack of retention of the radiolabeled d- and eHAND with an equal amount of GST5.68 protein shows that interaction with these bHLH factors is occurring specifically. (Figure 2). When GSTdHAND linked beads were incubated with the radiolabeled proteins, results similar to those obtained with eHAND were observed (Figure 2). As expected, GSTdHAND pulled down dHAND, eHAND, and E12 which demonstrates that full-length dHAND can form homodimers and heterodimers with eHAND and E12 (Figure 2). No significant retention of the radiolabeled proteins with GST5.68 occurred consistent with the yeast 2-hybrid results (Figure 2). Taken together, the yeast 2-hybrid results and biochemical pull-down results strongly suggest that the bHLH domains of both d- and eHAND have dimerization properties that allow for efficient homodimer formation and the formation of efficient dHAND/eHAND heterodimers.
Figure 2.
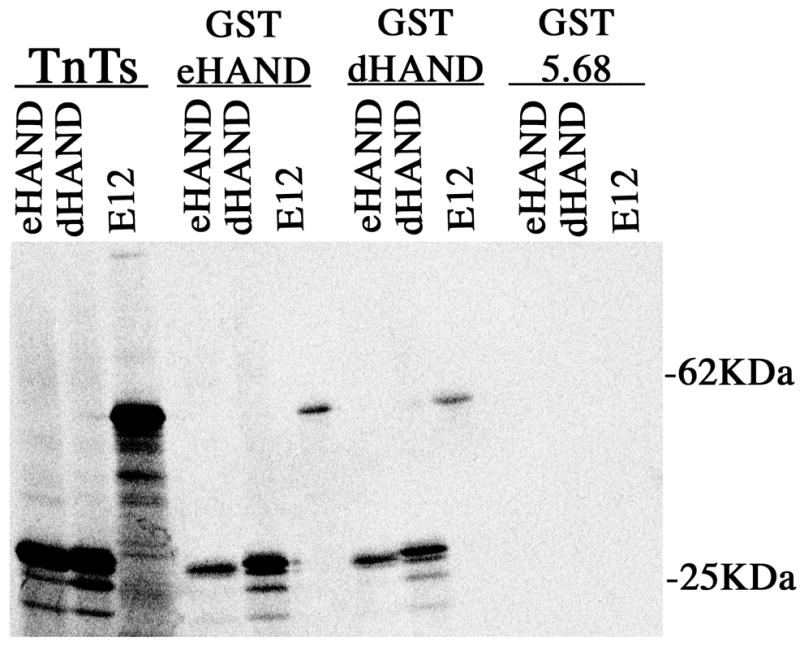
Biochemical pull-down analysis of HAND dimerization. GST fusion proteins for dHAND, eHAND, and 5.68 were bound to glutathione beads as described in methods. 35S labeled in vitro transcribed and translated proteins were then incubated with the bead bound protein, washed in high salt buffer and the bead mixtures were then run through a 10% SDS-PAGE gel. The gel was then dried and the retention of the labeled protein onto the beads was then visualized via phosphorimager.
To determine if HAND genes could interact in vivo, we looked at eHAND’s ability to homo/heterodimerize within a mammalian cell using a mammalian 2-hybrid assay. Bait plasmids for eHAND and Id were used against a panel of preys in COS cells (Figure 3). When the empty bait plasmid pBIND was cotransfected with our series of prey plasmids, no significant luciferase activity is observed (Figure 3). Analysis of pBIND Id shows no significant interaction with empty prey plasmid, however a strong activation of the luciferase reporter is seen when the Id bait is cotransfected with pSVE47VP16 (7.5 fold) and pACT MyoD (9.0 fold) (Figure 3). These results confirm previous findings that Id can interact with both of these factors and confirms that our interaction assay is working (Figure 2). Interaction of Id with eHAND and dHAND however showed background levels of luciferase activity confirming our yeast 2-hybrid results suggesting that HAND genes do not interact with Id to any significant degree (Figure 3). When pBIND eHAND is cotransfected with the empty prey vector pACT in our mammalian 2-hybrid assay basal levels of luciferase activity are observed (Figure 3). In agreement with our yeast and pull-down data, eHAND shows strong interaction when cotransfected with pSVE47VP16 further confirming the HAND genes ability to form heterodimers with the Class A family of bHLH genes. (Figure 3). When pBIND eHAND is co transfected with pACT eHAND or pACT dHAND, we observe a significant increase in luciferase activity indicating HAND homodimers and heterodimers can form efficiently within mammalian cells (Figure 3). Surprisingly, eHAND also showed a noticeable increase in luciferase activity when cotransfected with pACT MyoD (Figure 3). The biological significance of this interaction is unclear as eHAND and MyoD are not coexpressed during development, however the fact that eHAND can heterodimerize with another non-HAND Class B bHLH factor suggests that HAND genes may regulate transcription by dimerizing with both class A and class B bHLH partners.
