Abstract
Insertional mutagenesis continues to be a major concern in hematopoietic stem cell gene therapy. Non-conventional gene transfer vectors with more favorable integration features in comparison to conventional retrovirus and lentivirus vectors are being developed and optimized. Here we reported for the first time a systematic analysis of 198 ASLV insertion sites identified in rhesus long-term repopulating cells and a comparison of ASLV insertions to MLV (n=396) and SIV (n=289) insertions using the newly released rhesus genome databank. Despite a weak preference towards gene coding regions, ASLV integration is strikingly non-clustered, does not favor gene-rich regions, transcription start sites, or CpG islands. There was no propensity of insertions to be within or near proto-oncogenes, and most importantly, no insertions close to or within the Mds1-Evi1 locus, which is in strong contrast to significant overrepresentation of this insertion site for MLV vectors in the same transplantation model. Furthermore, ASLV LTRs do not have detectable promoter and enhancer activity in a quantitative luciferase assay to measure neighboring gene activation. The combination of these features is unique for ASLV and suggests that optimized vectors based on this virus could be useful and safe for gene transfer to hematopoietic stem and progenitor cells.
Keywords: retroviral integration site analysis, ASLV, RCAS, rhesus macaque, autologous transplantation, long-term repopulating hematopoietic cells
INTRODUCTION
A DNA copy of the genome of a retroviral vector stably integrates into the host genome. This is advantageous for gene transfer applications requiring sustained expression in proliferating targets, such as hematopoietic stem and progenitor cells. However, integration can have important effects on the engraftment, proliferation and survival of transduced cells because the integrated proviral DNA can either activate or disrupt host cell genes, resulting in immortalization 1, clonal dominance 2, and in worst case scenario, malignant transformation 3, 4. These genotoxic risks were assumed to be acceptably low during initial development of retroviral gene therapy vectors. However, four of ten patients from the French SCID–X1 gene therapy trial, and more recently one patient from the British SCID-X1 trial, developed T cell leukemias that appear to be caused by Moloney murine leukemia virus (MLV) vector integration near and activation of proto-oncogenes, most commonly Lmo2 3.
Genome-wide integration site analyses in cell lines and long-term repopulating hematopoietic stem and progenitor cells have demonstrated that MLV-based vectors preferentially integrate into regions near transcription start sites (TSS) and actively-expressed gene regions, locations where the strong enhancer and promoter elements in the MLV LTR can activate neighboring genes. HIV or SIV-derived lentiviral vectors have a strong preference for integration within genes 5–9. Recent reports also demonstrate that integrated MLV proviruses are frequently found clustered around particular sites in the genome, termed common insertion sites (CIS)10–12. MLV CIS are often in or near highly-expressed growth-promoting proto-oncogenes. This over-representation may reflect amplification of the cells with the vector-activated oncogenes rather than a preferential integration into these sites. Stem and progenitor cell targets may be particularly susceptible to these events as compared to terminally-differentiated cells, since they highly express genes that favor self-renewal and prevent differentiation. Regardless of the underlying mechanism, these integrations can have profound effects on hematopoiesis and the risk of leukemia or other manifestations of genotoxicity recently reported in human clinical gene therapy trials for chronic granulomatous disease (CGD) and SCID-X1 3, 13.
A genome-wide insertion site survey of avian sarcoma leukosis virus (ASLV) infected 293T cells showed that ASLV has a very weak preference for integrating in genes and no bias for transcriptional start sites (TSS) 6, 14. ASLV is replication-incompetent in mammalian cells and the promoter and enhancer in the LTR were selected for optimal expression in avian cells. These characteristics would suggest that viral vectors based ASLV could be less likely to cause oncogene activation in mammalian cells than MLV-based vectors. We recently reported that rhesus macaque HSPCs can be transduced using ASLV-derived RCAS (Replication Competent ALV LTR with a Splice acceptor) vectors expressing the GFP marker gene 15. Two rhesus macaques transplanted with autologous transduced CD34+ cells had 1–3% GFP-positive transduced cells in multiple hematopoietic lineages 18 months after transplantation.
The ASLV integration site profile in long-term repopulating HSPCs of these rhesus macaques is reported here. Using an optimized linear-amplification mediated PCR (LAM-PCR), we identified almost 300 unique insertion sites in CD34 positive cells or in the peripheral blood cells of these animals 4–18 months following transplantation. With the release of the rhesus macaque draft genome complete sequence in 2006 16, 17, we were able to analyze our data directly via comparison to rhesus instead of human sequences. The results obtained with ASLV integration were compared to sets of SIV and MLV insertions previously reported 7.
RESULTS
Characterization of in vivo integration profile of Avian Sarcoma Leukosis Virus Vectors in rhesus long-term repopulating cells
ASLV-derived RCAS vectors were produced by transient transfection of DF-1 chicken fibroblasts with RCANBPM2C(797-8)PolIIGFP plasmids as previously described15. Two rhesus macaques (RQ3592 and RQ4814) were transplanted with RCAS-transduced autologous CD34+ cells and followed for more than 18 months. A modification of the linear amplification-mediated PCR (LAM-PCR) method was used to identify ASLV provirus-genomic junctions. Following rhesus CD34+ cell transduction, we identified 98 insertion sites. The number of sites was limited by the amount of CD34+ cells that could be safely removed from the graft for analysis without impeding recovery. These insertions were not further analyzed or pooled with the in vivo data both because of the relatively small number of pre-transplant insertions and the concern that the insertions identified in a heterogeneous CD34+ population in vitro might not reflect the insertions in the long-term repopulating cells defined by engraftment and stable hematopoietic output in vivo.
