Abstract
The elastic rod theory is used together with all-atom normal mode analysis in implicit solvent to characterize the mechanical flexibility of duplex DNA. The bending, twisting, stretching rigidities extracted from all-atom simulations (on linear duplexes from 60 to 150 base pairs in length and from 94 base-pair minicircles) are in reasonable agreement with experimental results. We focus on salt concentration and sequence effects on the overall flexibility. Bending persistence lengths are about 20% higher than most experimental estimates, but the transition from low-salt to high-salt behavior is reproduced well, as is the dependence of the stretching modulus on salt (which is opposite to that of bending). CTG and CGG trinucleotide repeats, responsible for several degenerative disorders, are found to be more flexible than random DNA, in agreement with several recent studies, whereas poly(dA).poly(dT) is the stiffest sequence we have encountered. The results suggest that current all-atom potentials, which were parameterized on small molecules and short oligonucleotides, also provide a useful description of duplex DNA at much longer length scales.
1 Introduction
Advances in biomolecular simulations have allowed the studies of larger and more interesting molecules while conserving an all-atom representation of the system. The implicit treatment of solvation effects -avoiding the inclusion of thousands of water molecules even for a modest-size biomolecule - has played an important role in this development and often provides an accurate treatment of the solvation effects.1–3 In recent years the analytic generalized Born (GB) method has become increasingly popular over the more classical Poisson-Boltzmann approach due to its computational efficiency. Additionally, second derivatives of the solvation energy are easier to obtain for GB than for the Poisson-Boltzmann theory.4 Implicit models also have the advantage of allowing faster convergence in molecular dynamics simulations, hence, reducing the sampling time required; making them more affordable than explicit solvent simulations.5
In GB theory the electrostatic part of the solvation energy is given by:
| (1) |
where ri j is the distance between atom i and atom j, Ri and Rj are the effective Born radii of i and j, respectively. is usually defined as
| (2) |
although other alternatives have been investigated.6–8 Finally, εw is a dielectric constant (≈ 80 in water at 300K) and κ is the Debye-Huckel screening parameter used to reproduce electrostatic screening of salt. More details about the generalized Born model can be found elsewhere.5, 9 Whereas generalized Born theory has been widely adopted for molecular dynamics simulations it has only recently been extended to normal mode analysis by Brown et. al.4 Normal mode analysis provides several advantages over classical molecular dynamics even though it approximates the global potential by a harmonic function.10 First, it provides a clearer representation of the collective motions of biomolecules through a few of the lowest energy vibrational modes; second, it makes the evaluation of entropy contributions and other thermodynamic properties straightforward, and, finally it is more affordable when long timescales (sampling times) are required. For example, in the case of a 60-bp duplex of DNA the relaxation times for the lowest energy modes are close to 20 ns with a viscosity of about 0.6 cp, and should presumably become larger for longer duplexes. The recent implementation of analytical second derivatives of the Born energy in the molecular mechanics program package Nucleic Acid Builder (NAB)11 provides a new alternative to molecular dynamics in implicit solvent and was used to produce a picture of the vibrational motions for small nucleic acids.4 The same study can be extended to larger systems and may also be used to extract important information about the overall behavior of nucleic acids. These simulations have the advantages of allowing a sequence-dependent treatment as well as the study of salt concentration changes via a screening factor.
Here we examine the mechanical flexibility of DNA using comparisons to the elastic rod models for both linear and circular forms.12–15 Such a technique not only allows the benchmark of the GB method itself, but also provides information about the intrinsic stiffness of a given DNA sequence. DNA binding to nucleosomes is probably related to flexibility, and degenerative disorders such as fragile x syndrome, myotonic dystrophy, and Huntington disease are believed to be the results of an increase in flexibility of some given nucleotide repeats.16–18 There is, thus, considerable interest in fundamental, all atom-based approaches to estimating mechanical properties of DNA.
It is common practice, in both experimental and theoretical studies of nucleic acids, to compare results to a model where DNA is a homogeneous elastic rod, with bending, torsional and stretching rigidities as adjustable parameters. This analogy has been used in numerous studies to describe supercoiling of DNA19, 20 as well as DNA cyclization21. While such a representation can have obvious limitations for small sequences, it provides a useful picture for longer segments of DNA, focussing attention on general elastic properties, as opposed to a detailed analysis of the helical parameters of a given sequence. Since vibrational amplitudes are inversely proportional to frequencies, many of the long-range dynamical features of DNA are captured by its lowest energy vibrational modes. One can extract the rigidity constants governing the elasticity of the corresponding sequence by comparing the modes obtained from all-atom simulations to those coming from a continuum (elastic) model, where the characteristics of the rod (radius, length, mass …), are those of the optimized structure in the all-atom calculations. Such an approach was pioneered by Matsumoto and Gô, 15 who used a distance-dependent dielectric model with an all-atom force field and sequences as long as 36bp. The results were encouraging, but the rigidities reported were much higher than experimental and subsequent theoretical estimates. More recent extensions of this idea have relied on a knowledge-based force fields derived from the observed fluctuations at a local level in DNA duplexes.12–14 Here, we extend these ideas to all-atom simulations using modern solvent models, including the effects of added salt, and to much longer oligonucleotides.
2 Methods
2.1 All-atom simulations
All simulations were carried out with the molecular mechanics program package NAB (nucleic acid builder) using the Cornell et al. (parm94) force field.22 We used the pairwise approach of Hawkins et a123, 24 for the generalized Born model. The original model for salt effects in GB scaled the Debye-Huckel screening factor κ by a factor of 0.73, primarily to compensate for the lack of ion exclusion radius concept in the GB model. This could be important for short-range interactions but is less appropriate for long-range interactions of the type we are studying here where the asymptotic behavior of salt screening is more important. We thus did not scale κ for this study. The structures were minimized using a combination of conjugate gradients and Newton-Raphson minimization techniques to obtain an RMS gradient below 1 ×10−8 kcal/mol-Å. This level of convergence is necessary to avoid contamination from translational and rotational modes into true internal modes whose frequencies may be as low as 0.1 cm−1. The xmin minimizer from the biomolecular simulation program package AMBER25 was used to perform powerful but less costly Limited-memory Broyden-Fletcher-Goldfarb-Shanno (LBFGS) Truncated Newton Conjugate (TNC) minimizations. The diagonalization of the Hessian matrix was done using the ARPACK26 routines in combination with a Cholesky decomposition and inversion of the Hessian matrix -therefore providing a better separation of the eigenvalues- to enhance convergence. Using the ARPACK routines reduces the cost of the diagonalization from a few days to a few minutes for a 200 base pair DNA fragment. This technique is most appropriate here since only a few normal modes are necessary (fewer than 200). Another advantage of using the ARPACK package is the ease and efficiency of distributing the calculation on several processors.