Figure 3.
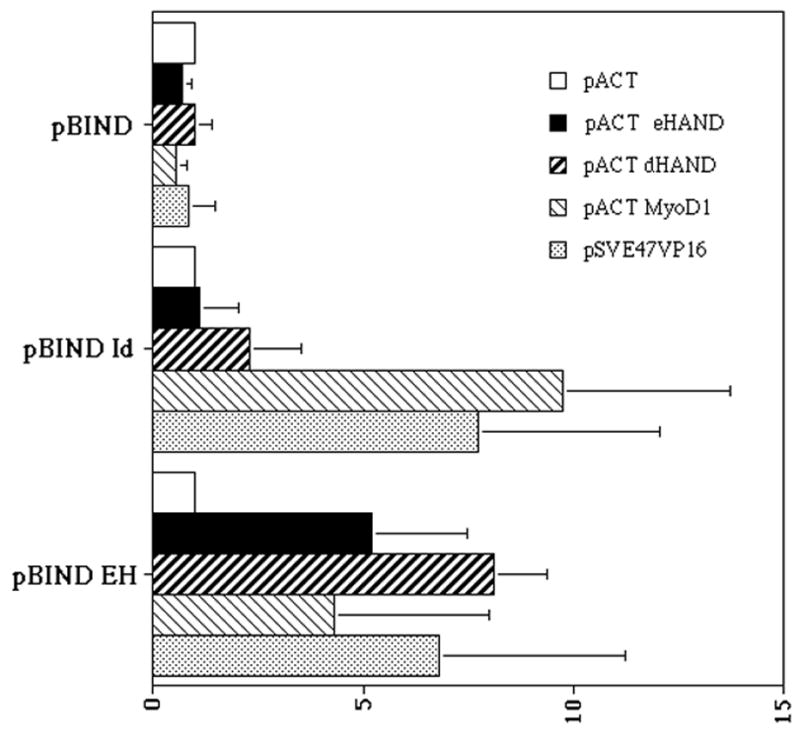
eHAND dimerization analysis by mammalian 2-hybrid. COS cells were transfected via lipofectin with (GAL)5LUC and the various bait and prey constructs as described in the methods. Cell extracts were prepared from 4 independent experiments and luciferase activity was measured as described in methods. Luciferase activity is expressed by fold with each bait/pACT control being set to 1.0 for comparison. Error bars denote standard error.
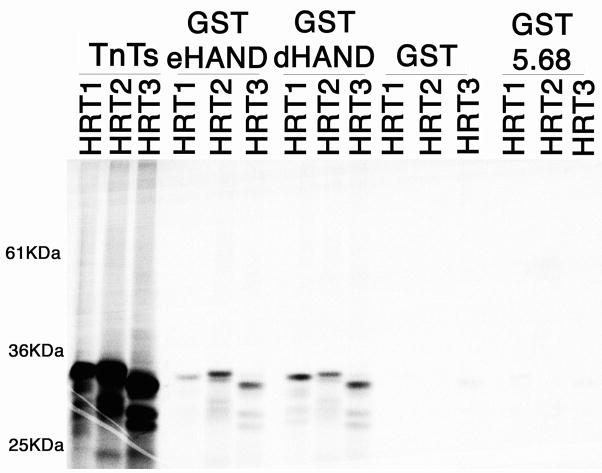
The observation that eHAND could form heterodimers with MyoD suggested to us that HAND genes might have a wide range of dimerization partners such that any coexpressed bHLH gene could potentially be a biologically important HAND partner. Recently the HRT family of bHLH transcription factors was identified and HRT1 and HRT2 were shown to be expressed within the early stages of the developing heart concurrent with both d- and eHAND as well as within the developing limb (16–18). To look at interaction, we preformed GST pull downs with all three HRT factors (Figure 4). Results show that both GSTdHAND and GSTeHAND retain significant amounts of radiolabeled HRT1, 2, and 3 when compared to levels of pulled-down HRT proteins observed with GST or GST 5.68 beads indicating efficient heterodimerization. Yeast 2-hybrid interactions confirm these observations (data not shown). Taken together these results suggest that d- and eHAND can dimerize with HRT factors and thus could effect transcription by directly binding DNA in an HRT/HAND complex.
