Abstract
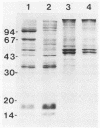
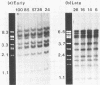
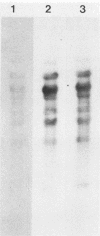
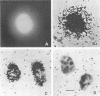
Viral DNA was found to be tightly associated with the nuclear matrix from HeLa cells lytically infected with human adenovirus type 5. The bound viral DNA, like cell DNA, was resistant to nonionic detergent and to extraction with high-salt (2 M NaCl) solution. However, whereas over 95% of the cell DNA was recovered in the matrix fraction, the amount of associated viral DNA varied during infection. Throughout the lytic cycle, the amount of matrix-associated adenovirus type 5 DNA increased until it reached a plateau level at 20 to 24 h after infection. At this stage, the matrix-bound DNA represented 87% of the total viral DNA; after this stage, additional newly synthesized viral DNA accumulated as non-matrix-associated DNA. DNase digestion studies revealed that all viral DNA sequences were equally represented in the matrix-bound DNA both early and late in infection; thus, unlike cell DNA, there seem to be no preferred attachment sites on the viral genome. An enrichment of viral DNA relative to cell DNA was found in the matrix-associated DNA after extensive DNase I digestion. This finding, together with an in situ hybridization study, suggests that the viral DNA is more intimately associated with the nuclear matrix than is cell DNA and probably does not exist in extended loops.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barrack E. R., Coffey D. S. The specific binding of estrogens and androgens to the nuclear matrix of sex hormone responsive tissues. J Biol Chem. 1980 Aug 10;255(15):7265–7275. [PubMed] [Google Scholar]
- Bellett A. J., Younghusband H. B. Replication of the DNA of chick embryo lethal orphan virus. J Mol Biol. 1972 Dec 30;72(3):691–709. doi: 10.1016/0022-2836(72)90185-4. [DOI] [PubMed] [Google Scholar]
- Benyajati C., Worcel A. Isolation, characterization, and structure of the folded interphase genome of Drosophila melanogaster. Cell. 1976 Nov;9(3):393–407. doi: 10.1016/0092-8674(76)90084-2. [DOI] [PubMed] [Google Scholar]
- Berezney R., Coffey D. S. Identification of a nuclear protein matrix. Biochem Biophys Res Commun. 1974 Oct 23;60(4):1410–1417. doi: 10.1016/0006-291x(74)90355-6. [DOI] [PubMed] [Google Scholar]
- Berezney R., Coffey D. S. Nuclear protein matrix: association with newly synthesized DNA. Science. 1975 Jul 25;189(4199):291–293. doi: 10.1126/science.1145202. [DOI] [PubMed] [Google Scholar]
- Buckler-White A. J., Humphrey G. W., Pigiet V. Association of polyoma T antigen and DNA with the nuclear matrix from lytically infected 3T6 cells. Cell. 1980 Nov;22(1 Pt 1):37–46. doi: 10.1016/0092-8674(80)90152-x. [DOI] [PubMed] [Google Scholar]
- Chin W. W., Maizel J. V., Jr The polypeptides of adenovirus. VIII. The enrichment of E3 (11,000) in the nuclear matrix fraction. Virology. 1977 Jan;76(1):79–89. doi: 10.1016/0042-6822(77)90284-7. [DOI] [PubMed] [Google Scholar]
- Cook P. R., Brazell I. A., Jost E. Characterization of nuclear structures containing superhelical DNA. J Cell Sci. 1976 Nov;22(2):303–324. doi: 10.1242/jcs.22.2.303. [DOI] [PubMed] [Google Scholar]
- Cook P. R., Brazell I. A. Mapping sequences in loops of nuclear DNA by their progressive detachment from the nuclear cage. Nucleic Acids Res. 1980 Jul 11;8(13):2895–2906. doi: 10.1093/nar/8.13.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook P. R., Brazell I. A. Supercoils in human DNA. J Cell Sci. 1975 Nov;19(2):261–279. doi: 10.1242/jcs.19.2.261. [DOI] [PubMed] [Google Scholar]