2.2 Linear DNA
This section is merely an overview of the elastic rod model that has been reviewed in several places.12, 15 The mechanical constants for a linear homogeneous rod are as follow:
| (3) |
where M is the total mass of the rod (molecule), IM is the moment of inertia per unit length, L is the length of the rod (or helical axis in the present case), νn is the vibrational frequency associated to mode number n. The polynomial roots Pn satisfy the condition LPn = 4.6941, 7.8548, 10.9955, 14.1372, 17.2788 (n=1,..5) for the differential equation defining the bending modes of an homogeneous rod.27 Additionally, a, the (bending) persistence length, can be defined as
In brief, we fit the all-atom vibrational frequencies to their analytical counterparts in order to evaluate the corresponding rigidities which are treated as adjustable parameters. Figs. 1 to 3 show all-atom normal modes that clearly exhibit bending, stretching and twisting behavior. The bending modes come in nearly-degenerate pairs in the all-atom calculation, corresponding to bending on the two directions perpendicular to the helix axis. The closer the degeneracy the less mixing there is with other internal modes. As an example, only the first three pairs of bending modes are considered in the 60bp case because for n > 4, there is more than 2–3% difference between the two frequencies. In the case of the stretching and twisting vibrational frequencies, discarding the contaminated modes is not as straightforward and one has to pay more attention to the eigenvectors. As the linear fragments get longer more modes can be included; roughly speaking, an additional mode can be included for each additional 30bps starting with 3 modes in the case of bending and stretching for 60bps (2 for twisting) to 6 modes for 150 bps (5 for twisting). The interactive essential dynamics plugin (IED),28 an extension of the visualization package VMD, was used to identify the character of each normal mode.
Figure 1.
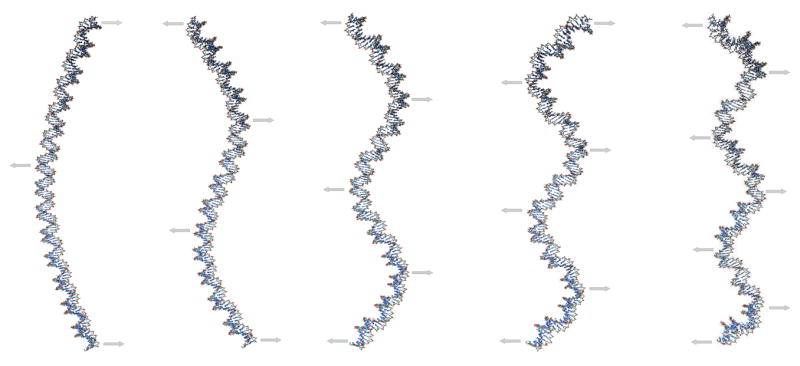
Low-frequency bending modes of a 150-bp linear segment of DNA. The arrows define the main motions associated with the corresponding vibration. There are nearly degenerate modes corresponding to each of these, but with displacements perpendicular to the plane of figure.
Figure 3.
Low-frequency twisting modes of a 150-bp linear segment of DNA. The arrows define the main motions associated with the corresponding vibration.
2.3 Circular DNA
As in the previous section, we present here a summary of the vibrational modes of circular elastic rod; details can be found elsewhere.13, 14, 29, 30 In the case of circular DNA there are three different types of motions: in-plane bending, out-of-plane bending, and torsion. The analytical solutions corresponding to these motions for a relaxed minicircle made from of a naturally straight elastic rod are as follow:
| (4) |
| (5) |
where R is the radius of the circle, ω = C/A, and the frequencies are measured in units of (16A/πρr6)1/2. When the circle is not torsionally relaxed these roots are no longer valid but one can add the excess twist and solve the dispersion equation directly.12 In the present example the relaxed 94-bp minicircle will correspond to a linking number Lk=8 while for the overtwisted representation Lk=9. [In solution, the average twist for DNA is about 34.5° corresponding to 10.4 bp per helix turn; therefore, a relaxed 94-bp minicircle would have a linking number Lk=9. However, the force field used here22 is known to underestimate DNA twist by about 4°; this translates to an overall twist of roughly 30.5° and about 12bp per helix turn. Thus, the in silico linking number for the relaxed 94-bp minicircle is close to Lk=8; meaning that for Lk=9 the circle is overtwisted by about one turn of helix.]
There are two types of bending in this model, corresponding to deformations in and out of the plane of the minicircle. The in-plane bending motions come in pairs, in the two directions perpendicular to the axis of the circle. Similarly, the out-of-plane bending is also two-fold and approximately degenerate in the GB simulations; the corresponding motions are shown in Fig. 4. The twisting motions are shown in Fig. 5; the motion corresponding to n=0 is a free rotation around the torus with no flexural deformations, meaning that there is no deformation of the circle (the radius of the helix is conserved throughout the motion); this mode is discarded in the fitting process even though it appears in the GB simulations.
Figure 4.
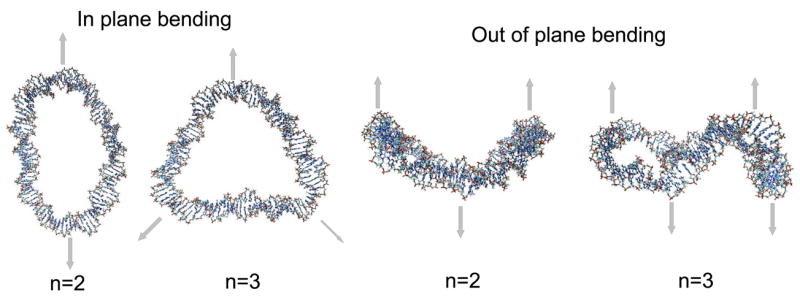
Low-frequency - in and out of plane bending - modes of circular DNA. The arrows define the main motions associated with the corresponding vibration. Bending motions corresponding to n=0 and n=1 do not exist in the analytical model,29, 30 so the first allowed bending comes for n=2.
Figure 5.
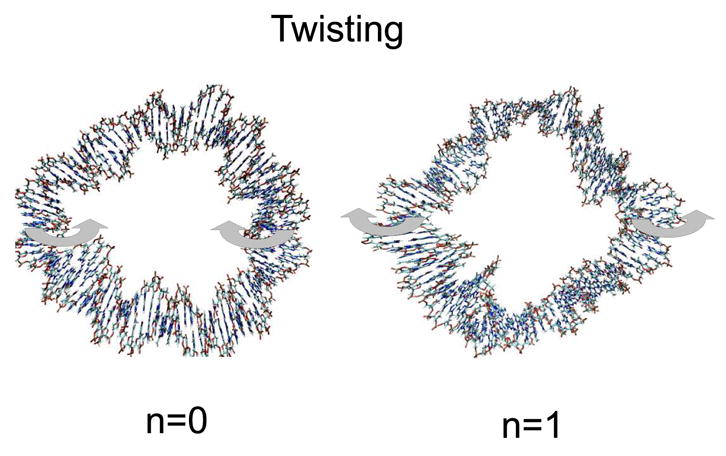
Low-frequency - torsional - modes of circular DNA. The arrows define the main motions associated with the corresponding vibration.