Figure 4.
Biochemical pull-down analysis of HAND dimerization with HRTs. GST fusion proteins for dHAND, eHAND, and 5.68 were bound to glutathione beads as described in methods. 35S labeled in vitro transcribed and translated HRT1, HRT2, and HRT3 were then incubated with the bead bound protein, washed in high salt buffer and the bead mixtures were then run through a 10% SDS-PAGE gel. The gel was then dried and the retention of the labeled protein onto the beads was then visualized via phosphorimager.
To examine the DNA binding properties of the HAND homodimers and heterodimers, gel mobility shift assays were performed using double stranded oligonucleotides containing an Ebox sequence that was reported to bind eHAND/E12 using in vitro transcribed and translated proteins (10). Results of these experiments recapitulated previously published results showing no HAND homodimer binding (data not shown, 10). Given the heterodimerization of MyoD with eHAND, we wanted to determine if HAND dimerization could negatively regulate MyoD/E12 DNA binding by titration of both E12 and MyoD (Figure 5). In this experiment, in vitro transcribed and translated MyoD and E12 were incubated with an Ebox sequence from the MCK enhancer previously shown to bind this heterodimer (3). As expected MyoD/E12 complexes shifted the migration of the labeled oligo and this migration was inhibited by the addition of excess cold oligo but not by the addition of a mutant oligo that did not contain an Ebox (Figure 5). Interestingly, addition of an equal amount of either eHAND or dHAND reduced the intensity of the shifted MyoD/E12 complex showing a disruption of DNA binding (Figure 5). Clearly in light of the mammalian 2-hybrid results showing MyoD/eHAND and E12/eHAND dimerization, the reduction of MyoD/E12 bound to DNA is a result of dHAND and eHAND directly competing for dimerization with both of these factors. Thus, HAND genes may act as negative regulators of certain class A and class B bHLH genes in a manner analogous to MISTI and Id family members.