Post-transplantation, LAM-PCR was performed on DNA purified from peripheral blood mononuclear cells (over 90% lymphocytes) and granulocytes sampled at 4, 6, 12 and 18 months. Despite the relatively low level of gene expression in the circulating cells of these animals long-term, all samples were polyclonal with at least 20 independent insertion sites identified at each time point.
All putative insertion sites identified by LAM-PCR were mapped using the newly-released rhesus macaque genome assembly (MMUL 1.0, Feb2006). The availability of the new sequence information made it possible to map 30–40% more insertions than could be mapped using the human genome sequence (see Materials and Methods). 198 insertions could be unequivocally mapped in long-term repopulating cells. Insertions occurred in all rhesus chromosomes, with the number of insertions per chromosome approximately proportional to chromosome size. Insertions were most frequent on chromosome 3 and chromosome 1, which are the two largest chromosomes in the rhesus, and least frequent on chromosomes 19, 21 and 22, which are small.
Distribution of integration sites relative to genes
We mapped the distribution of ASLV insertions with respect to genes. For designation of rhesus genes, we used the ensembl genebuild Aug 2007 (http://www.ensembl.org), database version 45.10e, which comprises 38,185 predicted genes. This source is significantly better annotated for rhesus genes than the more commonly used UCSC BLAT RefSeq gene annotation. Out of 198 unique insertions identified in granulocyte or lymphocyte samples 4–18 months post-transplantation, 85 (42.9%) were within ensembl gene coding sequences. We compared this dataset to 10,000 sets of randomly-selected in silico integration sites. Each set contains 198 sequences, with each sequence matched by in silico characteristics to an ASLV integration site. That is, each control sequence, which is picked randomly from the rhesus genome, is the same length as the ASLV insertion site sequence, is adjacent to the restriction enzyme site used to clone the ASLV insertion site and has a unique hit by BLAST to the rhesus genome.
Compared to our control 1,980,000 computer generated random insertion sites (33.8% within genes, Table 1), there was a mild but significant increase for ASLV insertions occurring within genes (p=0.0029), suggesting some degree of non-random integration. These results are consistent with the previous report of ASLV insertion patterns in Hela cells, which also demonstrated a weak integration preference towards human RefSeq gene coding regions 6, 14. We compared ASLV to our previously-reported MLV and SIV insertions identified in long-term repopulating cells in the identical rhesus autologous transplantation model, reanalyzing the MLV and SIV insertions using the rhesus instead of the human genome (Table 1). 45.71% of MLV and 77.16% of SIV insertions were within ensembl gene coding regions, in comparison to 33.78% for random in silico controls. Thus both ASLV and MLV have a weak preference for integrating within genes, whereas SIV has a much stronger bias (p<0.001).
TABLE 1.
Distribution of ASLV integrations with respect to transcription units and CpG islands: comparison with MLV and SIV
| ASLV(n=198) | SIV(n=289) | MLV(n=396) | random | |
|---|---|---|---|---|
| Transcription units | 42.93%a,b | 77.16% | 45.71% | 33.78% |
| exons | 0.51% | 4.84% | 2.02% | 1.43% |
| introns | 42.42% | 72.32% | 43.69% | 32.35% |
| within 10kb upstream of TSS | 8.59%c | 5.88% | 10.61%g | 5.53% |
| within 30kb upstream of TSS | 17.68%d | 21.11% | 23.74%h | 14.16% |
| within 10kb downstream of TES | 6.57%e | 2.42% | 4.29% | 4.11% |
| within CpG islands | 1.52%f | 0.00% | 0.76% | 0.89% |
TSS: Transcription Start Sites; TES: Transcription End Sites; random are 1,980,000 randomly in silico controls generated as described in Materials and Methods,
values are significantly different from random integration (p=0.0029) by chi-square test
values are significantly different from SIV integration
values are indistinguishable from random integration
values are significantly different from random integration by chi-square test
To understand the pattern of ASLV insertion sites relative to gene density, we examined ensembl genes within each 1-Mbp window of the rhesus genome and subdivided genomic regions into three categories: low gene density regions comprised of 0–10 ensembl genes per 1-Mbp, middle gene density regions comprised of 11–20 ensembl genes per 1-Mbp and high gene density regions comprised of more than 20 ensembl genes per 1-Mbp. We calculated the frequency of ASLV insertions within each category and compared the results with the MLV and SIV datasets and the computer simulated random dataset. As shown in Fig. 1, 66.67% of ASLV insertions are within low gene density regions, versus 33.22% for SIV insertions (p<0.001) and 24.75% of ASLV insertions are within middle gene density regions, similar to random insertions. In contrast, MLV and SIV both had significant bias for inserting into middle gene density regions. Only 8.6% of ASLV insertions were in high gene density regions, again similar to that of random insertions. MLV and SIV both had strong preferences for inserting into regions of high gene density, with SIV integrating into these regions four times more often than ASLV or the random in silico sets (p<0.001).
Fig. 1.