- Herman R., Weymouth L., Penman S. Heterogeneous nuclear RNA-protein fibers in chromatin-depleted nuclei. J Cell Biol. 1978 Sep;78(3):663–674. doi: 10.1083/jcb.78.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge L. D., Mancini P., Davis F. M., Heywood P. Nuclear matrix of HeLa S3 cells. Polypeptide composition during adenovirus infection and in phases of the cell cycle. J Cell Biol. 1977 Jan;72(1):194–208. doi: 10.1083/jcb.72.1.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz M. S. Intermediates in the synthesis of type 2 adenovirus deoxyribonucleic acid. J Virol. 1971 Nov;8(5):675–683. doi: 10.1128/jvi.8.5.675-683.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner R. L., Kelly T. J., Jr The structure of replicating adenovirus 2 DNA molecules. Cell. 1977 Dec;12(4):1007–1020. doi: 10.1016/0092-8674(77)90165-9. [DOI] [PubMed] [Google Scholar]
- Long B. H., Huang C. Y., Pogo A. O. Isolation and characterization of the nuclear matrix in Friend erythroleukemia cells: chromatin and hnRNA interactions with the nuclear matrix. Cell. 1979 Dec;18(4):1079–1090. doi: 10.1016/0092-8674(79)90221-6. [DOI] [PubMed] [Google Scholar]
- McCready S. J., Godwin J., Mason D. W., Brazell I. A., Cook P. R. DNA is replicated at the nuclear cage. J Cell Sci. 1980 Dec;46:365–386. doi: 10.1242/jcs.46.1.365. [DOI] [PubMed] [Google Scholar]
- Nelkin B. D., Pardoll D. M., Vogelstein B. Localization of SV40 genes within supercoiled loop domains. Nucleic Acids Res. 1980 Dec 11;8(23):5623–5633. doi: 10.1093/nar/8.23.5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PHILIPSON L. Adenovirus assay by the fluorescent cell-counting procedure. Virology. 1961 Nov;15:263–268. doi: 10.1016/0042-6822(61)90357-9. [DOI] [PubMed] [Google Scholar]
- Pardoll D. M., Vogelstein B., Coffey D. S. A fixed site of DNA replication in eucaryotic cells. Cell. 1980 Feb;19(2):527–536. doi: 10.1016/0092-8674(80)90527-9. [DOI] [PubMed] [Google Scholar]
- Pardoll D. M., Vogelstein B. Sequence analysis of nuclear matrix associated DNA from rat liver. Exp Cell Res. 1980 Aug;128(2):466–470. doi: 10.1016/0014-4827(80)90083-x. [DOI] [PubMed] [Google Scholar]
- Pardue M. L., Gall J. G. Nucleic acid hybridization to the DNA of cytological preparations. Methods Cell Biol. 1975;10:1–16. doi: 10.1016/s0091-679x(08)60727-x. [DOI] [PubMed] [Google Scholar]
- Pettersson U., Sambrook J. Amount of viral DNA in the genome of cells transformed by adenovirus type 2. J Mol Biol. 1973 Jan;73(1):125–130. doi: 10.1016/0022-2836(73)90164-2. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Sussenbach J. S., van der Vliet P. C. Viral DNA synthesis in isolated nuclei from adenovirus-infected KB cells. FEBS Lett. 1972 Mar;21(1):7–10. doi: 10.1016/0014-5793(72)80149-2. [DOI] [PubMed] [Google Scholar]
- Thomas G. P., Mathews M. B. DNA replication and the early to late transition in adenovirus infection. Cell. 1980 Nov;22(2 Pt 2):523–533. doi: 10.1016/0092-8674(80)90362-1. [DOI] [PubMed] [Google Scholar]
- Tyndall C., Younghusband H. B., Bellett A. J. Some adenovirus DNA is associated with the DNA of permissive cells during productive or restricted growth. J Virol. 1978 Jan;25(1):1–10. doi: 10.1128/jvi.25.1.1-10.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B., Pardoll D. M., Coffey D. S. Supercoiled loops and eucaryotic DNA replicaton. Cell. 1980 Nov;22(1 Pt 1):79–85. doi: 10.1016/0092-8674(80)90156-7. [DOI] [PubMed] [Google Scholar]
- Winston V. D., Bolen J. B., Consigli R. A. Isolation and characterization of polyoma uncoating intermediates from the nuclei of infected mouse cells. J Virol. 1980 Mar;33(3):1173–1181. doi: 10.1128/jvi.33.3.1173-1181.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vliet P. C., Kwant M. M. Role of DNA polymerase gamma in adenovirus DNA replication. Nature. 1978 Nov 30;276(5687):532–534. doi: 10.1038/276532a0. [DOI] [PubMed] [Google Scholar]