Abstract
Plasma liver-enzyme tests are widely used in the clinic for the diagnosis of liver diseases and for monitoring the response to drug treatment. There is considerable evidence that human genetic variation influences plasma levels of liver enzymes. However, such genetic variation has not been systematically assessed. In the present study, we performed a genome-wide association study of plasma liver-enzyme levels in three populations (total n = 7715) with replication in three additional cohorts (total n = 4704). We identified two loci influencing plasma levels of alanine-aminotransferase (ALT) (CPN1-ERLIN1-CHUK on chromosome 10 and PNPLA3-SAMM50 on chromosome 22), one locus influencing gamma-glutamyl transferase (GGT) levels (HNF1A on chromosome 12), and three loci for alkaline phosphatase (ALP) levels (ALPL on chromosome 1, GPLD1 on chromosome 6, and JMJD1C-REEP3 on chromosome 10). In addition, we confirmed the associations between the GGT1 locus and GGT levels and between the ABO locus and ALP levels. None of the ALP-associated SNPs were associated with other liver tests, suggesting intestine and/or bone specificity. The mechanisms underlying the associations may involve cis- or trans-transcriptional effects (some of the identified variants were associated with mRNA transcription in human liver or lymphoblastoid cells), dysfunction of the encoded proteins (caused by missense variations at the functional domains), or other unknown pathways. These findings may help in the interpretation of liver-enzyme tests and provide candidate genes for liver diseases of viral, metabolic, autoimmune, or toxic origin. The specific associations with ALP levels may point to genes for bone or intestinal diseases.
Main Text
Plasma liver-enzyme tests are widely used in the clinic to identify patients with liver diseases, to monitor the course and severity of these diseases and the effect of therapies, and to detect drug-induced liver injury.1,2 These tests also have substantial epidemiologic significance that extends beyond the liver, given that they have been shown to be prospective risk factors for type 2 diabetes, cardiovascular disease, and all-cause mortality in multiple large studies.3–6 Therefore, it is of interest to identify genes or loci affecting these markers in order to establish whether such loci are also associated with these clinical endpoints.
Plasma liver-enzyme levels are influenced by environmental and genetic factors. The estimated heritabilities range from 33% for alanine-aminotransferase (ALT) to 61% for gamma-glutamyl transferase (GGT).7,8 So far, only a limited number of genes that influence liver-enzyme levels have been identified, mostly those responsible for Mendelian liver diseases such as mutations in the HFE gene in hemochromatosis (MIM 235200).9,10 A thorough understanding of the genetic determinants of plasma liver enzymes is important for proper interpretation of these tests. Indeed, such information could assist in our understanding of the interindividual differences in the propensity for development of liver dysfunction in the presence of toxins or conditions such as metabolic syndrome. As such, identification of genes associated with liver-enzyme levels could reveal previously unsuspected candidate genes for liver diseases of viral, metabolic, autoimmune, or toxic origin.
The goal of the present study was to identify, by using a genome-wide association (GWA) approach, genes influencing plasma levels of ALT and aspartate-aminotransferase (AST), two markers of hepatocyte injury and liver fat accumulation, and of alkaline phosphatase (ALP) and GGT, used primarily as indicators of biliary or cholestatic diseases and heavy alcohol consumption.
In the discovery phase, we carried out independent GWA studies in three population-based cohorts, the CoLaus Study from Lausanne Switzerland11,12 (n = 5636), the InCHIANTI Study from Tuscany Italy13 (n = 1200), and a subset of the LOLIPOP Study from West London UK14,15 (n = 879) (Table 1). The clinical characteristics of the participants are described in Table 2. The mean levels of liver-enzyme tests varied somewhat between populations, presumably because of slight differences in the demographics of the populations under study and methodological differences in the assays (Table S1 available online). Accordingly, we used study-specific criteria for GWA-genotyping quality control11,14 and liver-enzyme-level analyses (Appendix A). Additional genotypes were imputed on the basis of the HapMap Phase II data with the software IMPUTE.16 For these imputed data, we performed association analysis with SNPTEST, using the full posterior probability genotype distribution for each study separately (adding in relevant covariables). Only SNPs with a posterior probability score >0.90, high genotype information content (proper_info >0.5), and minor allele frequency >0.01 were considered for these imputed association analyses.
Table 1.
Description of Six Collections Used in GWAS on Plasma Liver-Enzyme Levels
| CoLaus | InCHIANTI | LOLIPOP | ||||
|---|---|---|---|---|---|---|
| Sample size (n) | 5636 | 1200 | 879 | 1006 | 1005 | 2693 |
| Location | Lausanne, Switzerland | Tuscany, Italy | London, UK | London, UK | London, UK | London, UK |
| Ethnicity | European white | European white | European white | Indian Asian | European white | Indian Asian |
| Study Design | Population-based | Population-based | Population-based enriched with coronary artery disease | Nested metabolic syndrome case and control | Nested metabolic syndrome case and control | Nested metabolic syndrome case and control |
| Data Usage | GWA Discovery | GWA Discovery | GWA Discovery | Replication | Replication | Replication |
| Liver enzymes | AST,a ALT,b GGT,c and ALPd | AST, ALT, GGT, ALP | ALT, GGT, ALP | ALT, GGT, ALP | ALT, GGT, ALP | ALT, GGT, ALP |
| Genotyping Platform | Affymetrix 500k | Illumina HumanHap550 | Affymetrix 500k | Perlegen, custom array | Perlegen, custom array | Illumina HumanHap300 |
| Number of SNPs genotyped | 370,697 | 496,032 | 387,549 | 248,537 | 266,722 | 317,968 |
AST, aspartate aminotransferase.
ALT, alanine aminotransferase.
GGT, gamma-glutamyltransferase.
ALP, alkaline phosphatase.
Table 2.