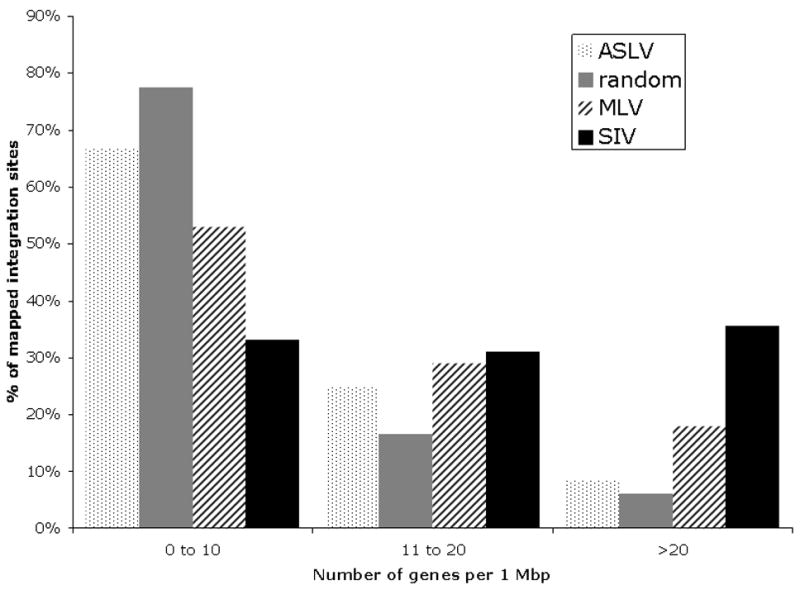
Distribution of ASLV integration sites relative to gene density within 1-Mbp windows compared to MLV and SIV integration sites, and to in silico-generated random controls. Each bar corresponds to the percentage of integrations within the corresponding gene density region.
Distribution of integration sites relative to transcription start site (TSS) and CpG islands
Previous studies reported that MLV has a strong preference for integrating near transcription start sites (TSS) 6–9. We analyzed integrations within 10kb of transcription units. As is shown in Table 1, 17 out of 198 (8.59%) ASLV insertions were within 10kb upstream of TSS, which is not significantly different (p=0.0518) from random insertions (5.53%). 13 out of 198 (6.57%) ASLV insertions were within 10kb downstream of transcription end sites (TES), comparable to 4.11% for random insertions (p=0.0647). With expansion of the window to 30kb upstream of TSS, still well within the range for enhancer effects, 35 out of 198 (17.68%) insertions were within 30kb upstream of TSS, again comparable to random insertions (14.16%). This suggests that ASLV does not favor TSS or TES. 42 out of 396 (10.61%) MLV insertions were within 10kb upstream of TSS, indicating a modest bias towards this region, whereas SIV integrations were indistinguishable from random insertions (5.88% for SIV versus 5.53% for random). These finding are similar but not identical to our previous published analysis of the same MLV and SIV insertions mapped via comparison to the human genome. The lower percentage of MLV insertions near TSS using the rhesus sequence for mapping may be due to a less complete annotation of rhesus genes.
In 5kb, 10kb and 30kb windows surrounding TSS (Fig 2A), MLV integrations were increased both up and downstream of TSS with a decreasing frequency of integrations as the distance from the TSS increased. ASLV integrations were more unevenly distributed around TSS. When analyzed in a 5kb bin size, ASLV shows a mild preference for integrating 5kb upstream but not downstream of TSS. However, combining both upstream and downstream of TSS using different window sizes, ASLV had a weaker bias toward this region than MLV.
Fig. 2.
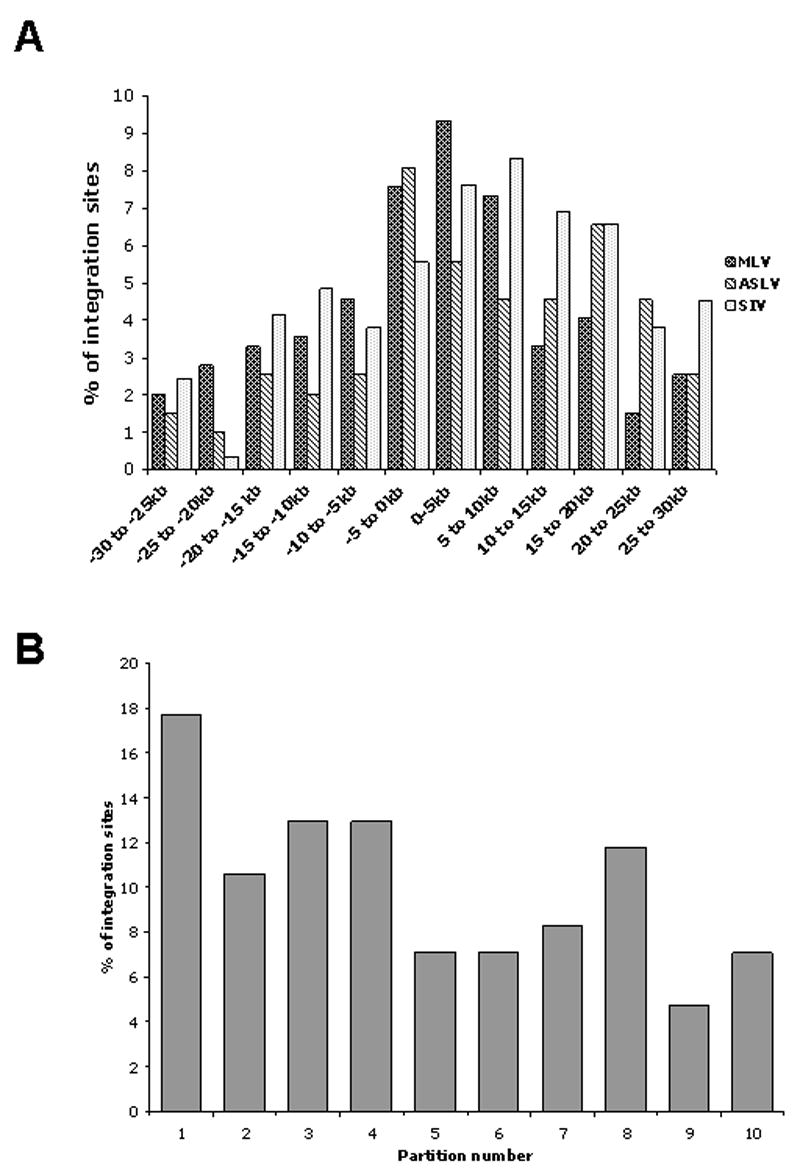
Distribution of integration sites relative to a 30kb window around transcription start sites (TSS) and transcription units. (A) Distribution of integration sites relative to a 30kb window around transcription start sites (TSS). White bars -ASLV integrations, gray bars- MLV integrations; black bars-SIV integrations. (B) Distribution of ASLV integration sites relative to transcription units in comparison with 10, 000 random integration sites. Gene coding regions were arbitrarily divided into ten equal units and the black bars show the percentage of integrations within each unit. The grey line indicates the results of 10, 000 random integration sites.