3 Results
3.1 Salt concentration dependence for linear DNA
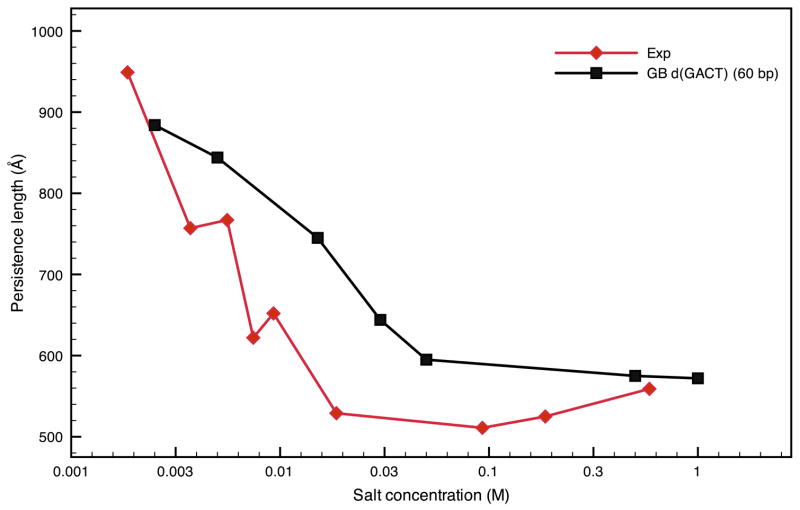
The method described in this work for extracting rigidities of DNA fragments has been primarily used for 60 bp-sequences, because such sequences offer an acceptable compromise between computational cost (especially for optimizing the structures) and being long enough to promote long-range bending motions. Longer sequences were also investigated in order to assess the dependence on sequence length. Here, we examine the sequence d(GTGACTGACTGACTG)n, paired with its Watson-Crick complementary sequence in a B-form duplex. This sequence is referred to by its main nucleotide repeat GACTn for the sake of succinctness, and has been the subject of previous computational studies.4, 31, 32 It appears to be a good choice to describe the behavior of DNA in general since it will be shown later to be of average rigidity. The salt dependence of the persistence lengths as well as the stretching rigidities are compared to experimental estimates33, 34 in Figs. 6 and 7 and in Table 1. The overall salt dependence of the persistence length (Fig. 6) is well reproduced by the GB results, although the transition from low-salt to high-salt behavior occurs at a higher concentration than is suggested by experiment, and the computed bending rigidities are somewhat too large. Both of these may be a result of the very simple dependence on salt given in Eq. 1, and more elaborate models have been proposed,35, 36 and will be examined in future studies.
Figure 6.
Comparison of the persistence lengths from experiment33 and extracted from the elastic rod model for the d(GACT)4 sequence as a function of salt concentration at 300K.
Figure 7.
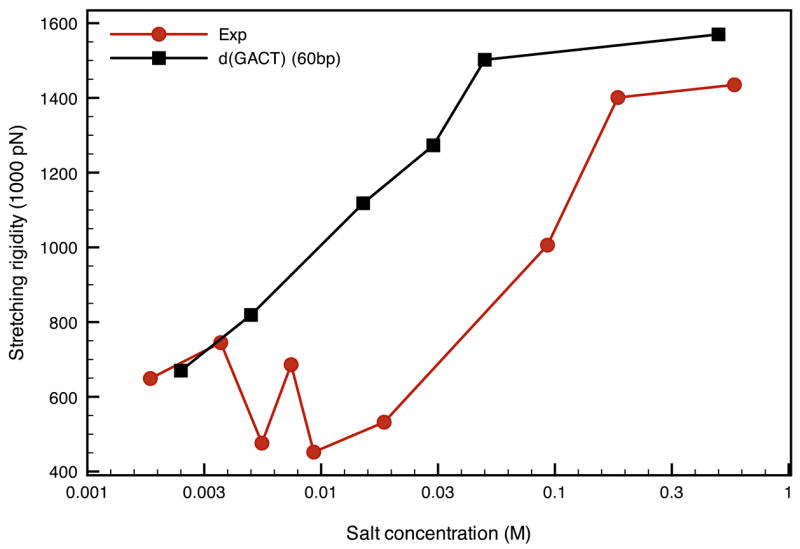
Comparison of the stretching moduli from experiment33 and extracted from the elastic rod model for the d(GACT)4 sequence as a function of salt concentration at 300K.
Table 1.
Lowest bending (B), stretching (S) and twisting (T) frequencies in cm−1 for the sequence GACTn, n = 4,6,8,10 with a salt concentration of 0.05M and T=300K.
| Sequences | GACT4 | GACT6 | GACT8 | GACT10 |
|---|---|---|---|---|
| B1 | 0.114–0.116 (0.100) | 0.053-0.053 (0.047) | 0.030-0.030 (0.027) | 0.019-0.019 (0.017) |
| B2 | 0.294–0.295 (0.275) | 0.140–0.142 (0.130) | 0.081-0.081(0.075) | 0.052-0.052 (0.048) |
| B3 | 0.522–0.532 (0.539) | 0.263–0.264 (0.254) | 0.153–0.156 (0.147) | 0.099–0.100 (0.094) |
| B4 | — | 0.408–0.413 (0.420) | 0.246–0.247 (0.243) | 0.159–0.163 (0.156) |
| B5 | — | — | 0.353–0.360 (0.363) | 0.233–0.234 (0.233) |
| B6 | — | — | — | 0.317–0.323 (0.325) |
| S1 | 0.664 (0.619) | 0.439 (0.406) | 0.324 (0.307) | 0.263 (0.248) |
| S2 | 1.289 (1.237) | 0.852 (0.812) | 0.646 (0.614) | 0.526 (0.496) |
| S3 | 1.807 (1.856) | 1.242 (1.218) | 0.955 (0.921) | 0.769 (0.744) |
| S4 | — | 1.577 (1.623) | 1.232 (1.228) | 1.010 (0.992) |
| S5 | — | — | 1.496 (1.535) | 1.242 (1.240) |
| S6 | — | — | — | 1.446 (1.488) |
| T1 | 0.443 (0.413) | 0.302 (0.276) | 0.228 (0.206) | 0.183 (0.164) |
| T2 | 0.811 (0.826) | 0.589 (0.553) | 0.442 (0.412) | 0.358 (0.328) |
| T3 | — | 0.796 (0.829) | 0.636(0.618) | 0.522 (0.491) |
| T4 | — | — | 0.789 (0.824) | 0.660 (0.655) |
| T5 | — | — | — | 0.781 (0.819) |
The results are similar for stretching (Fig. 7), where the general trend is again well reproduced by the GB simulations. Note the opposite salt-dependence of the stretching and bending moduli: the bending persistence length increases with salt concentration whereas the stretching moduli decreases. This is not consistent with simple elasticity theory,33 where both the bending and the twisting moduli are proportional to the Young’s modulus. Nevertheless, this property of simple elasticity theory clashes with both our results and conclusions from single molecule stretching studies. This could be one of the limitations of comparing DNA to an isotropic elastic rod where it is clear that effects other than purely macroscopic mechanisms are involved in the behavior of the persistence length and stretching rigidity. A possible explanation was that the decrease in the stretching modulus could be due to local melting of A.T rich regions,33 but in our study this phenomenon occurs without any presence of melting.