Clinical Characteristics of the Participants in the Discovery Data Sets from CoLaus, InCHIANTI, and LOLIPOP Population-Based Studies
| CoLausa | InCHIANTIa | LOLIPOPa | |
|---|---|---|---|
| n | 5636 | 1200 | 879b |
| Sex (% F) | 52.8 | 55.2 | 23.0 |
| Age (years) | 53.2 ± 10.8 | 68.4 ± 15.9 | 52.8 ± 10.4 |
| Fasting glucose (mmol/L) | 5.56 ± 1.16 | 5.24 ± 1.36 | 5.50 ± 1.54 |
| Insulin (μIU/mL) | 8.76 ± 6.25 | 10.9 ± 6.15 | 10.3 ± 10.7 |
| BMI (kg/m2) | 25.8 ± 4.6 | 27.1 ± 4.1 | 27.9 ± 4.8 |
| Waist (cm) | 89.3 ± 13.4 | NA | 96.7 ± 13.0 |
| Hip (cm) | 101.0 ± 9.3 | NA | 103.0 ± 9.0 |
| Total cholesterol (mmol/L) | 5.60 ± 1.04 | NA | 5.32 ± 1.07 |
| LDL-cholesterol (mmol/L) | 3.34 ± 0.92 | 3.43 ± 0.90 | 3.25 ± 0.93 |
| Triglycerides (mmol/L) | 1.40 ± 1.20 | 1.42 ± 0.88 | 1.61 ± 1.16 |
| HDL-cholesterol (mmol/L) | 1.64 ± 0.44 | 1.43 ± 0.39 | 1.36 ± 0.35 |
| Albumin (g/L) | 44.2 ± 2.5 | 42.3 ± 3.2 | 43.6 ± 2.9 |
| Smoking status (% smokers) | 11.8 | 18.8 | 64.4 |
| Alcohol consumption (%)c | 77.3 | NA | 75.7 |
| Type 2 Diabetes (%) | 6.6 | 11.1 | 11.3 |
| AST (U/L) | 29.7 ± 13.5 | 20.2 ± 0.4 | NA |
| ALT (U/L) | 27.6 ± 19.5 | 18.1 ± 0.5 | 29.1 ± 16.4 |
| ALP (U/L) | 63.4 ± 20.4 | 204.2 ± 71.5 | 80.9 ± 38.5 |
| GGT (U/L) | 32.1 ± 39.9 | 26.2 ± 28.7 | 40.2 ± 44.8 |
Results are expressed as mean ± SD.
Subset from the LOLIPOP population-based study enriched with CAD cases.
Alcohol consumption was defined as alcohol intake ≥1 unit per week.
Quantile-quantile plots (Figure 1) revealed the presence of a substantial number of SNPs associated with ALT, GGT, and ALP levels at a genome-wide significance level (p value < 10−7). No SNP with plasma levels of AST was associated with genome-wide significance. It is not clear why AST was uninformative in this case. Highly significantly associated SNPs were located within discrete regions of the genome (Figure 2), including two loci for ALT (10q24.2 and 22q13.31), two loci for GGT (12q24.31 and 22q11.23), and five loci for ALP (1p36.12, 6p22.2, 9q34.13, 10q21.2-21.3, and 19q13.33).
Figure 1.
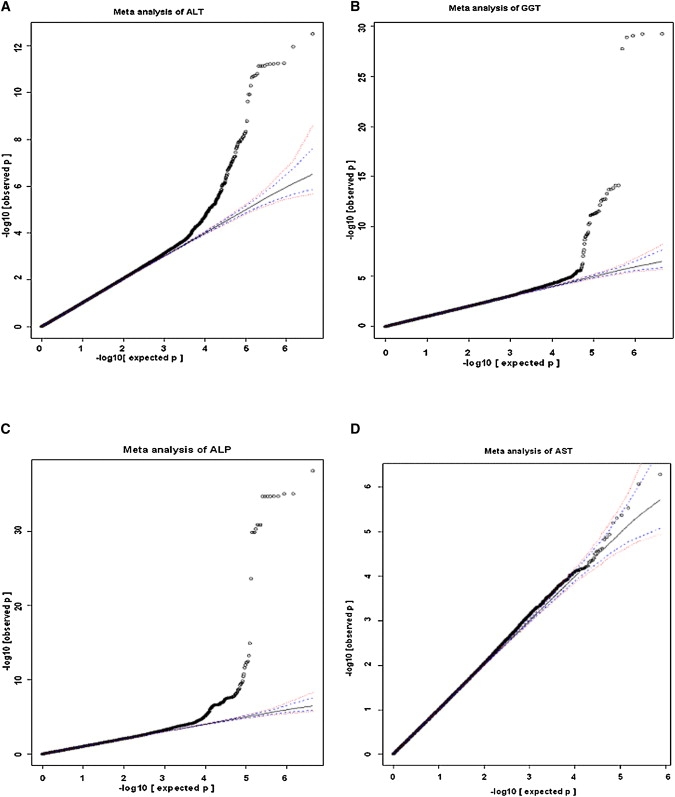
Quantile-Quantile Plot for GWA for ALT, GGT, ALP, and AST in a Meta-analysis from CoLaus, InCHIANTI, and LOLIPOP Discovery Study Data Sets with Imputed Autosomal SNPs
The blue and red dotted lines correspond to 95% CI and 99% CI, respectively. Each panel shows data for the enzymes as follows: (A), ALT; (B), GGT; (C), ALP; and (D), AST.
Figure 2.