We next examined whether specific regions within genes were more likely sites of integration than others. We analyzed the distribution of the integrations within genes by dividing the distance of each integration site from the TSS to the TES into ten units of equal length. The frequencies of insertions within each unit were calculated. As diagrammed in Fig 2B, ASLV integrations were distributed over the entire length of the genes, with no preference for any part of the transcription unit.
CpG islands are sequences rich in CpG dinucleotides and are often found in the vicinity of TSS. ASLV integrations showed no preference for CpG islands, with only 3 of 198 insertions (1.52 %) in CpG islands, indistinguishable from the random sets (0.89%).
No clustering of common insertion sites (CISs)
To search for common insertion sites (CIS), we used the stringent CIS definition of Suzuki et al.18. Two, three or four insertions were considered to be CIS if they fell within a 30-kbp, 50-kbp or 100-kbp window, respectively. CIS of fifth or higher order were defined by a 200-kbp window. Using the definitions, we found no CIS for the ASLV integration sites. The two closest sites were 39,453 base pairs apart. The next closest were 111,494 base pairs apart. We expanded the analysis to look for ASLV integration sites within 1 Mb windows. There were a total of 24 integration sites within 1 Mb of another independent integration site, and these can be divided into 11 clusters. We analyzed whether the clustering of ASLV sites is different from computer simulated random integrants. Performing 5000 random datasets, each with 198 integrants, we found a mean of 26 random integration sites in each set, with a range of 4–49, indicating that ASLV clustering is indistinguishable from random clustering in our sample. We reanalyzed our MLV and SIV insertions using the same criteria and found significant clustering for both MLV and SIV. Out of 396 MLV insertions, 41 insertions (10.35%) fell within 30 kb of other independent insertions, thereby forming 20 independent clusters. The largest clusters were in or near Mds-Evi1, as previously reported 1. There were 25 (7.27%) out of 289 SIV insertions in 10 independent clusters. Of these, 9 clusters were within 30 kb windows and the other cluster was within a 50kb window. The mean span of clusters is 9,034bps for MLV and 11,033 bps for SIV. The two closest integrants for SIV are SIV_RQ3556_6p21.32e and SIV_RQ3556_6p21.32d, which are only 874 bps apart. The two closest integrants for MLV are rq2428-17q25.3 and rq2277-17q25.3, which are only 220 bps apart. No overlapping clusters were identified between the MLV and SIV datasets. Both MLV and SIV integrations show a higher degree of clustering than ASLV integrations, and, in terms of clustering, SIV integration is not significantly different from that of MLV.
We previously reported clustering of MLV insertions in or near the Mds1-Evi1 proto-oncogene locus (14 out of a total of 702 insertions) in the same rhesus model. We have not found evidence of ASLV insertions into the Mds1-Evi1 locus at any time point, significantly different from the MLV result (P=0.04, Fisher’s exact test).
Recent studies suggest that MLV and HIV have a tendency to integrate into genes involved in cellular growth and proliferation. The distribution of MLV integrations is especially biased for genes associated with oncogenic transformation. We compared the ASLV integration sites with sites entered into the retrovirus-tagged cancer gene database (RTCGD) 19, and found no overlap between ASLV integration sites and these genes (Supplemental table 1).