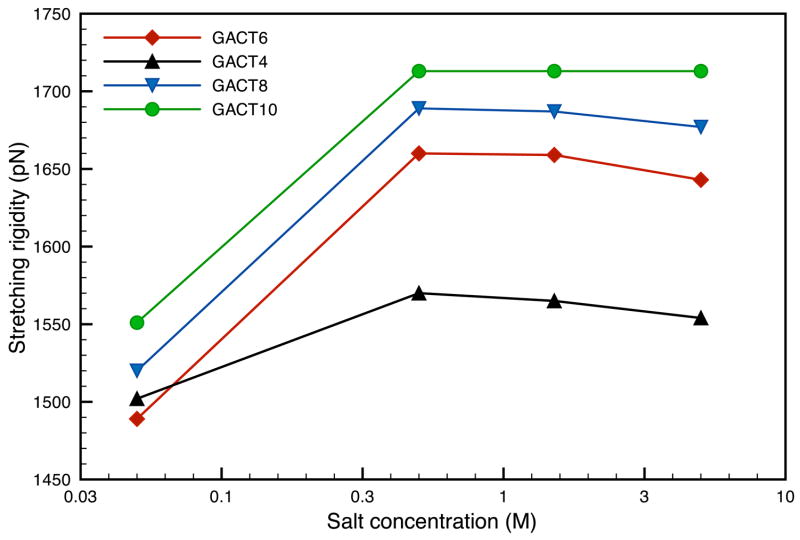
We also looked at longer sequences with up to 150 base pairs; the corresponding results are shown in Figs 8–10. The persistence length depends on the sequence length, and saturates at about 120bp; a possible explanation is the fact that for smaller sequences (less than 90bp), the calculated bending frequencies do not match the analytical frequencies as well as they do for larger sequences. This could mean that the elastic rod model is a more appropriate approximation for sequences longer than 100bp. A similar pattern was observed with comparable calculations by Matsumoto and Go, 15 though the persistence lengths reported there were much larger. This trend disappears for fragments longer than 100bp and the persistence lengths for the GACT sequence converge in the vicinity of 700 Å; a somewhat larger value than the 500–550 Å widely reported for random DNA. A similar behavior is observed with the stretching moduli (Figure 9) but to a lesser extent and the stretching rigidities quickly converge to a value of about 1700 pN, a value close to the 1500pN usually reported in the literature. In this case the stretching rigidities are close in value throughout the range of concentration investigated here.
Figure 8.
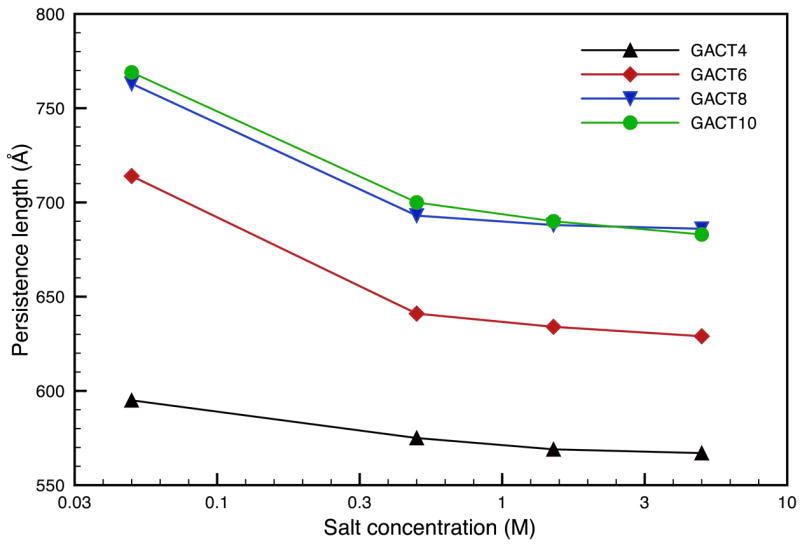
Comparison of the persistence lengths for various GACT repeat sequences as a function of salt concentration at 300K..
Figure 10.
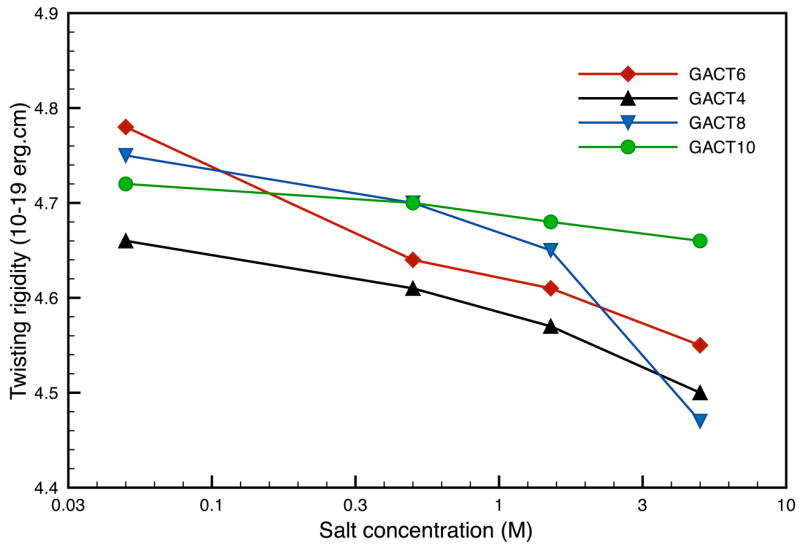
Comparison of the twisting moduli for various GACT repeat sequences as a function of salt concentration at 300K.
Figure 9.
Comparison of the stretching moduli for various GACT repeat sequences as a function of salt concentration at 300K.