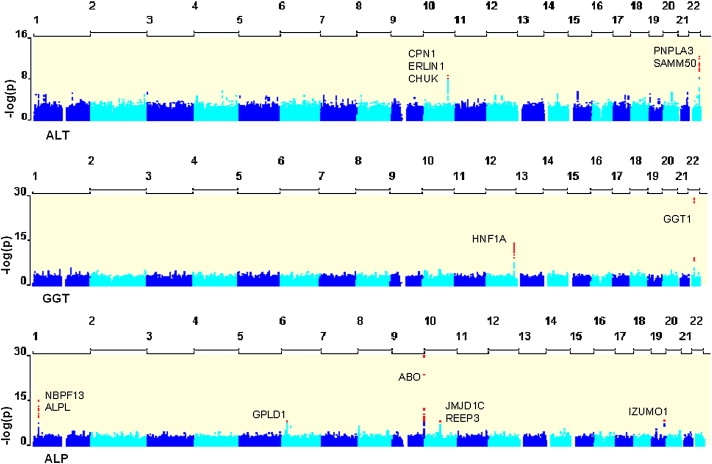
GWA Analysis of ALT, GGT, and ALP
Represented here are three Manhattan plots showing the degree of association (−log10 p value) between imputed SNPs included in the meta-analysis of three discovery collections totaling 7715 individuals. Red dots correspond to the SNPs with p value ≤ 10−8.
A total of 74 Affymetrix-genotyped SNPs (24 for ALT, 8 for GGT, and 42 for ALP, Table S2) with a p value ≤ 10−5 in the discovery phase was subsequently examined in 3699 Indian Asian and 1005 European white participants from the LOLIPOP study (Table 1). After joint analysis of discovery and replication data, 32 SNPs (10 for ALT, 6 for GGT, and 16 for ALP, Table S3 and Figures S1–S8) reached genome-wide significance with directionally consistent signals (Figures S9–S16 for the leading SNPs) among the discovery and replication cohorts. The remaining 42 SNPs were not considered as replicated because of either lack of high-quality imputation (Appendix A) of the presence of inconsistent direction of the effect across studies or because they did not reach genome-wide significance level (p ≤ 10−8). It is also conceivable that absence of replication was due to LD differences between the European white and Indian Asian cohorts. Out of the 32 replicated SNPs, 19 SNPs were considered to be independent (r2 < 0.4) (Table 3). These SNPs were located within two loci for ALT, two loci for GGT, and four loci for ALP.
Table 3.
Independent SNPs Associated with Liver-Enzyme Levels with Genome-wide Significance in Combined GWAS Analysis of Discovery and Replication Data Sets
| SNPa | CHR | Positionb | Nearest Gene | Region | Minor Allelec | MAFd |
Three Discovery Collections (n = 7,715) |
Three Replication Collections (n = 4,704) |
Six Collections (n = 12,419) |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pooled Beta-Coefficient (standard error) | Combined p Value | Pooled Beta-Coefficient (standard error) | Combined p Value | Pooled Beta-Coefficient (standard error) | Combined p Value | |||||||
| ALT | ||||||||||||
| rs11597390 | 10 | 101851425 | CPN1 | intergenic | A | 0.36 | −0.044 (0.008) | 2.9 × 10−8 | −0.023 (0.013) | 0.08 | −0.039 (0.007) | 1.5 × 10−8 |
| rs11597086 | 10 | 101943695 | CHUK | Intronic | C | 0.43 | −0.038 (0.007) | 3.6 × 10−7 | −0.030 (0.012) | 0.01 | −0.036 (0.006) | 1.8 × 10−8 |
| rs11591741 | 10 | 101966491 | CHUK | intronic | C | 0.43 | −0.038 (0.007) | 4.5 × 10−7 | −0.029 (0.012) | 0.02 | −0.035 (0.006) | 3.0 × 10−8 |
| rs2281135 | 22 | 42657471 | PNPLA3 | intronic | T | 0.18 | 0.065 (0.010) | 8.2 × 10−12 | 0.051 (0.012) | 1.3 × 10−5 | 0.060 (0.007) | 8.4 × 10−16 |
| rs2143571 | 22 | 42716587 | SAMM50 | intronic | A | 0.19 | 0.046 (0.009) | 9.4 × 10−7 | 0.038 (0.012) | 1.8 × 10−3 | 0.043 (0.007) | 7.2 × 10−9 |
| GGT | ||||||||||||
| rs1169313 | 12 | 119905390 | HNF1A | intronic | C | 0.38 | −0.007 (0.001) | 3.2 × 10−8 | −0.003 (0.001) | 7.0 × 10−5 | −0.005 (0.001) | 1.8 × 10−10 |
| rs4820599 | 22 | 23314767 | GGT1 | intronic | G | 0.31 | 0.006 (0.001) | 3.9 × 10−6 | 0.008 (0.002) | 1.8 × 10−6 | 0.007 (0.001) | 4.0 × 10−11 |
| ALP | ||||||||||||
| rs1780324 | 1 | 21567063 | NBPF3-ALPL | intergenic | T | 0.43 | 0.032 (0.005) | 1.4 × 10−10 | 0.031 (0.007) | 1.0 × 10−5 | 0.031 (0.004) | 7.0 × 10−15 |
| rs9461011 | 6 | 24548330 | GPLD1 | intronic | G | 0.23 | 0.033 (0.006) | 4.4 × 10−8 | 0.030 (0.009) | 0.001 | 0.032 (0.005) | 1.9 × 10−10 |
| rs9467160 | 6 | 24549725 | GPLD1 | intronic | A | 0.24 | 0.033 (0.006) | 2.2 × 10−8 | 0.035 (0.009) | 1.3 × 10−4 | 0.034 (0.005) | 1.2 × 10−11 |
| rs8176720 | 9 | 133162427 | ABO | synonymous | G | 0.32 | 0.031 (0.005) | 1.4 × 10−8 | 0.031 (0.008) | 7.0 × 10−5 | 0.031 (0.004) | 4.3 × 10−12 |
| rs641959 | 9 | 133163253 | ABO | intronic | G | 0.26 | 0.035 (0.006) | 6.8 × 10−10 | 0.024 (0.008) | 0.004 | 0.031 (0.005) | 2.1 × 10−11 |
| rs514708 | 9 | 133163297 | ABO | intronic | T | 0.26 | 0.035 (0.006) | 6.2 × 10−10 | 0.024 (0.008) | 0.004 | 0.031 (0.005) | 1.9 × 10−11 |
| rs672316 | 9 | 133167679 | ABO | intronic | C | 0.27 | 0.032 (0.006) | 1.4 × 10−8 | 0.023 (0.008) | 0.006 | 0.029 (0.005) | 4.4 × 10−10 |
| rs657152 | 9 | 133168819 | ABO | intronic | T | 0.39 | −0.057 (0.005) | 4.6 × 10−29 | −0.029 (0.007) | 4.9 × 10−5 | −0.047 (0.004) | 1.7 × 10−30 |
| rs474279 | 9 | 133169171 | ABO | intronic | T | 0.24 | 0.030 (0.006) | 3.4 × 10−7 | 0.023 (0.008) | 0.006 | 0.028 (0.005) | 8.3 × 10−9 |
| rs552148 | 9 | 133183035 | ABO | upstream | T | 0.24 | 0.032 (0.006) | 3.1 × 10−8 | 0.022 (0.008) | 0.008 | 0.029 (0.005) | 1.2 × 10−9 |
| rs12355784 | 10 | 64791571 | JMJD1C | intronic | A | 0.48 | 0.026 (0.005) | 4.7 × 10−7 | 0.025 (0.007) | 2.7 × 10−4 | 0.025 (0.004) | 5.0 × 10−10 |
| rs10761779 | 10 | 64944933 | REEP3 | intergenic | G | 0.49 | 0.025 (0.005) | 3.9 × 10−7 | 0.024 (0.007) | 4.5 × 10−4 | 0.025 (0.004) | 6.9 × 10−10 |
Independent SNPs (r2 < 0.4) were selected from each loci.