Quantitation of neighboring gene activation by ASLV, MLV and HIV LTRs
The promoter/enhancer activity of the ASLV LTR depends on both the species and the cell type used in the assay. To assess the relative strength of ASLV provirus LTR enhancer activity on adjacent gene expression as compared to LTRs from other integrating vectors derived from MLV, HIV or human foamy virus (FV), the proviral forms of each vector were cloned in both forward and reverse orientations into the pACT5 plasmid, 795 bps upstream of a minimal promoter-IRES-luciferase expression cassette 20 (Figure 3A). The MLV construct contained intact LTRs, as compared to the HIV and FV constructs, which contained significant U3 region deletions. These configurations were chosen as most representative versions of each vector under preclinical or clinical development. Enhancer activity was measured by comparing luciferase activity for plasmids with the inserted viral DNA to the base plasmid containing only the minimal promoter 19. Figure 3B shows luciferase activity measured in three independent experiments after transfection of primitive hematopoietic K562 cells with the constructs and normalization for transfection efficiency. Insertion of the RCAS ASLV DNA in either the forward or reverse orientation did not increase luciferase expression over the control plasmid containing only the minimal promoter, suggesting that the ASLV LTR has little or no enhancer activity in K562 cells. The non-deleted MLV LTR, in contrast, resulted in a significant increase in luciferase expression whether in forward or reverse orientation, indicating strong enhancer activity and/or read-through transcription from MLV LTRs (P=0.002). FV and SIV LTR constructs also had only basal levels of luciferase activity, indistinguishable from ASLV LTR constructs.
Fig. 3.
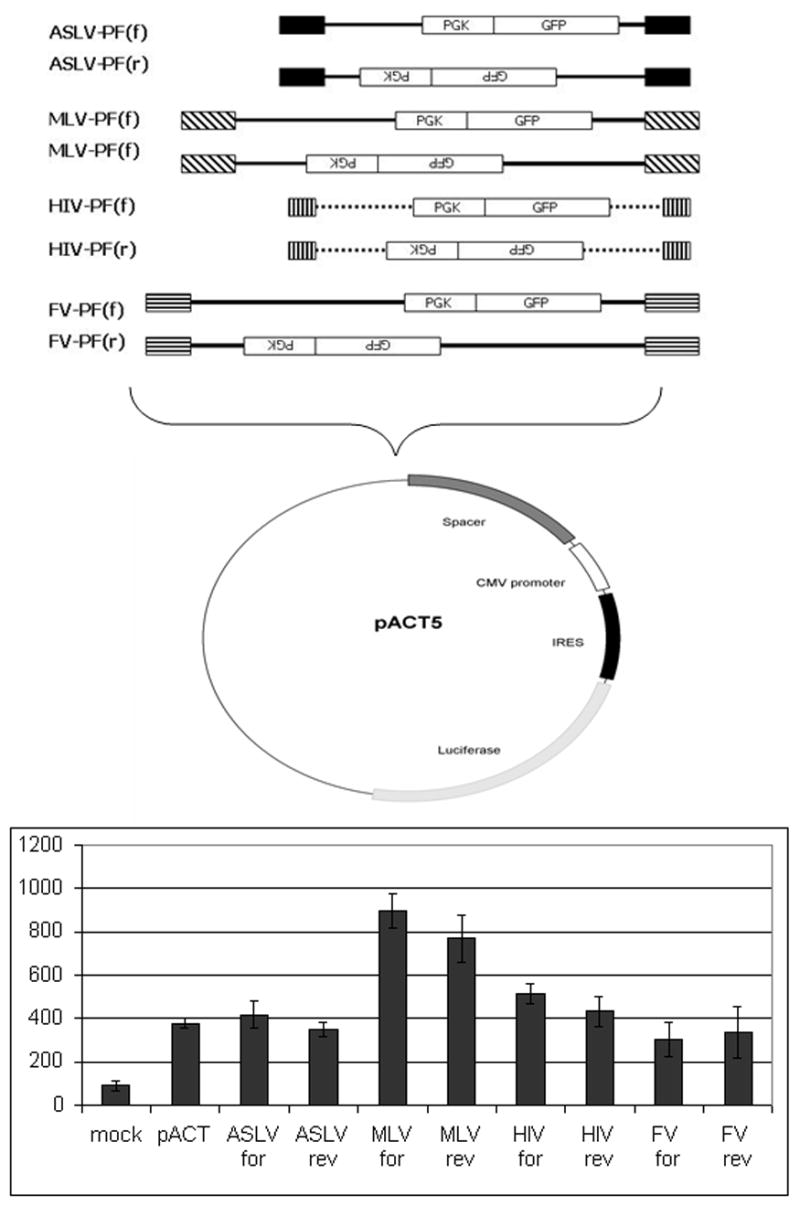
Activation of adjacent gene expression by proviral LTRs. (A) ASLV, MLV, HIV and FV LTR constructs. All LTR constructs contain identical phosphoglycerate kinase promoter (PGK)-GFP expression cassettes. Both HIV and FV LTRs have deletions in U3 regions. MLV LTRs are non-deleted. The LTRs are cloned upstream of an IRES-luciferase expression cassette placed downstream of a minimal promoter. Vector proviruses can activate luciferase expression either through vector enhancer elements acting on the minimal promoter, or read-through transcription from a vector promoter. (B) One day following nucleofection of K562 cells with the plasmids indicated, luciferase activity from K562 cell lysates was measured. The mean values +/−standard errors from 3 independent experiments are shown. Y axis denotes luciferase activity measured in cpm (counts per minute) after correction for transfection efficiency. Mock-no plasmid, pACT-plasmid containing only a minimal promoter, remaining bars have an ASLV, MLV or HIV LTR inserted 795 bps upstream of the minimal promoter, in either forward (for) or reverse (rev) orientation.