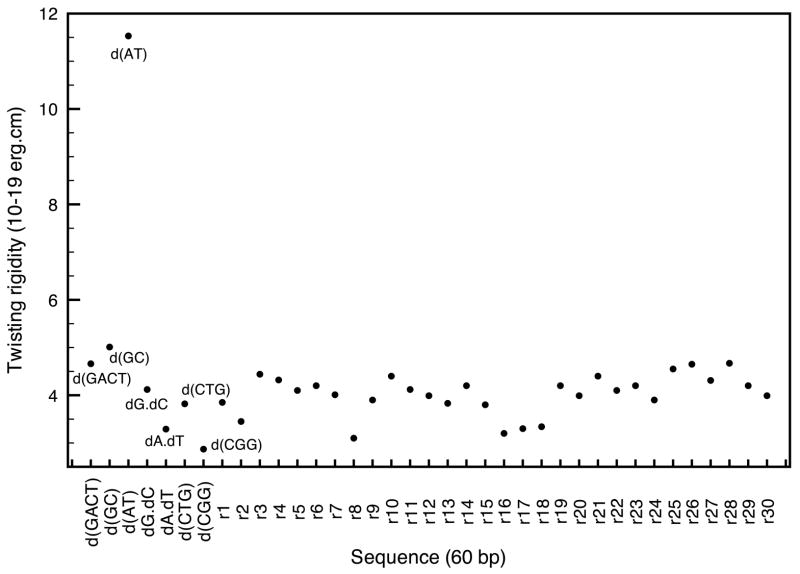
The twisting motions were harder to identify and the quality of the fits to the elastic model is lower than for the bending and stretching modes. However, the average twisting modulus for the GACT sequence, 4.7 10−19erg.cm (Fig. 10), agrees well with the latest experimental estimates of 4.5 10−19erg.cm.37, 38 Note that the twisting rigidities depend less on sequence length than was the case for other mechanical constants. As noted above, the frequency fits get better as the sequence length increases, hence, indicating that DNA behaves more like an elastic rod for longer sequences. The macroscopic interactions take over the local phenomena more present in smaller sequences.
3.2 Sequence dependence of mechanical properties for linear DNA
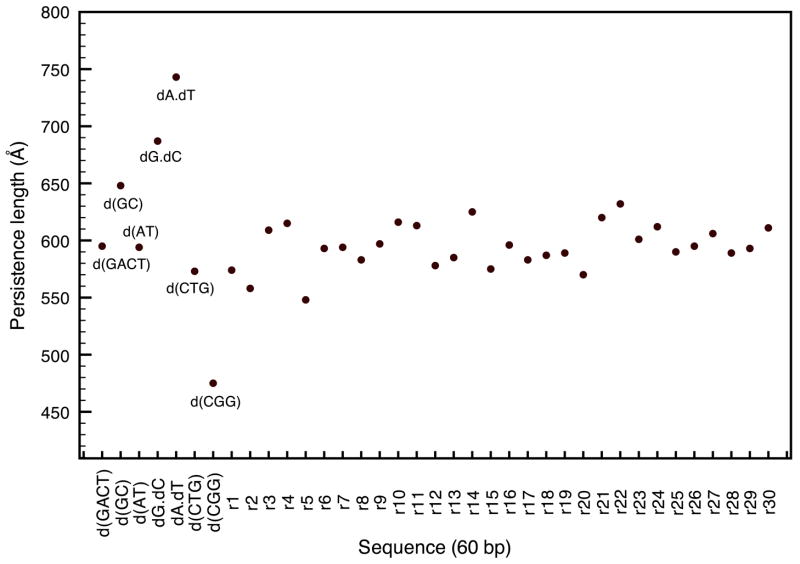
An area where mechanical properties of DNA are important is the interactions between nucleic acid and histone proteins in the nucleosome, and, for DNA packaging in general. Many degenerative diseases are believed to be caused by an increase of flexibility in DNA therefore preventing transcription. Several sequences have been linked to fragile x syndrome, myotonic dystrophy, and Huntington disease; and recent studies have shown an enhancement in the flexibility of DNA segments containing these triplet repeats.16–18 One of the advantages of the microscopic method described here is its ability to take sequence effects into account and study the flexibility for different DNA sequences. Figure 11 shows the persistence lengths for various sequences that include the well-known CTG, CGG triplet repeats and other sequences. We also computed the persistence length of thirty randomly generated sequences of 60 bp each; the GC ratio chosen for the random sequences was assumed to be the average GC content in the human genome (42 percent).39, 40 The homopolymers poly(dA).poly(dT), poly(dG).poly(dC) (referred to as dA.dT and dG.dC) are the most rigid of the set while heteropolymers poly(dAT) and poly(dGC) offer more flexibility.41–43 Similar conclusions are found in many experimental studies on nucleosome formations,44, 45 and both homopolymers are known to activate transcription due to their relative stiffness. The sequence - d(GACT)- used in the previous section was an appropriate choice as its persistence length is average.
Figure 11.
Comparison of the persistence lengths for various DNA sequences with a salt concentration of 0.05M and T=300K.
Among the most interesting repeats we looked at are CTG and CGG; both of them have been shown to exhibit a greater flexibility than random DNA with, in most studies, CTG being more flexible than CGG. In our results, CTG is located on the lower end of the persistence length spectrum, and CGG has a persistence length 25 percent smaller than average. The extreme flexibility of the CGG repeats was also tested by replacing the nine central base pairs of the homopolymer dA.dT by three CGG repeats, which enhanced its flexibility by 20–25 percent. The robustness of the method was also tested by comparing the persistence lengths of 90 (575 Å vs. 710 Å) and 120 (664 Å vs. 853 Å) base-pair segments of CGG repeats and GACT repeats in order confirm that the relative flexibility of the CGG repeat is still present for longer sequences and is not affected by the sequence-length dependence shown in the previous section.
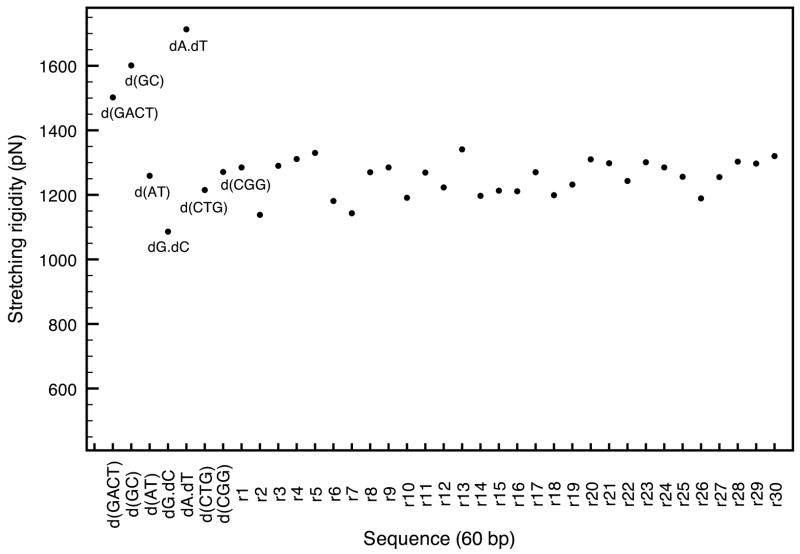
The results of this section also agree favorably with a study by Lankas et. al,46 in that the four DNA oligomers, poly(dA).poly(dT), poly(dG).poly(dC), poly(dAT), poly(dGC), are found to have similar intrinsic flexibility in both studies, especially for the stretching rigidities; these can be found in table 2 and figure 12. In recent single-molecule studies, the melting transition happens for a lower stretching force in poly(dAT) (1259 pN) than for poly(dGC) (1601 pN),47 which is consistent with the relative stretching rigidities in this work where poly(dAT) is much more flexible with respect to stretching than poly(dGC). As in the previous section, identifying the twisting modes is not as straightforward but the results are yet again within experimental estimates (Table 2 and figure 13). The average value for a random sequence is in the vicinity of 4.02 10−19erg.cm while the most flexible sequence so far (CGG) has a twisting modulus of 2.87 10−19erg.cm. In summary, it is clear that sequences including CGG and CTG triplet repeats are more flexible than random DNA. In both cases, at least two of their rigidity constants are smaller than that of random DNA. In contrast, stiffer repeats such as dA.dT and dG.dC have, in average, larger rigidity constants than random DNA.