Figure 5.
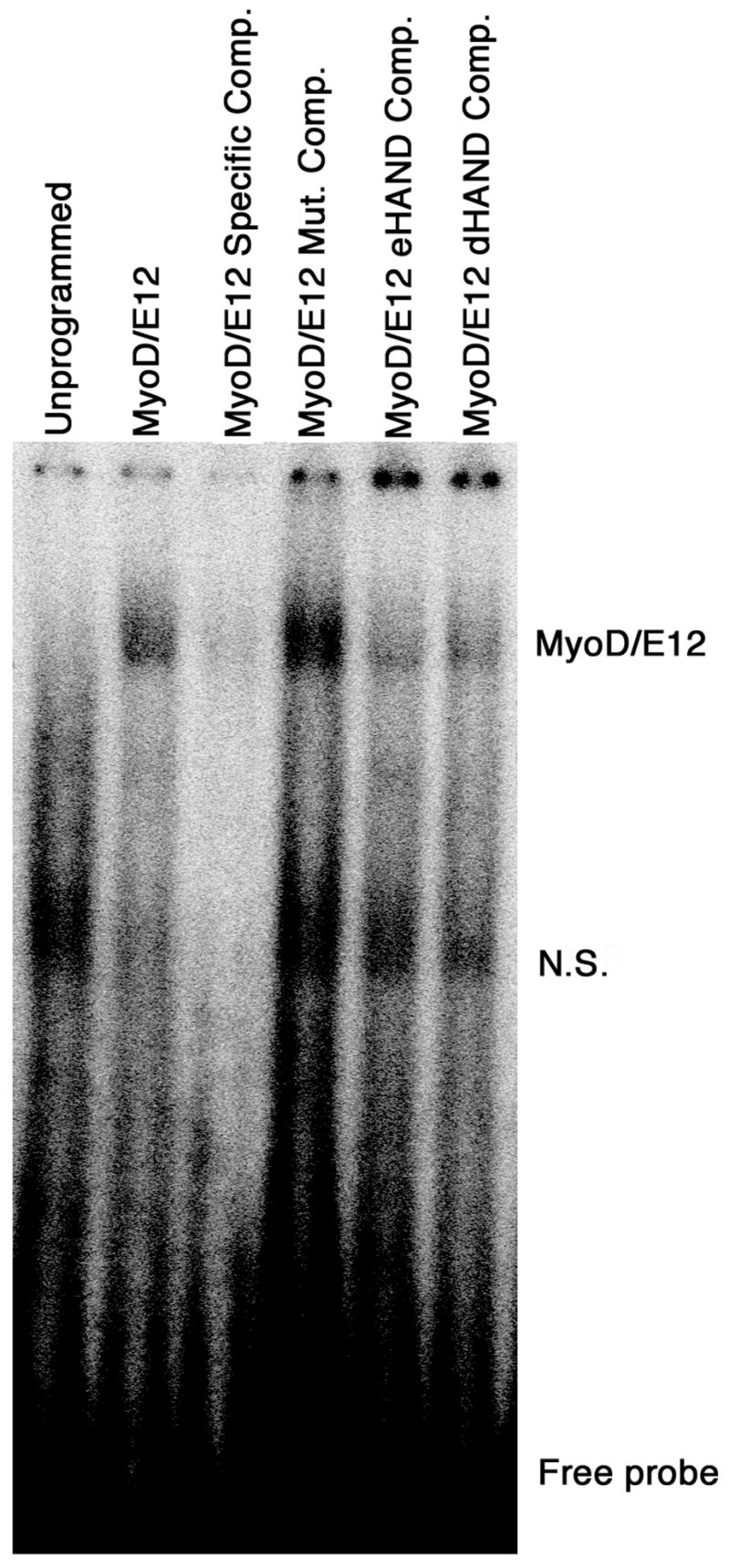
Disruption of MyoD/E12 DNA binding by dHAND and eHAND. Gel retardation assays using synthetic MCK Ebox sequences and in vitro translated and transcribed proteins. Approximately 40 fMol of radio labeled probe was incubated with unprogrammed reticulocyte lysate (unprogrammed) and lysates programmed with MyoD and E12 (MyoD/E12). Specific-competition (Specific Comp.) with 50pMol of unlabeled MCK oligonucleotide or competition with 50pmol of mutant oligo (Mut. Comp.) was used to show DNA binding is specific. Equal volume of reticulocyte lysates programmed with eHAND (eHAND Comp.) or dHAND (dHAND Comp.) was addend to the MyoD/E12 complex incubations. (N.S.) Non-specific complex observed in the unprogrammed lysates.
Discussion
In our efforts to find novel HIPs that interact with eHAND using the yeast 2-hybrid system, we have uncovered the ability of eHAND and dHAND to form homodimers and heterodimers in vivo. This is an important finding as HAND genes, based on expression patterns and their initial isolation in yeast 2-hybrid screens using E12 as bait, have been assumed to be biologically active only when dimerized with E-proteins. Unlike the myogenic bHLH genes, HAND genes are fully capable of forming homodimers and heterodimers with themselves in mammalian cells, suggesting that the various dimeric forms of the HAND genes can control the transcription of a diverse set of presently unknown downstream genes. The skeletal muscle bHLH gene MISTI has been shown to form homodimers and heterodimers with MyoD. (26). The MyoD/MISTI heterodimer is thought to form an inactive complex, blocking MyoD from forming E-protein heterodimers and thus inhibiting its activity. Our data show that HAND genes can function in the same manner as MISTI and although HAND/MyoD interaction appears to be biologically irrelevant, it does strongly suggest that HAND genes could sequester yet to be discovered, coexpressed bHLH factors by a similar mechanisms.