DISCUSSION
In an effort to develop safer retroviral gene therapy vectors, we focused on ASLV-based vectors. This virus is of avian origin, does not replicate in mammalian cells, and has an almost random integration profile in cell lines6, 14. Despite the relatively low gene transfer efficiency in our pilot in vivo study, we achieved stable polyclonal reconstitution of hematopoesis with ASLV vector-containing cells expressing a GFP transgene, now out over three years following transplantation. In the current study we mapped the sites of ASLV vector DNA insertions in granulocytes and lymphocytes 4–18 months post-transplantation and found a mild but significant predilection for integration in gene coding regions, but no preference for integrations near transcription start sites or in CpG islands. These results are consistent with the results of previous studies of ASLV integration in human and avian cell lines 14. These comparable results suggest that the ASLV integration machinery interacts similarly with chromatin in three different species (human, rhesus and avian) and in different target cell types (epithelial and hematopoietic). A direct comparison of the integration sites for the ASLV vector and the integration sites we previously reported for MLV and SIV vectors, utilizing an identical rhesus transplantation model and re-analyzing all three vector insertion datasets using the recent rhesus draft genome, suggests that ASLV may be a relatively safe vector in this system.
Integrated proviruses can activate adjacent proto-oncogene expression either directly by promoter insertion or indirectly through the effects of the enhancer(s) in the LTR 21, 22. In principle, these risks may be minimized by avoiding strong enhancers such as the MLV LTR, using self-inactivating vectors with deletion of the enhancer region in the LTR or preventing read-through transcription by improved polyadenylation 21–24. The ASLV LTR promoter/enhancer is active in some mammalian cells. However, the vector was selected for expression in avian cells 25. Although a provirus can be established in transduced mammalian cells, completing the first half of the viral life cycle, infected mammalian cells do not produce infectious virions. Proviral activation of adjacent proto-oncogenes depends on the activity of the LTR promoter and enhancer in the target cell and host species. For this reason, we tested the activity of the LTR in K562 cells, a primitive hematopoietic human cell line. In our assays, the ASLV RCAS LTRs do not detectably activate a luciferase reporter gene, irrespective of LTR orientation. In contrast, the MLV LTRs have significant enhancer activity in these cells using the same assay.
Common insertion sites (CISs) 18, 26, in or near proto-oncogenes, have recently been identified following MLV transduction of human 5, 10, 11, rhesus 1 and murine long-term repopulating cells 27. The high incidence of CIS suggests that insertions may influence HSPC engraftment, survival and/or proliferation. One study found a significant preference for clustering of insertions in or near proto-oncogenes in CD34+ cells at the end of transduction prior to transplantation, suggesting that these gene regions are particularly susceptible to MLV integration12. Identification of CIS is a powerful approach that has been used by a number of laboratories to identify genes contributing to oncogenesis following infection of mice with replication-competent MLV 19. By comparing mapped locations of the integrants in tumors to randomly generated integrations from 100,000 Monte Carlo trials, cutoffs were defined for determining whether two or more clustered integrations happen simply by chance versus being implicated in the pathogenesis of the tumor 19. Despite some disputes regarding the exact definitions of CIS 18, these criteria have generally been used to define CIS and statistical significance can be calculated depending on the size of the insertion dataset. Using 30-kbp, 50-kbp or 100-kbp window cutoffs for 2, 3 or 4 insertions, respectively, no CIS were identified for ASLV in the two rhesus macaques. This finding differs significantly (p<0.001) from the clustering we found for MLV and SIV integrations in the same transplantation model.
We found no ASLV insertion into the Mds1-Evi1 locus at any time point, in contrast to significant over-representation of this insertion site for MLV vectors in the same transplantation model 1. This locus was even more markedly over-represented in a human clinical trial for chronic granulomatous disease, utilizing an MLV vector with a particularly strong LTR enhancer. In that human trial, hematopoietic clones containing insertions in or near the Mds1-Evi1 locus expanded in vivo and became completely dominant, eventually resulting in loss of effective transgene expression and myelodysplastic hematopoiesis in both patients 13, 28. In vitro studies suggested that activation of this locus can immortalize myeloid progenitor cells 29, 30. In our samples there were no ASLV insertions near or within genes in the RTCGD database of proto-oncogenes previously implicated in experimental tumorigenesis with replicating viruses. Whether the lack of CIS, clustering or an over-representation of Mds1-Evi1 or other worrisome clones is an inherent result of the insertion preference of ASLV, or the result of the relatively weak enhancer activity of the proviral LTR on neighboring genes is unclear. The fact that inclusion of a strong MLV promoter-enhancer as the internal promoter in our SIV vector did not result in clonal dominance or any Mds1-Evi1 clones in vivo in our rhesus model suggests that complex interactions between the enhancer itself and its location in the context of an integrated provirus determine genotoxicity.
In summary, ASLV-derived vectors can be used to transduce rhesus long-term repopulating cells. The data we have suggests that, compared to an MLV vector, the ASLV vectors are much less likely to cause insertional activation of oncogenes in the primate model. Further development of lentivirus, human foamy virus and enhancer-deleted self-inactivating (SIN) MLV vectors is ongoing. These vectors are expected to replace standard MLV vectors in HSC clinical gene therapy trials. Results in our rhesus model suggest that ASLV vectors should also be considered for further development. Endogenous ASLV sequences will need to be deleted in order to increase the size of the transgenes that can be inserted and packaged. Regulatory agencies will likely require that the vectors be made replication-incompetent, even in avian cells. Further improvement in gene transfer efficiency to HSPCs could be pursued, for instance via vector concentration or alternative envelope pseudotypes. Our preliminary results are very encouraging, particularly regarding the relative lack of genotoxicity, and stimulate continued efforts with ASLV vectors.