Position is of NCBI Build 35.
Minor allele corresponds to forward strand and is of NCBI Build 35.
MAF, Minor allele frequency. It is based on CoLaus study.
The ALT-associated 10q24.2 region (Figure S1) spans three genes. CHUK (IKK-α) is a ubiquitously expressed serine threonine protein kinase that modulates the NF-κB-transcription-factor-dependent activation of several genes implicated in insulin resistance. Its intronic SNP rs11591741 has been reported to be associated with expression of its upstream gene CWF19L1 (CWF19-like 1) in human liver (p = 1.47 × 10−39),17 probably through its transcriptional regulatory effect. CPN1 encodes arginine carboxypeptidase-1, a liver-expressed plasma metallo-protease that protects the body from potent vasoactive and inflammatory peptides containing C-terminal arginine or lysine (such as kinins or anaphylatoxins), which are released into the circulation.18,19 Defects in CPN1 are the cause of carboxypeptidase N deficiency (MIM 212070). ERLIN1 encodes a member of the prohibitin family of proteins that define lipid-raft-like domains of the endoplasmic reticulum.20 The second association peak for ALT on chromosome 22q13.31 (Figure S2) encompasses a 57 kb fragmented linkage disequilibrium (LD) region encompassing SAMM50 and PNPLA3. SAMM50 is a subunit of mitochondrial SAM translocase complex for importation of proteins such as metabolite-exchange anion-selective channel precursors,21,22 whose N-terminal domain is essential for the biogenesis of mitochondria. The variation of imputed SNP rs3761472 (p = 2.7 × 10−9) causes N-terminal Asp110Glu substitution in SAMM50, which may lead to mitochondrial dysfunction and impaired cell growth. PNPLA3 (ADPN) is a liver-expressed transmembrane protein with phospholipase activity.23 It has been shown to be significantly upregulated during adipocyte differentiation, and in response to fasting and feeding, indicating its role in facilitating both energy mobilization and lipid storage in adipose and liver.24 PNPLA3 mRNA expression was elevated in subcutaneous and visceral adipose tissue of obese subjects.25 The lead SNP rs2281135 is in complete LD (r2 = 1) with intronic SNPs rs1010022 and rs2072907, two obesity-associated tagSNPs that showed significant differences in adipose PNPLA3 mRNA expression and adipocyte lipolysis from experimental data.25 Two imputed nonsynonymous SNPs within PNPLA3 (rs738409 [Ile148Met], p = 3.7 × 10−10, and rs2294918 [Lys434Glu], p = 6.0 × 10−4), represent putative exonic splicing silencer elements and might play a role in gene regulation.26 Homozygous carriers of the GG genotype for rs2281135 had a 34% greater risk (OR 1.34 [1.13–1.60]) of having ALT levels above upper limits of normal (defined as 36 U/L in females, and 60 U/L in males) in the CoLaus study. The effect of the lead SNPs at both CPN1 and PNPLA3 loci was not specific for ALT because these SNPs were also associated with plasma levels of AST (Table 4), suggesting that these genes are generally predisposed to hepatocyte dysfunction.
Table 4.