The Hairy-related transcription factors, HRT’s, (also known as HESR’s and HEY’s) are expressed within the somatic mesoderm, central nervous system, kidney and nasal epithelium and like the Hairy and Enhancer of Split (HES) bHLH factors, HRT’s have been shown to depend on notch signaling (16–18). HRT1 and HRT2 are also coexpressed with the HAND genes during both limb and cardiac development. Members of the HES family of bHLH factors repress transcription via binding a cis acting element termed a Nbox and are thought to establish boundaries of expression within the tissue they are expressed. As HRTs respond to the same signaling pathway as HES factors, these genes are thought to play a similar biological role (16,17). Very recently expression studies of dHAND within the developing limbs of mice, chick and fish show that dHAND plays and important role in limb anterior-posterior patterning (27–29). The finding that HAND genes play a direct role in anterior-posterior patterning taken together with our data that shows HAND genes can interact with HRT genes, which are postulated to establish discrete boundaries of gene expression that set up embryonic patterning, strongly suggests that HAND and HRT gene function in controlling anterior-posterior patterning may be through heterodimerization and possibly independent of E protein dimerization.
The ability of E proteins to form heterodimers with class B genes established the paradigm of how these ubiquitously expressed genes play a role in tissue-specific transcription. Myogenic bHLH family members which are expressed exclusively in skeletal muscle give the E proteins their specificity to implement skeletal muscle specification and differentiation. HAND genes are expressed within heart, neural crest, lateral and extraembryonic mesoderm, developing sympathetic nerves and maternally derived deciduum. When considering this complex expression profile, it is difficult to imagine how these genes could control specific gene expression in these diverse tissues solely by heterodimerization with E proteins. It has been recently reported that E12/E47 is not expressed within trophoblast giant cells, a cell type that has been shown to require eHAND for proper development (13, 15, 30). Thus, eHAND homodimers and/or an eHAND heterodimer with an unidentified bHLH protein regulate transcription in an E-protein independent environment. The observation that d and eHAND can interact with MyoD and HRT’s suggests that HAND genes are not limited in their choice of dimeric partners. Therefore, it is likely that transcription of HAND-regulated genes within different tissues as well as affinity of DNA binding of cis-acting targets may be mediated by the bHLH partner, as is the case with E proteins.
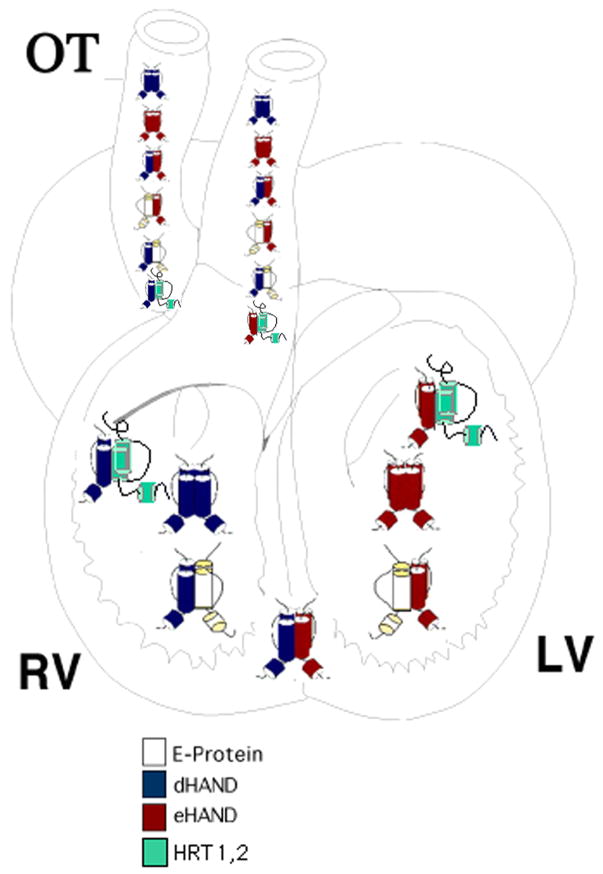
Speculation on the role of HAND dimerization in the developing heart becomes extremely compelling. Most interesting the sided expression of dHAND in the right ventricle and eHAND in the left ventricle represents an anterior-posterior expression that is made left-right through cardiac looping (31). In light of dHAND’s role in the developing limb, it suggests that HAND genes may play a role in heart patterning. As dHAND is expressed in the developing right ventricle and eHAND is expressed in the developing left ventricle and with both genes being coexpressed at the ventricle boundary and within the outflow tract, a transcription factor gradient is formed (Figure 6). dHAND homodimers would form preferentially within the right ventricle, eHAND homodimers within the left ventricle, and the boundary of the developing ventricles and outflow tract would have a mixture of dHAND and eHAND allowing for dHAND/eHAND heterodimers (Figure 6). Considering the ubiquitous expression of E proteins within the heart, as well as HRT factors and yet to be discovered cardiac bHLH factors, an extremely diverse population of HAND transcriptional dimers is possible.