MATERIALS AND METHODS
Sample collection, processing and retrieval of vector integration sites by linear amplification mediated PCR (LAM-PCR)
The production of ASLV-derived RCAS vector particles via transfection of DF-1 chicken fibroblasts, the transduction of autologous rhesus macaque CD34+ cells and transplantation of these cells following ablative conditioning with total body irradiation were previously described 15. Peripheral blood samples from rhesus macaques RQ3592 and RQ4814 used for analyses of integration events into rhesus long-term repopulating cells (LTRCs) were obtained at 4, 6, 12 and 18 months post-transplantation. Granulocytes and lymphocytes were purified via ficoll-hypaque density gradient separation (LSM, Organon Teknika Corp., Durham, NC).
For identification of proviral-genomic junction sequences, the LAM-PCR method was performed as previously described 7, 31 with modifications. Biotinylated linear LTR primer RCASLTRa-bio: 5′biotin-TGCTTACCACAGATATCCTG-3′ (IDT, Coralville, IA) was used for initial amplification of vector-proviral junction sequences. After bead purification and hexanucleotide priming, the resulting double stranded DNA was digested with TasI, HinPI, or TaqαI, and annealed to a corresponding linker cassette. Exponential nested PCR was performed using the following primer sets: RCAS LTR-R1: CCTTACTTCCACCAATCG, RCASLTR-R2: CGGTGCTTTTTCTCTCCT, LCI: GACCCGGGAGATCTGCAG, LCIII: AGTGGCACAGCAGTTAGG. All PCR reactions were performed on a Gradient Cycler (Applied Biosystems, Foster City, CA).
LAM-PCR amplicons were purified after separation on agarose gels, cloned into the TOPO TA vector (Invitrogen) and sequenced. Alignment of the cloned integration sequences to the rhesus genome was carried out using the rhesus macaque genome database (assembly MMUL 1.0, Feb2006) 16. Insertions were scored as valid using the following criteria: (1) greater than 95% homology to published sequence over at least a 95% span of the entire non-vector sequence (2) presence of the predicted vector LTR-genome junction (3) length of at least 40 bps (4) an unequivocal best unique hit using BLAT and the UCSC Genome Browser gateway (www.genome.ucsc.edu) for comparison to the draft rhesus genome. Unmappable sequences were either too short (<40 bp), had multiple hits and could not be localized due to repetitive sequences or had non-matching base pairs immediately adjacent to the LTR junction. For gene annotation and mapping, ensembl genebuild Aug 2007 (http://www.ensembl.org), database version 45.10e comprising 38,185 predicted gene transcripts was used. Compared with RefSeq, this database is significantly better annotated for the rhesus macaque.
A random dataset for comparison was generated by computer simulation of 10,000 insertions into the rhesus genome at the same distance from the corresponding restriction site for each of the 198 mapped unique ASLV sequences (1,980,000 in total). The MLV and SIV integration sequences from our previous studies 7 were reanalyzed using the rhesus macaque genome database (assembly MMUL 1.0, Feb2006) and ensembl genebuild Aug 2007. All ASLV insertion sequences along with reanalyzed SIV and MLV insertion sequences will be available upon request.
Two, three or four insertions were considered to be common integration sites if they fell within a 30-kb, 50-kb or 100-kb window from each other, respectively 19. The genomic window for CISs of fifth order and higher was set to 200 kb. Computer simulations (10,000 runs) were performed to calculate the likelihood of random, coincidental insertions. CISs of different orders were analyzed independently of each other.
Statistical significance was determined by chi-square test. P values <0.01 were considered statistically significant.
Enhancer activity assay
The pACT5 plasmid was used as described to measure neighboring gene activation by vector proviruses 20. pACT5 contains a 795-bp spacer from intron 2 of the human HPRT gene, followed by a minimal CMV promoter, an internal ribosome entry site element, the firefly luciferase gene, and a synthetic polyadenylation site. Proviral sequences were cloned into the pACT5 plasmid in both forward and reverse orientations relative to the luciferase gene. The MLV construct contained an intact wild-type LTR derived from LXSHD 32. The HIV construct was derived from pRRL-CMV-GFP-SIN, which contains a 400-bp deletion in the U3 region of the LTR33. The HFV construct also contains a deletion of the U3 LTR region 34. K562 human chronic myeloid leukemia cells were grown in Iscove’s modified Dulbecco’s medium (IMDM) with glutamine, supplemented with 10% heat-inactivated fetal bovine serum (FBS; HyClone, Logan, UT), penicillin (100 U/ml) and streptomycin (100 μg/ml) at 37°C in a 5% CO2 incubator. Nucleofection was done using the Amaxa Nucleofector (Amaxa Inc. Gaithersburg, Maryland) according to manufacturer’s instructions. In brief, K562 cells were centrifuged at 200g for 10 minutes and re-suspended in suspension solution V at a concentration of 1×107 cells/ml. 106 K562 cells were mixed with 10 μl of a DNA sample containing 2 μg of control plasmid pACT5 or the molar equivalent of each provirus-LTR-containing plasmid, then nucleofected in a 2.0 mm electroporation cuvette using program T-16. All transfections were done in 6 well plates. The MaxiGFP vector was used for a GFP-positive control. A luciferase reporter PGL3 was used as a positive control for luciferase expression. After nucleofection, cells were transferred to 1.0 ml of prewarmed IMDM plus 10% FBS in a microfuge tube, then added to an additional 1.0 ml of prewarmed medium. A portion (33.3%) of the cells were collected 24 hours following nucleofection, centrifuged at 200g for 10 minutes and resuspended in 100 μl of 1× passive lysis buffer (Promega, Madison, Wisconsin). A freeze-thaw cycle was performed to ensure complete lysis of the cells. Luciferase activity was measured according to the manufacturer’s instructions (Luciferase Reporter Assay System E4030, Promega, Madison, Wisconsin) on the Perkin Elmer Victor3 MultiLabel Plate Reader (Perkin Elmer, Waltham, Massachusetts).