Association of the Lead SNPs with Plasma Levels of Liver Enzymes in the CoLaus Study
| SNP | Nearest Gene |
ALT |
AST |
ALP |
GGT |
||||
|---|---|---|---|---|---|---|---|---|---|
| Beta (% 95 CI) | p Value | Beta (% 95 CI) | p Value | Beta (% 95 CI) | p Value | Beta (% 95 CI) | p Value | ||
| rs11597390 | CPN1 | −0.043 (−0.062−0.023) | 1.4 × 10−5 | −0.022 (−0.035−0.009) | 0.0009 | −0.011 (−0.023−0.001) | 0.08 | −0.004 (−0.008−0.0001) | 0.04 |
| rs2281135 | PNPLA3 | 0.064 (0.041–0.087) | 2.6 × 10−8 | 0.035 (0.020–0.050) | 5.7 × 10−6 | −0.005 (−0.019−0.010) | 0.51 | −0.001 (−0.006−0.003) | 0.61 |
| rs1169313 | HNF1A | 0.002 (−0.016–0.019) | 0.86 | 0.001 (−0.011–0.012) | 0.92 | 0.013 (0.002–0.024) | 0.03 | −0.006 (−0.009−0.002) | 0.0009 |
| rs4820599 | GGT1 | −0.002 (−0.021−0.017) | 0.82 | 0.003 (−0.009–0.016) | 0.6 | 0.006 (−0.006–0.018) | 0.36 | 0.003 (−0.001–0.007) | 0.09 |
| rs1780324 | NBPF3-ALPL | 0.01 (−0.007–0.027) | 0.26 | 0.015 (0.003–0.026) | 0.01 | 0.031 (0.020–0.042) | 5.6 × 10−8 | 0.003 (−0.001–0.006) | 0.14 |
| rs9467160 | GPLD1 | 0.003 (−0.018–0.024) | 0.76 | −0.003 (−0.017−0.011) | 0.66 | 0.031 (0.018–0.044) | 5.6 × 10−6 | 0.001 (−0.003–0.005) | 0.65 |
| rs657152 | ABO | 0.012 (−0.007–0.030) | 0.21 | −0.001 (−0.013−0.011) | 0.88 | −0.061 (−0.072−0.050) | 2.5 × 10−25 | 0.001 (−0.002–0.005) | 0.57 |
| rs12355784 | JMJD1C | −0.003 (−0.021−0.015) | 0.72 | 0.001 (−0.011–0.013) | 0.83 | 0.026 (0.014–0.037) | 1.6 × 10−5 | 0.003 (−0.001–0.007) | 0.1 |
When examining the association with GGT levels, we identified a series of SNPs within 300 kb on either side of the gene encoding GGT1 on chromosomal region 22q11.23 (Figure S3). As such, these SNPs met the definition of cis-acting SNPs.27 The typed rs4820599 (p = 4.0 × 10−11) is in high LD with previously reported variant rs5751901 (r2 = 0.71) associated with serum GGT13 and with rs6519519 (r2 = 0.96), a SNP associated with transcript abundance in lymphoblastoid cells;28 such a finding suggests that the genetic association observed here is due to differential expression of the gene. Another association peak was observed on chromosomal region 12q24.31 (Figure S4) encompassing HNF1A (TCF-1), the hepatic nuclear factor predominantly expressed in the human liver. This transcriptional regulatory protein plays a prominent role in the activation of a large family of hepatocyte-specific genes involved in hepatocyte differentiation and liver development.29,30 Mutations within this gene are associated with type III form of maturity-onset diabetes of the young (MODY3 [MIM 600496])31 and hepatic adenomas, which are frequently accompanied by an elevation in plasma GGT level (MIM 142330).32 SNP rs7953249 (p = 5.1 × 10−8) was also closely associated in the CoLaus study with plasma C-reactive protein levels (p = 4.4 × 10−10), consistent with recently reported observations.33 It is conceivable that variants within this gene, including imputed rs2464196 (Ser487Asn, p = 3.2 × 10−12) located within the C-terminal transactivation domain of HNF1A, may broadly affect the transcriptional effect of this nuclear factor.
The strongest association with ALP levels was observed within the ABO locus on chromosome 9q34.13 (Figure S5). Association between ALP levels and the ABO blood group has been reported previously.34 This association was specific to plasma ALP (Table 4) with SNP rs657152 (p = 1.7 × 10−30) accounting for 2% of the total variance of ALP levels in the CoLaus study (Table S4). The mechanism underlying this association remains unclear and may be due to genetically determined variations in the proportion of isoenzymes among different blood types because it has been shown that the appearance of intestinal ALP in the plasma, particularly after fatty meals, is associated with ABO blood group and secretor status.35 In addition, we detected a strong association with the ALPL locus (Figure S6), which encodes nonspecific alkaline phosphatase found in liver, bone, kidney, and other tissues and is responsible for hypophosphatasia.36 Association of orthologous ALPL gene with serum ALP levels has been reported in mice37 but not in humans. One cis-acting SNP rs1780324 within ALPL had been shown to markedly affect the gene expression in lymphoblastoid cells.28 One trans-acting signal was identified within GPLD1 (glycosylphosphatidylinositol specific phospholipase D1) gene (Figure S7). GPLD1 hydrolyzes the inositol phosphate linkage in proteins anchored by phosphatidylinositol glycans (GPI-anchor), thus releasing these proteins from the membrane. Elevated serum levels of GPLD1 and hepatic mRNA expression have been reported in nonalcoholic fatty liver disease (NAFLD).38 Finally, one additional trans-acting SNP was localized in the JMJD1C (TRIP8)-REEP3 (receptor accessory protein 3) region. JMJD1C encodes thyroid-hormone-receptor interactor 8, a hormone-dependent transcription factor that regulates expression of a variety of specific target genes39 (Figure S8). The observation that none of the ALP-associated genes were crossassociated with other liver enzymes (Table 4) may indicate that the association signal was generated by variations in bone- or intestine- rather than liver-metabolic pathways.40
The findings reported here were generated on a limited number of genome-wide scans, and it is anticipated that adding more scans with enriched liver phenotypes within the discovery and replication phase will further expand the number of genes and loci associated with plasma liver-enzyme levels in the population.
At this stage, the discovery of five trans-acting loci and one cis-acting locus influencing plasma levels of liver enzymes may point to mechanisms regulating these enzymes and could assist in the interpretation of liver tests in the clinic. Most importantly, these genes represent candidates for susceptibility to liver diseases. Analysis between variants within these genes and NAFLD, alcohol-, viral, autoimmune, or toxin-induced liver injury is warranted.