Figure 6.
Model of potential HAND gene dimers within the heart. The model schematized here shows E12 (white), dHAND (blue), eHAND (red), and HRT1 and 2 (green) positioned within the heart where expression of these genes has been determined. In the developing heart, dHAND is expressed predominately within the right ventricle (RV) whereas eHAND expression is extremely low. Thus, cardiomyocytes with this region could form dHAND homodimers, dHAND/HRT and dHAND/E12 heterodimers. In the left ventricle (LV), eHAND expression is high whereas dHAND expression is low thus eHAND homodimers, eHAND/HRT, and eHAND/E12 heterodimers are possible. At the ventricle boundary and in the outflow tract (OT), both HAND genes are expressed making eHAND/dHAND heterodimer formation possible in addition to homodimers, HAND/HRT, and HAND/E12 complexes.
The ability of HAND proteins to form homo and heterodimers with themselves and other class B bHLH factors allows for both direct transcriptional activation/repression via DNA binding and the negative regulation of itself and other bHLH factors via the formation of non-binding dimer complexes. Recently, eHAND was shown to inhibit the DNA binding of MASH2 to the MCK Ebox by competing with MASH2 for E12 (30). In biochemical pull down analysis we show that eHAND can heterodimerize with MASH1 (unpublished results). In light of this data and interaction of eHAND with MyoD and the HRTs, it is possible that eHAND can heterodimerize with MASH2 and thus the reduction of DNA binding of the MASH2/E12 complex is a dual competition of eHAND for both bHLH factors. This type of negative regulation is observed in the Id family of HLH genes (32). The Id HLH proteins lack a basic region that is used to contact DNA and can form heterodimers with E proteins locking E12/E47 in inactive complexes. When considering the variations in gene expression and protein modifications such as phosphorylation that can alter protein-protein interactions, a resulting shift of dimerization choices of the HAND genes may represent the mechanism by which these proteins regulate function.
Although reports on the dHAND and eHAND knockout mice show down regulation of several genes, only a report on the regulation of the Adenylosuccinate Synthetase I gene shows direct regulation by dHAND or eHAND, and the exact cis-acting elements within the Adenylosuccinate Synthetase gene promoter that respond to HAND activation have yet to be identified (33). bHLH genes have been shown to bind to either Ebox or Nbox sequences, but it has not been established if other sequences can be utilized for DNA-binding. In detailed analysis of the DNA binding characteristics of the myogenic bHLH genes and E-proteins using binding site selection protocols, it has been established that each member of the bHLH dimer recognizes a specific 1/2 site within the Ebox and that different combinations of these factors selectively bind different Eboxes (34, 35). Casting experiments done on eHAND show that eHAND/E12 heterodimers bind a degenerate Ebox NNTCTG and that eHAND homodimers can not bind this sequence (10). Due to the lack of any cis-acting elements known to be transcriptionally regulated by d- or eHAND the question of whether HAND homodimers, heterodimers, and/or HAND/E-protein heterodimers can regulate expression remains unanswered. In the previous DNA binding analysis eHAND was combined with an E-protein in the casting procedure. To Determine DNA binding sequences for HAND homodimers and E-protein independent heterodimers casting experiments will need to preformed to determine if any HAND dimer pair has the ability to bind DNA. It is possible that HAND homodimers and dHAND/eHAND heterodimers recognize a non-Ebox DNA target sequence. This situation is seen with the bHLH factor HES-1 which does not bind efficiently to an Ebox but recognizes a CACNAG motif termed a Nbox (32, 36). Interestingly eHAND, like HES-1, contains a proline within its basic domain which is atypical for bHLH family members. However, we do not see DNA binding of eHAND to oligonucleotides containing Nbox sequences with or without E12 (data not shown). Taken together our results show, that unlike most class B bHLH factors, eHAND and dHAND have expanded dimerization specificity that allows them to form a variety of dimeric complexes in vivo. This observation suggests that HAND genes can function to negatively regulate themselves as well as other bHLH proteins by directly competing for bHLH partners. By forming homo and heterodimers, HAND genes can alter their own DNA binding recognition and the DNA binding properties of other bHLH genes revealing a complex mechanism for HAND gene regulation of biological function.