Table 2.
Mechanical constants for various DNA sequences with a salt concentration of 0.05M and T=300K. The values reported in italics and bold correspond to stiffer and more flexible characteristics than random DNA, respectively
| Sequences | Persistence Length (Å) | Stretching rigidity (pN) | Twisting rigidity (10−19erg.cm) |
|---|---|---|---|
| GACT | 595 | 1502 | 4.66 |
| poly d(AT) | 594 | 1259 | 11.53 |
| poly d(GC) | 648 | 1601 | 5.01 |
| poly d(A) . poly d(T) | 743 | 1713 | 3.29 |
| poly d(G) . poly d(C) | 697 | 1086 | 4.12 |
| CTG | 573 | 1215 | 3.82 |
| CGG | 475 | 1271 | 2.87 |
| Random | 594 | 1251 | 4.02 |
| Random min | 548 | 1138 | 3.10 |
| Random max | 632 | 1341 | 4.67 |
| Standard deviation | 19 | 55 | 0.41 |
Figure 12.
Comparison of stretching moduli for various DNA sequences with a salt concentration of 0.05M and T=300K.
Figure 13.
Comparison of twisting moduli for various DNA sequences with a salt concentration of 0.05M and T=300K.
3.3 Circular DNA
In an influential study by Cloutier and Widom,48 particular sequences of DNA were found to form DNA minicircles with expectations 400 fold higher than predicted. In this section we examine whether this increase in cyclization rate could be explained in part by the elastic properties of these sequences. We chose the 94bp sequence with the highest J value (TA94) (i.e. the sequence with the highest closure probability). This sequence increases nucleosome positioning by a few orders of magnitude compared to random DNA. The reason for this enhancement is not clear; sharp bending might be responsible for the increase in flexibility as it was pointed out in a study by Lankas et. al.49
The bending motions that deform the circle within the plane of the molecule are twofold degenerate in the GB simulations and correspond to deformations along two perpendicular axis in the plane of the minicircle (Fig. 4 and table 3). Three pairs of in-plane bending motions were used in the fitting process. The frequencies corresponding to these motions are not exactly degenerate due to the fact that the mini-circle is not fully symmetrical in sequence. However, they are sufficiently close in value to consider them as equivalent in the model. The GB frequencies are in good agreement with the analytical model for both in-plane and out-of-plane bending motions for a relaxed and torsionally strained minicircle. However, the nearly-degenerate vibrational frequencies are not as close in energy in the case of the overtwisted circle, especially for larger values of n. Only two twisting modes were identified; corresponding to n = 1 and n = 2 in the analytical model (figure 5 and table 3). The rigidities extracted for both the relaxed and overtwisted circles are shown in Table 4. The bending persistence lengths are 571Å, 610 Å, and 628 Å for the relaxed, overtwisted, and linear configurations of TA94, respectively. Hence, even forcing DNA into a small circle seems to have only a mild effect on its intrinsic bending propensities. By contrast, the twisting rigidity for our linear model is much larger than that of random DNA (6.74 10−19erg.cm), whereas it is essentially the same as random DNA for both circular models (4.74 10−19erg.cm and 4.18 10−19erg.cm for the relaxed and overtwisted configuration, respectively).
Table 3.
Lowest in plane bending, out of plane bending, and twisting frequencies in cm−1 for the 94-base-pair minicircle (TA94) with a salt concentration of 0.05M and T=300K.
| Relaxed minicircle | Overtwisted minicircle | |||||
|---|---|---|---|---|---|---|
| Modes | GB frequencies | Analytical frequencies | GB frequencies | Analytical frequencies | ||
| IPB2 | 0.165 | 0.172 | 0.121 | 0.166 | 0.178 | 0.116 |
| IPB3 | 0.394 | 0.452 | 0.366 | 0.443 | 0.471 | 0.377 |
| IPB4 | 0.672 | 0.724 | 0.697 | 0.704 | 0.778 | 0.734 |
| OP2 | 0.217 | 0.231 | 0.211 | 0.211 | 0.248 | 0.235 |
| OP3 | 0.525 | 0.545 | 0.496 | 0.520 | 0.542 | 0.551 |
| OP4 | 0.793 | 0.851 | 0.864 | 0.802 | 0.901 | 0.958 |
| T1 | 0.663 | 0.529 | 0.654 | 0.556 | ||
| T2 | 0.995 | 1.057 | 1.148 | 1.270 | ||
Table 4.
Mechanical constants extracted from the results presented in Table 3
| Relaxed minicircle | Overtwisted minicircle | Linear | |
|---|---|---|---|
| Persistence length (Å) | 571 | 610 | 628 |
| Twisting rigidity (10−19erg.cm) | 4.74 | 4.18 | 6.74 |
Based on the previous results, and considering the observed length dependence of the method, it appears that the sequence used in this section ranks within the most flexible sequences encountered (628 Å for TA94 vs. 732 Å for d(GACT), considered in this study to behave like random DNA). A comparison of the persistence lengths for several known sequences can be found in table 5. The conclusions of Cloutier and Widom48 on the persistence length and twisting rigidity is in accord with our observations with our linear DNA model; the persistence length is found to be shorter than the one of random DNA, whereas the twisting rigidity is higher. Note that this is also true in the case of our circular DNA model, this is an encouraging sign that the force field and methods used in this work reproduce the different aspects of macromolecular motions of nucleic acids accurately. The high flexibility of this sequence could partly explain its large J value, though, it is clear that other factors such as kinks or extreme bending are also involved in the formation of nucleosomes. Further studies on DNA binding to nucleosome core proteins in in progress.
Table 5.