Acknowledgments
The CoLaus study was sponsored in part by GlaxoSmithKline, a pharmaceutical company. X.Y., D.W., N.L., K.S., H.S., L.C., and V.M. are full-time employees of GlaxoSmithKline. The InCHIANTI study was supported in part by the Intramural Research Program of the National Institute on Aging, National Institutes of Health, and Department of Health and Human Services, project number 1 Z01 AG000965-02. LOLIPOP was supported by the British Heart Foundation Grant SP/04/002. We thank the individuals who volunteered to participate in the CoLaus Study, as well as the staff at Lausanne University Hospital and within GlaxoSmithKline who made this collection possible. The computations for CoLaus imputation were performed in part at the Vital-IT center for high performance computing of the Swiss Institute of Bioinformatics. We thank Jacques Beckmann for his comments.
Appendix A
Material and Methods
Study Populations. The CoLaus study is a population-based sample consisting of 5694 Lausanne residents aged 35 to 75 years. The study design and protocol have been described previously.11,12,42
The London Life Sciences Population (LOLIPOP) study is an ongoing population-based cohort study of ∼30,000 Indian Asian and European white men and women, aged 35–75 years, recruited from the lists of 58 general practitioners in West London, United Kingdom.14,15 The participants included in the present study are a subset of the LOLIPOP cohort study. These subsets were assembled for specific experiments for identification of genetic variants underlying metabolic syndrome and coronary artery disease (CAD). The samples were genotyped on separate platforms (Table 1).
The InCHIANTI study is a population-based sample that includes 1200 individuals of <65 age and 1155 individuals of age ≥65 years. Details of the study design and protocol have been described previously.13 A total of 1200 subjects who had both WGS data and liver-enzyme levels measured were included in this study.
Genome-wide Association Statistics. Linear-regression analyses were done on natural log-transformed ALT, ALP, and AST and power-transformed GGT independently in each study. Regression analyses were done with the PLINK software package43 adjusted by age, gender, and geographical principle components analyzed by EIGENSOFT software,44 plus smoking and alcohol intake if they were significant covariates for the trait.
Imputation. For CoLaus, InCHIANTI, and LOLIPOP discovery set genotypes were imputed separately on the basis of the HapMap Phase II build35 data with the software IMPUTE.16 For these imputed data, we performed association analysis with SNPTEST, using the full posterior-probability genotype distribution for each study separately (adding in relevant covariables). Only SNPs with a posterior probability score >0.90, high genotype information content (proper_info >0.5), and MAF >0.01 were considered for the imputed association analyses. SNP imputation for LOLIPOP substudies used the MACH program (v1.0). For European data sets, CEPH haplotypes in HapMap were used as reference haplotypes. Indian Asian data sets were imputed on the basis of a combination (mixed) of HapMap populations, given that this showed greater concordance with real genotypes, compared with use of any one HapMap population. Imputation analyses were based on HapMap build35, dbSNP build 125. Only SNPs with RSQR ≥0.3 were considered for the meta-analysis.
Meta-analysis. For the initial GWA screen, analyses were performed within study, with study-specific criteria for GWA-genotyping QC and analyses (as described above). We meta-analyzed these summary data by using a fixed-effects model and inverse-variance weighted averages of beta-coefficients to obtain a combined estimate of the overall beta-coefficient and its standard error. Among-study heterogeneity was assessed with the χ2 test.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
Affymetrix genotyping protocol, http://www.affymetrix.com/support/technical/whitepapers/brlmmwhitepaper.pdf
EIGENSOFT software, http://genepath.med.harvard.edu/∼reich/EIGENSTRAT.htm
IMPUTE, http://www.stats.ox.ac.uk/∼marchini/software/gwas/gwas.html
International Hapmap Project, http://www.hapmap.org
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim
PLINK software package, http://pngu.mgh.harvard.edu/purcell/plink/
SNPTEST, http://www.stats.ox.ac.uk/∼marchini/software/gwas/gwas.html
WGAviewer,41 http://www.genome.duke.edu/centers/pg2/downloads/wgaviewer.php
References
- 1.Lazo M., Selvin E., Clark J.M. Brief communication: Clinical implications of short-term variability in liver function test results. Ann. Intern. Med. 2008;148:348–352. doi: 10.7326/0003-4819-148-5-200803040-00005. [DOI] [PubMed] [Google Scholar]
- 2.Wilke R.A., Lin D.W., Roden D.M., Watkins P.B., Flockhart D., Zineh I., Giacomini K.M., Krauss R.M. Identifying genetic risk factors for serious adverse drug reactions: Current progress and challenges. Nat. Rev. Drug Discov. 2007;6:904–916. doi: 10.1038/nrd2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forlani G., Di Bonito P., Mannucci E., Capaldo B., Genovese S., Orrasch M., Scaldaferri L., Di Bartolo P., Melandri P., Dei, Cas A. Prevalence of elevated liver enzymes in Type 2 diabetes mellitus and its association with the metabolic syndrome. J. Endocrinol. Invest. 2008;31:146–152. doi: 10.1007/BF03345581. [DOI] [PubMed] [Google Scholar]