Acknowledgments
The authors would like to thank Tania Fernandez, Kunal Patel, and Cedric Wheelock for excellent technical assistance, Drs. G. James, B. Black, and E. Olson for their generosity with reagents Drs. M. Steinhelper, B. Black, A. Rawls, and J. Wilson-Rawls for helpful comments and review of the manuscript. This work was supported by grants from the American Heart Association and National Institutes of Health (R01 HLA61677-01A1).
References
- 1.Lee JE. Current Opinion in Neurobiology. 1997;7(1):13–20. doi: 10.1016/s0959-4388(97)80115-8. [DOI] [PubMed] [Google Scholar]
- 2.Molkentin JD, Olson EN. Current Opinion in Genetics & Development. 1996;6(4):445–53. doi: 10.1016/s0959-437x(96)80066-9. [DOI] [PubMed] [Google Scholar]
- 3.Klein ENOaWH. Genes and Development. 1994;8:1–8. doi: 10.1101/gad.8.1.1. [DOI] [PubMed] [Google Scholar]
- 4.Hu JS, Olson EN, Kingston RE. Mol Cell Biol. 1992;12(3):1031–42. doi: 10.1128/mcb.12.3.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neuman T, Keen A, Knapik E, Shain D, Ross M, Nornes HO, Zuber MX. European Journal of Neuroscience. 1993;5(4):311–8. doi: 10.1111/j.1460-9568.1993.tb00498.x. [DOI] [PubMed] [Google Scholar]
- 6.Olson EN, Klein WH. Genes Dev. 1994;8(1):1–8. doi: 10.1101/gad.8.1.1. [DOI] [PubMed] [Google Scholar]
- 7.Chakraborty T, Brennan TJ, Li L, Edmondson D, Olson EN. Mol Cell Biol. 1991;11(7):3633–41. doi: 10.1128/mcb.11.7.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Staudinger J, Perry M, Elledge SJ, Olson EN. J Biol Chem. 1993;268(7):4608–11. [PubMed] [Google Scholar]
- 9.Cserjesi P, Brown D, Lyons GE, Olson EN. Dev Biol. 1995;170(2):664–78. doi: 10.1006/dbio.1995.1245. [DOI] [PubMed] [Google Scholar]
- 10.Hollenberg SM, Sternglanz R, Cheng PF, Weintraub H. Mol Cell Biol. 1995;15(7):3813–22. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cross JC, Flannery ML, Blanar MA, Steingrimsson E, Jenkins NA, Copeland NG, Rutter WJ, Werb Z. Development. 1995;121(8):2513–23. doi: 10.1242/dev.121.8.2513. [DOI] [PubMed] [Google Scholar]
- 12.Srivastava D, Cserjesi P, Olson EN. Science. 1995;270(5244):1995–9. doi: 10.1126/science.270.5244.1995. [DOI] [PubMed] [Google Scholar]
- 13.Firulli AB, McFadden DG, Lin Q, Srivastava D, Olson EN. Nat Genet. 1998;18(3):266–70. doi: 10.1038/ng0398-266. [DOI] [PubMed] [Google Scholar]
- 14.Srivastava D, Thomas T, Lin Q, Kirby ML, Brown D, Olson EN. Nat Genet. 1997;16(2):154–60. doi: 10.1038/ng0697-154. [DOI] [PubMed] [Google Scholar]
- 15.Riley P, Anson-Cartwright L, Cross JC. Nat Genet. 1998;18(3):271–275. doi: 10.1038/ng0398-271. [DOI] [PubMed] [Google Scholar]
- 16.Nakagawa O, Nakagawa M, Richardson JA, Olson EN, Srivastava D. Dev Biol. 1999;216:72–84. doi: 10.1006/dbio.1999.9454. [DOI] [PubMed] [Google Scholar]
- 17.Kokubo H, Lun Y, Johnson RL. Biochem Biophys Res Commun. 1999;260(2):459–465. doi: 10.1006/bbrc.1999.0880. [DOI] [PubMed] [Google Scholar]