Mechanical constants for several 94bp sequences with a salt concentration of 0.05M and T=300K.
| Sequences | Persistence Length (Å) | Stretching rigidity (pN) | twisting rigidity (10−19erg.cm) |
|---|---|---|---|
| dA.dT | 862 | 1899 | 3.63 |
| d(GACT) | 732 | 1489 | 4.78 |
| d(CGG) | 570 | 1397 | 3.07 |
| TA94 | 628 | 1385 | 6.85 |
4 Conclusions
Normal mode analysis in implicit solvent using the generalized Born model provides a powerful tool to describe collective molecular motions. The method developed in this paper allows a better understanding of how the flexibility of nucleic acids is affected by salt concentration and their lengths and sequences. The use of an all-atom force field allows us to both test the force field model itself (showing, for example, limitations in our current description of salt effects,) and to explore sequence dependences and to compare linear to circular structures. Our future plans include a systematic study of sequences that are found to bind (or not to bind) to nucleosomes, and which may provide new insights into DNA flexibility and its connections to transcription. Another extension under active investigation adds solvent frictional effects to the normal mode approximation, as so-called Langevin modes.50 Such a method could potentially give an even more accurate picture of these molecular motions and direct knowledge of several important properties including relaxation times, rotational and translational diffusion constants. The initial results presented here suggest that many interesting features of duplex DNA, at a length scale of hundred of Angstroms, can be modeled reasonably well using force fields originally developed by comparisons to results for very short olignonucleotides. This provides a new window on ways to understand the behavior of DNA for much longer sequences.
Figure 2.

Low-frequency stretching modes of a 150-bp linear segment of DNA. The arrows define the main motions associated with the corresponding vibration.
Acknowledgments
The authors would like to thank Filip Lankas for providing some of the pdb files (minicircles) and for helpful discussions. This work was supported by NIH grant RR12255 and was also supported in part by the National Science Foundation through TeraGrid resources provided by NCSA and TACC.
References
- 1.Cramer CJ, Truhlar DG. Implicit solvation models: Equilibria, structure, spectra, and dynamics. Chem Rev. 1999;99:2161–2200. doi: 10.1021/cr960149m. [DOI] [PubMed] [Google Scholar]
- 2.Feig M, Brooks CL. Recent advances in the development and application of implicit solvent models in biomolecule simulations. Curr Opin Struct Biol. 2004;14:217–224. doi: 10.1016/j.sbi.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 3.Bashford D, Case DA. Generalized born models of macromolecular solvation effects. Annu Rev Phys Chem. 2000;51:129–152. doi: 10.1146/annurev.physchem.51.1.129. [DOI] [PubMed] [Google Scholar]
- 4.Brown RA, Case DA. Second derivatives in generalized born theory. J Comput Chem. 2006;27:1662–1675. doi: 10.1002/jcc.20479. [DOI] [PubMed] [Google Scholar]
- 5.Tsui V, Case DA. Theory and applications of the generalized born solvation model in macro-molecular simulations. Biopolymers. 2000;56:275–291. doi: 10.1002/1097-0282(2000)56:4<275::AID-BIP10024>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 6.Jayaram B, Liu Y, Beveridge DL. A modification of the generalized born theory for improved estimates of solvation energies and pk shifts. J Chem Phys. 1998;109:1465–1471. [Google Scholar]
- 7.Onufriev A, Case DA, Bashford D. Effective born radii in the generalized born approximation: The importance of being perfect. J Comput Chem. 2002;23:1297–1304. doi: 10.1002/jcc.10126. [DOI] [PubMed] [Google Scholar]
- 8.Sigalov G, Scheffel P, Onufriev A. Incorporating variable dielectric environments into the generalized born model. J Chem Phys. 2005;122:094511. doi: 10.1063/1.1857811. [DOI] [PubMed] [Google Scholar]
- 9.Tsui V, Case DA. Molecular dynamics simulations of nucleic acids with a generalized born solvation model. J Am Chem Soc. 2000;122:2489–2498. [Google Scholar]
- 10.Case DA. Normal-mode analysis of protein dynamics. Curr Opin Struct Biol. 1994;4:285–290. [Google Scholar]
- 11.Macke TA, Case DA. In: Modeling unusual nucleic acid structures. in molecular modeling of nucleic acids. Leontes NB, SantaLucia J Jr, editors. Washington, DC: American Chemical Society; 1998. [Google Scholar]
- 12.Matsumoto A, Olson WK. Sequence-dependent motions of DNA: A normal mode analysis at the base-pair level. Biophys J. 2002;83:22–41. doi: 10.1016/S0006-3495(02)75147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsumoto A, Tobias I, Olson WK. Normal-mode analysis of circular DNA at the base-pair level. 1. Comparison of computed motions with the predicted behavior of an ideal elastic rod. J Chem Theory Comput. 2005;1:117–129. doi: 10.1021/ct049950r. [DOI] [PubMed] [Google Scholar]
- 14.Matsumoto A, Tobias I, Olson WK. Normal-mode analysis of circular DNA at the base-pair level. 2. Large-scale configurational transformation of a naturally curved molecule. J Chem Theory Comput. 2005;1:130–142. doi: 10.1021/ct049949s. [DOI] [PubMed] [Google Scholar]
- 15.Matsumoto A, Go N. Dynamic properties of double-stranded DNA by normal mode analysis. J Chem Phys. 1999;110:11070–11075. [Google Scholar]
- 16.Wang YH, Griffith J. Expanded CTG triplet blocks from the myotonic-dystrophy gene create the strongest known natural nucleosome positioning elements. Genomics. 1995;25:570–573. doi: 10.1016/0888-7543(95)80061-p. [DOI] [PubMed] [Google Scholar]
- 17.Godde JS, Wolffe AP. Nucleosome assembly on CTG triplet repeats. J Bio Chem. 1996;271:15222–15229. doi: 10.1074/jbc.271.25.15222. [DOI] [PubMed] [Google Scholar]
- 18.Bacolla A, Gellibolian R, Shimizu M, Amirhaeri S, Kang S, Ohshima K, Larson JE, Harvey SC, Stollar BD, Wells RD. Flexible DNA: Genetically unstable CTG.CAG and CGG.CCG from human hereditary neuromuscular disease genes. J Bio Chem. 1997;272:16783–16792. doi: 10.1074/jbc.272.27.16783. [DOI] [PubMed] [Google Scholar]
- 19.Bouchiat C, Mezard M. Elasticity model of a supercoiled DNA molecule. Phys Rev Lett. 1998;80:1556–1559. [Google Scholar]