- 4.Lee D.H., Silventoinen K., Hu G., Jacobs D.R., Jr., Jousilahti P., Sundvall J., Tuomilehto J. Serum gamma-glutamyltransferase predicts non-fatal myocardial infarction and fatal coronary heart disease among 28,838 middle-aged men and women. Eur. Heart J. 2006;27:2170–2176. doi: 10.1093/eurheartj/ehl086. [DOI] [PubMed] [Google Scholar]
- 5.Monami M., Bardini G., Lamanna C., Pala L., Cresci B., Francesconi P., Buiatti E., Rotella C.M., Mannucci E. Liver enzymes and risk of diabetes and cardiovascular disease: Results of the Firenze Bagno a Ripoli (FIBAR) study. Metabolism. 2008;57:387–392. doi: 10.1016/j.metabol.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 6.Kazemi-Shirazi L., Endler G., Winkler S., Schickbauer T., Wagner O., Marsik C. Gamma glutamyltransferase and long-term survival: Is it just the liver? Clin. Chem. 2007;53:940–946. doi: 10.1373/clinchem.2006.081620. [DOI] [PubMed] [Google Scholar]
- 7.Bathum L., Petersen H.C., Rosholm J.U., Hyltoft Petersen P., Vaupel J., Christensen K. Evidence for a substantial genetic influence on biochemical liver function tests: Results from a population-based Danish twin study. Clin. Chem. 2001;47:81–87. [PubMed] [Google Scholar]
- 8.Whitfield J.B., Zhu G., Nestler J.E., Heath A.C., Martin N.G. Genetic covariation between serum gamma-glutamyltransferase activity and cardiovascular risk factors. Clin. Chem. 2002;48:1426–1431. [PubMed] [Google Scholar]
- 9.Adams P.C., Barton J.C. Haemochromatosis. Lancet. 2007;370:1855–1860. doi: 10.1016/S0140-6736(07)61782-6. [DOI] [PubMed] [Google Scholar]
- 10.Stepec S., Makuc J., Markovic S., Medica I., Peterlin B. Distribution of HFE gene mutations in Slovenian patients with hereditary hemochromatosis. Ann. Hematol. 2008;87:667–669. doi: 10.1007/s00277-008-0463-2. [DOI] [PubMed] [Google Scholar]
- 11.Loos R.J., Lindgren C.M., Li S., Wheeler E., Zhao J.H., Prokopenko I., Inouye M., Freathy R.M., Attwood A.P., Beckmann J.S. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat. Genet. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sandhu M.S., Waterworth D.M., Debenham S.L., Wheeler E., Papadakis K., Zhao J.H., Song K., Yuan X., Johnson T., Ashford S. LDL-cholesterol concentrations: A genome-wide association study. Lancet. 2008;371:483–491. doi: 10.1016/S0140-6736(08)60208-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Melzer D., Perry J.R., Hernandez D., Corsi A.M., Stevens K., Rafferty I., Lauretani F., Murray A., Gibbs J.R., Paolisso G. A genome-wide association study identifies protein quantitative trait loci (pQTLs) PLoS Genet. 2008;4:e1000072. doi: 10.1371/journal.pgen.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chambers J.C., Elliott P., Zabaneh D., Zhang W., Li Y., Froguel P., Balding D., Scott J., Kooner J.S. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat. Genet. 2008;40:716–718. doi: 10.1038/ng.156. [DOI] [PubMed] [Google Scholar]
- 15.Kooner J.S., Chambers J.C., Aguilar-Salinas C.A., Hinds D.A., Hyde C.L., Warnes G.R., Gomez Perez F.J., Frazer K.A., Elliott P., Scott J. Genome-wide scan identifies variation in MLXIPL associated with plasma triglycerides. Nat. Genet. 2008;40:149–151. doi: 10.1038/ng.2007.61. [DOI] [PubMed] [Google Scholar]
- 16.Marchini J., Howie B., Myers S., McVean G., Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 17.Schadt E.E., Molony C., Chudin E., Hao K., Yang X., Lum P.Y., Kasarskis A., Zhang B., Wang S., Suver C. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skidgel R.A., Bennett C.D., Schilling J.W., Tan F.L., Weerasinghe D.K., Erdos E.G. Amino acid sequence of the N-terminus and selected tryptic peptides of the active subunit of human plasma carboxypeptidase N: Comparison with other carboxypeptidases. Biochem. Biophys. Res. Commun. 1988;154:1323–1329. doi: 10.1016/0006-291x(88)90284-7. [DOI] [PubMed] [Google Scholar]
- 19.Matthews K.W., Mueller-Ortiz S.L., Wetsel R.A. Carboxypeptidase N: A pleiotropic regulator of inflammation. Mol. Immunol. 2004;40:785–793. doi: 10.1016/j.molimm.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 20.Browman D.T., Resek M.E., Zajchowski L.D., Robbins S.M. Erlin-1 and erlin-2 are novel members of the prohibitin family of proteins that define lipid-raft-like domains of the ER. J. Cell Sci. 2006;119:3149–3160. doi: 10.1242/jcs.03060. [DOI] [PubMed] [Google Scholar]
- 21.Kutik S., Stojanovski D., Becker L., Becker T., Meinecke M., Kruger V., Prinz C., Meisinger C., Guiard B., Wagner R. Dissecting membrane insertion of mitochondrial beta-barrel proteins. Cell. 2008;132:1011–1024. doi: 10.1016/j.cell.2008.01.028. [DOI] [PubMed] [Google Scholar]
- 22.Kozjak V., Wiedemann N., Milenkovic D., Lohaus C., Meyer H.E., Guiard B., Meisinger C., Pfanner N. An essential role of Sam50 in the protein sorting and assembly machinery of the mitochondrial outer membrane. J. Biol. Chem. 2003;278:48520–48523. doi: 10.1074/jbc.C300442200. [DOI] [PubMed] [Google Scholar]
- 23.Wilson P.A., Gardner S.D., Lambie N.M., Commans S.A., Crowther D.J. Characterization of the human patatin-like phospholipase family. J. Lipid Res. 2006;47:1940–1949. doi: 10.1194/jlr.M600185-JLR200. [DOI] [PubMed] [Google Scholar]
- 24.Moldes M., Beauregard G., Faraj M., Peretti N., Ducluzeau P.H., Laville M., Rabasa-Lhoret R., Vidal H., Clement K. Adiponutrin gene is regulated by insulin and glucose in human adipose tissue. Eur. J. Endocrinol. 2006;155:461–468. doi: 10.1530/eje.1.02229. [DOI] [PubMed] [Google Scholar]