- 18.Steidl C, Leimeister C, Klaunt B, Maier M, Nanda I, Dixon D, Clarke R, Schmid MMG. Genomics. 2000;66(2):195–203. doi: 10.1006/geno.2000.6200. [DOI] [PubMed] [Google Scholar]
- 19.Xin XQ, Nelson C, Collins L, Dorshkind K. Journal of Immunology. 1993;151(10):5398–407. [PubMed] [Google Scholar]
- 20.Staub O, Dho S, Henry P, Correa J, Ishikawa T, McGlade J, Rotin D. EMBO Journal. 1996;15(10):2371–80. [PMC free article] [PubMed] [Google Scholar]
- 21.Dobrosotskaya I, Guy RK, James GL. J of Biol Chem. 1997;272(50):31589–97. doi: 10.1074/jbc.272.50.31589. [DOI] [PubMed] [Google Scholar]
- 22.James P, Halladay J, Craig EA. Genetics. 1996;144(4):1425–36. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith DB, Johnson KS. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 24.Rhodes KJ, Monaghan MM, Barrezueta NX, Nawoschik S, Bekele-Arciri Z, Matos MF, Nakahira K, Schechter LE, Trimmer JS. J Neuroscience. 1996;16:4846–4860. doi: 10.1523/JNEUROSCI.16-16-04846.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Molkentin JD, Firulli AB, Black BL, Martin JF, Hustad CM, Copeland N, Jenkins N, Lyons G, Olson EN. Mol Cell Biol. 1996;16(7):3814–24. doi: 10.1128/mcb.16.7.3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lemcercier C, To RQ, Carrasco KKinieczny SF. EMBO J. 1998;17(5):1412–1422. doi: 10.1093/emboj/17.5.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charite JMcfadden DG, Olson EN. Development. 2000;127:2461–2470. doi: 10.1242/dev.127.11.2461. [DOI] [PubMed] [Google Scholar]
- 28.Fernandez-Teran M, Piedra ME, Kathiriya IS, Srivastava D, Rodriguez-Rey JC, Ros MA. Development. 2000;127:2133–2142. doi: 10.1242/dev.127.10.2133. [DOI] [PubMed] [Google Scholar]
- 29.Yelon D, Ticho B, Halpern ME, Ruvinsky I, Ho RK, Silver LM, Stainier DYR. Development. 2000;127:2573–2582. doi: 10.1242/dev.127.12.2573. [DOI] [PubMed] [Google Scholar]
- 30.Scott IC, Anson-Cartwright L, RIiley P, Reda D, Cross JC. Mol Cell Biol. 2000;20(2):530–541. doi: 10.1128/mcb.20.2.530-541.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olson EN, Srivastava D. Science. 1996;272(5262):671–6. doi: 10.1126/science.272.5262.671. [DOI] [PubMed] [Google Scholar]
- 32.Massari ME, Murre C. Mol Cell Biol. 2000;20(2):429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis AL, Xia Y, Datta SK, McMillin J, Kellems RE. J Biol Chem. 1999;274(20):14188–14197. doi: 10.1074/jbc.274.20.14188. [DOI] [PubMed] [Google Scholar]
- 34.Blackwell TK, Weintraub H. Science. 1990;250:1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- 35.Writght WE, Binder M, Funk W. Mol Cell Biol. 1991;11(8):4101–4110. doi: 10.1128/mcb.11.8.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishibashi M, Ang SL, Shiota K, Nakanishi S, Kageyama R, Guillemot F. Genes and Dev. 1995;9:3136–3148. doi: 10.1101/gad.9.24.3136. [DOI] [PubMed] [Google Scholar]