- 20.Benham CJ. Elastic Model of Supercoiling. PNAS. 1977;74:2397–2401. doi: 10.1073/pnas.74.6.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuurer PB, Manning RS, Maddocks JH. DNA rings with multiple energy minima. Biophys J. 2000;79:116–136. doi: 10.1016/S0006-3495(00)76277-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A 2nd generation force-field for the simulation of proteins, nucleic-acids, and organic-molecules. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- 23.Hawkins CJ, Cramer GD, Truhlar DG. Pairwise solute descreening of solute charges from a dielectric medium. Chem Phys Lett. 1995;246:122–129. [Google Scholar]
- 24.Hawkins CJ, Cramer GD, Truhlar DG. Parametrized models of aqueous free energies of solvation based on pairwise descreening of solute atomic charges from a dielectric medium. J Phys Chem. 1996;100:19824–19839. [Google Scholar]
- 25.Case DA, Cheatham TE, Darden T, Gohlke H, Luo R, Merz KM, Onufriev A, Simmerling C, Wang B, Woods RJ. The amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lehoucq RB, Sorensen DC, Yang C. SIAM; Philadelphia, PA: 1999. [Google Scholar]
- 27.Strutt (Lord Rayleigh) JW. The theory of sound. 2. Maxmillan; London: 1894. [Google Scholar]
- 28.Mongan J. Interactive essential dynamics. J Comp-Aided Mol. 2004;18:433–436. doi: 10.1007/s10822-004-4121-z. [DOI] [PubMed] [Google Scholar]
- 29.Coleman BD, Lembo M, Tobias I. A new class of flexure-free torsional vibrations of annular rods. Meccanica. 1996;31:565–575. [Google Scholar]
- 30.Tobias I. A theory of thermal fluctuations in DNA miniplasmids. Biophys J. 1998;74:2545–2553. doi: 10.1016/S0006-3495(98)77962-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beveridge DL, Barreiro G, Byun KS, Case DA, Cheatham TE, Dixit SB, Giudice E, Lankas F, Lavery R, Maddocks JH. Molecular Dynamics Simulations of the 136 Unique Tetranucleotide Sequences of DNA Oligonucleotides. I. Research Design and Results on d(CpG) Steps. Biophys J. 2004;87:3799–3813. doi: 10.1529/biophysj.104.045252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dixit SB, Beveridge DL, Case DA, Cheatham TE, III, Giudice E, Lankas F, Lavery R, Maddocks JH, Osman R, Sklenar H, Thayer KM, Varmai P. Molecular dynamics simulations of the 136 unique tetranucleotide sequences of DNA oligonucleotides. II. Sequence context effects on the dynamical structures of the 10 unique dinucleotide steps. Biophys J. 2005;89:3721–3740. doi: 10.1529/biophysj.105.067397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baumann CG, Smith SB, Bloomfield VA, Bustamante C. Ionic effects on the elasticity of single DNA molecules. Proc Natl Acad Sci. 1997;94:6185–6190. doi: 10.1073/pnas.94.12.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wenner JR, Williams MC, Rouzina I, Bloomfield VA. Salt dependence of the elasticity and overstretching transition of single DNA molecules. Biophys J. 2002;82:3160–3169. doi: 10.1016/S0006-3495(02)75658-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tjong H, Zhou HX. GBr6: A Parameterization-Free, Accurate, Analytical Generalized Born Method. J Phys Chem B. 2007;111:3055–3061. doi: 10.1021/jp066284c. [DOI] [PubMed] [Google Scholar]
- 36.Tjong H, Zhou HX. GBr6NL: A generalized Born method for accurately reproducing solvation energy of the nonlinear Poisson-Boltzmann equation. J Chem Phys. 2007;126:195102. doi: 10.1063/1.2735322. [DOI] [PubMed] [Google Scholar]
- 37.Bryant Z, Stone MD, Gore J, Smith SB, Cozzarelli NR, Bustamante C. Structural transitions and elasticity from torque measurements on DNA. Nature. 2003;424:338–341. doi: 10.1038/nature01810. [DOI] [PubMed] [Google Scholar]
- 38.Moroz JD, Nelson P. Entropic elasticity of twist-storing polymers. Macromolecules. 1998;31:6333–6347. [Google Scholar]
- 39.Furey TS, Haussler D. Integration of the cytogenetic map with the draft human genome sequence. Hum Mol Genet. 2003;12:1037–1044. doi: 10.1093/hmg/ddg113. [DOI] [PubMed] [Google Scholar]
- 40.Derse D, Crise B, Li Y, Princler G, Lum N, Stewart C, Mcgrath CF, Hughes SH, Munroe DJ, Wu XL. Human t-cell leukemia virus type 1 integration target sites in the human genome: Comparison with those of other retroviruses. J Virol. 2007;81:6731–6741. doi: 10.1128/JVI.02752-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kundu S, Bhattacharya D, Thakur AR, Majumdar R. Nucleosomal positioning and genetic divergence study based on DNA flexibility map. J Biomol Struct Dyn. 2001;18:527–533. doi: 10.1080/07391102.2001.10506685. [DOI] [PubMed] [Google Scholar]
- 42.Jithesh PV, Singh P, Joshi RR. Molecular dynamics studies of trinucleotide repeat DNA involved in neurodegenerative disorders. J Biomol Struct Dyn. 2001;19:479–495. doi: 10.1080/07391102.2001.10506756. [DOI] [PubMed] [Google Scholar]
- 43.Baldi P, Brunak S, Chauvin Y, Pedersen AG. Structural basis for triplet repeat disorders: a computational analysis. Bioinformatics. 1999;15:918–929. doi: 10.1093/bioinformatics/15.11.918. [DOI] [PubMed] [Google Scholar]
- 44.Zhu ZW, Thiele DJ. A specialized nucleosome modulates transcription factor access to a c-glabrata metal responsive promoter. Cell. 1996;87:459–470. doi: 10.1016/s0092-8674(00)81366-5. [DOI] [PubMed] [Google Scholar]
- 45.Nelson HCM, Finch JT, Luisi BF, Klug A. The structure of an oligo(dA).oligo(dT) tract and its biological implications. Nature. 1987;330:221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- 46.Lankas F, Sponer J, Hobza P, Langowski J. Sequence-dependent elastic properties of DNA. J Mol Bio. 2000;299:695–709. doi: 10.1006/jmbi.2000.3781. [DOI] [PubMed] [Google Scholar]
- 47.Rief M, Clausen-Schaumann H, Gaub HE. Sequence-dependent mechanics of single DNA molecules. Nat Struct Biol. 1999;6:346–349. doi: 10.1038/7582. [DOI] [PubMed] [Google Scholar]
- 48.Cloutier TE, Widom J. Spontaneous sharp bending of double-stranded DNA. Mol Cell. 2004;14:355–362. doi: 10.1016/s1097-2765(04)00210-2. [DOI] [PubMed] [Google Scholar]
- 49.Lankas F, Lavery R, Maddocks JH. Kinking occurs during molecular dynamics simulations of small DNA minicircles. Structure. 2006;14:1527–1534. doi: 10.1016/j.str.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 50.Lamm G, Szabo A. Langevin modes of macromolecules. J Chem Phys. 1986;85:7334–7348. [Google Scholar]