- 25.Johansson L.E., Hoffstedt J., Parikh H., Carlsson E., Wabitsch M., Bondeson A.G., Hedenbro J., Tornqvist H., Groop L., Ridderstrale M. Variation in the adiponutrin gene influences its expression and associates with obesity. Diabetes. 2006;55:826–833. doi: 10.2337/diabetes.55.03.06.db05-1075. [DOI] [PubMed] [Google Scholar]
- 26.Yuan H.Y., Chiou J.J., Tseng W.H., Liu C.H., Liu C.K., Lin Y.J., Wang H.H., Yao A., Chen Y.T., Hsu C.N. FASTSNP: An always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic Acids Res. 2006;34:W635–W641. doi: 10.1093/nar/gkl236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stranger B.E., Forrest M.S., Clark A.G., Minichiello M.J., Deutsch S., Lyle R., Hunt S., Kahl B., Antonarakis S.E., Tavare S. Genome-wide associations of gene expression variation in humans. PLoS Genet. 2005;1:e78. doi: 10.1371/journal.pgen.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dixon A.L., Liang L., Moffatt M.F., Chen W., Heath S., Wong K.C., Taylor J., Burnett E., Gut I., Farrall M. A genome-wide association study of global gene expression. Nat. Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 29.Cereghini S., Yaniv M., Cortese R. Hepatocyte dedifferentiation and extinction is accompanied by a block in the synthesis of mRNA coding for the transcription factor HNF1/LFB1. EMBO J. 1990;9:2257–2263. doi: 10.1002/j.1460-2075.1990.tb07396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frain M., Swart G., Monaci P., Nicosia A., Stampfli S., Frank R., Cortese R. The liver-specific transcription factor LF-B1 contains a highly diverged homeobox DNA binding domain. Cell. 1989;59:145–157. doi: 10.1016/0092-8674(89)90877-5. [DOI] [PubMed] [Google Scholar]
- 31.Bellanne-Chantelot C., Carette C., Riveline J.P., Valero R., Gautier J.F., Larger E., Reznik Y., Ducluzeau P.H., Sola A., Hartemann-Heurtier A. The type and the position of HNF1A mutation modulate age at diagnosis of diabetes in patients with maturity-onset diabetes of the young (MODY)-3. Diabetes. 2008;57:503–508. doi: 10.2337/db07-0859. [DOI] [PubMed] [Google Scholar]
- 32.Bluteau O., Jeannot E., Bioulac-Sage P., Marques J.M., Blanc J.F., Bui H., Beaudoin J.C., Franco D., Balabaud C., Laurent-Puig P. Bi-allelic inactivation of TCF1 in hepatic adenomas. Nat. Genet. 2002;32:312–315. doi: 10.1038/ng1001. [DOI] [PubMed] [Google Scholar]
- 33.Reiner A.P., Barber M.J., Guan Y., Ridker P.M., Lange L.A., Chasman D.I., Walston J.D., Cooper G.M., Jenny N.S., Rieder M.J. Polymorphisms of the HNF1A gene encoding hepatocyte nuclear factor-1 alpha are associated with C-reactive protein. Am. J. Hum. Genet. 2008;82:1193–1201. doi: 10.1016/j.ajhg.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whitfield J.B., Martin N.G. Determinants of variation in plasma alkaline phosphatase activity: A twin study. Am. J. Hum. Genet. 1983;35:978–986. [PMC free article] [PubMed] [Google Scholar]
- 35.Walker B.A., Eze L.C., Tweedie M.C., Evans D.A. The influence of ABO blood groups, secretor status and fat ingestion on serum alkaline phosphatase. Clin. Chim. Acta. 1971;35:433–444. doi: 10.1016/0009-8981(71)90218-x. [DOI] [PubMed] [Google Scholar]
- 36.Brun-Heath I., Lia-Baldini A.S., Maillard S., Taillandier A., Utsch B., Nunes M.E., Serre J.L., Mornet E. Delayed transport of tissue-nonspecific alkaline phosphatase with missense mutations causing hypophosphatasia. Eur. J. Med. Genet. 2007;50:367–378. doi: 10.1016/j.ejmg.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 37.Foreman J.E., Blizard D.A., Gerhard G., Mack H.A., Lang D.H., Van Nimwegen K.L., Vogler G.P., Stout J.T., Shihabi Z.K., Griffith J.W. Serum alkaline phosphatase activity is regulated by a chromosomal region containing the alkaline phosphatase 2 gene (Akp2) in C57BL/6J and DBA/2J mice. Physiol. Genomics. 2005;23:295–303. doi: 10.1152/physiolgenomics.00062.2005. [DOI] [PubMed] [Google Scholar]
- 38.Chalasani N., Vuppalanchi R., Raikwar N.S., Deeg M.A. Glycosylphosphatidylinositol-specific phospholipase d in nonalcoholic Fatty liver disease: A preliminary study. J. Clin. Endocrinol. Metab. 2006;91:2279–2285. doi: 10.1210/jc.2006-0075. [DOI] [PubMed] [Google Scholar]
- 39.Lee J.W., Choi H.S., Gyuris J., Brent R., Moore D.D. Two classes of proteins dependent on either the presence or absence of thyroid hormone for interaction with the thyroid hormone receptor. Mol. Endocrinol. 1995;9:243–254. doi: 10.1210/mend.9.2.7776974. [DOI] [PubMed] [Google Scholar]
- 40.Christenson R.H. Biochemical markers of bone metabolism: An overview. Clin. Biochem. 1997;30:573–593. doi: 10.1016/s0009-9120(97)00113-6. [DOI] [PubMed] [Google Scholar]
- 41.Ge D., Zhang K., Need A.C., Martin O., Fellay J., Urban T.J., Telenti A., Goldstein D.B. WGAViewer: Software for genomic annotation of whole genome association studies. Genome Res. 2008;18:640–643. doi: 10.1101/gr.071571.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Firmann M., Mayor V., Vidal P.M., Bochud M., Pecoud A., Hayoz D., Paccaud F., Preisig M., Song K.S., Yuan X. The CoLaus study: A population-based study to investigate the epidemiology and genetic determinants of cardiovascular risk factors and metabolic syndrome. BMC Cardiovasc. Disord. 2008;8:6. doi: 10.1186/1471-2261-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