Supplementary Material
Acknowledgments
The authors would like to thank Stephanie Sellers for assistance with CD34+ cell purification and culture, and the staff of the 5 Research Court primate facility for excellent animal care. This work is supported by intramural research program of NHLBI, NHGRI and NCI of the National Institutes of Health.
References
- 1.Calmels B, Ferguson C, Laukkanen MO, Adler R, Faulhaber M, Kim HJ, et al. Recurrent retroviral vector integration at the Mds1/Evi1 locus in nonhuman primate hematopoietic cells. Blood. 2005;106:2530–2533. doi: 10.1182/blood-2005-03-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fehse B, Roeder I. Insertional mutagenesis and clonal dominance: biological and statistical considerations. Gene Ther. 2007 doi: 10.1038/sj.gt.3303052. [DOI] [PubMed] [Google Scholar]
- 3.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 4.Seggewiss R, Pittaluga S, Adler RL, Guenaga FJ, Ferguson C, Pilz IH, et al. Acute myeloid leukemia is associated with retroviral gene transfer to hematopoietic progenitor cells in a rhesus macaque. Blood. 2006;107:3865–3867. doi: 10.1182/blood-2005-10-4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsukahara T, Agawa H, Matsumoto S, Matsuda M, Ueno S, Yamashita Y, et al. Murine leukemia virus vector integration favors promoter regions and regional hot spots in a human T-cell line. Biochem Biophys Res Commun. 2006;345:1099–1107. doi: 10.1016/j.bbrc.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC, et al. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:E234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hematti P, Hong BK, Ferguson C, Adler R, Hanawa H, Sellers S, et al. Distinct genomic integration of MLV and SIV vectors in primate hematopoietic stem and progenitor cells. PLoS Biol. 2004;2:e423. doi: 10.1371/journal.pbio.0020423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bushman F, Lewinski M, Ciuffi A, Barr S, Leipzig J, Hannenhalli S, et al. Genome-wide analysis of retroviral DNA integration. Nat Rev Microbiol. 2005;3:848–858. doi: 10.1038/nrmicro1263. [DOI] [PubMed] [Google Scholar]
- 9.Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 10.Schwarzwaelder K, Howe SJ, Schmidt M, Brugman MH, Deichmann A, Glimm H, et al. Gammaretrovirus-mediated correction of SCID-X1 is associated with skewed vector integration site distribution in vivo. J Clin Invest. 2007;117:2241–2249. doi: 10.1172/JCI31661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deichmann A, Hacein-Bey-Abina S, Schmidt M, Garrigue A, Brugman MH, Hu J, et al. Vector integration is nonrandom and clustered and influences the fate of lymphopoiesis in SCID-X1 gene therapy. J Clin Invest. 2007;117:2225–2232. doi: 10.1172/JCI31659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cattoglio C, Facchini G, Sartori D, Antonelli A, Miccio A, Cassani B, et al. Hot spots of retroviral integration in human CD34+ hematopoietic cells. Blood. 2007;110:1770–1778. doi: 10.1182/blood-2007-01-068759. [DOI] [PubMed] [Google Scholar]
- 13.Ott MG, Schmidt M, Schwarzwaelder K, Stein S, Siler U, Koehl U, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- 14.Narezkina A, Taganov KD, Litwin S, Stoyanova R, Hayashi J, Seeger C, et al. Genome-wide analyses of avian sarcoma virus integration sites. J Virol. 2004;78:11656–11663. doi: 10.1128/JVI.78.21.11656-11663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu J, Ferris A, Larochelle A, Krouse AE, Metzger ME, Donahue RE, et al. Transduction of rhesus macaque hematopoietic stem and progenitor cells with avian sarcoma and leukosis virus vectors. Hum Gene Ther. 2007;18:691–700. doi: 10.1089/hum.2006.175. [DOI] [PubMed] [Google Scholar]
- 16.Han K, Konkel MK, Xing J, Wang H, Lee J, Meyer TJ, et al. Mobile DNA in Old World monkeys: a glimpse through the rhesus macaque genome. Science. 2007;316:238–240. doi: 10.1126/science.1139462. [DOI] [PubMed] [Google Scholar]
- 17.Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, Mardis ER, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 18.Wu X, Luke BT, Burgess SM. Redefining the common insertion site. Virology. 2006;344:292–295. doi: 10.1016/j.virol.2005.08.047. [DOI] [PubMed] [Google Scholar]
- 19.Akagi K, Suzuki T, Stephens RM, Jenkins NA, Copeland NG. RTCGD: retroviral tagged cancer gene database. Nucleic Acids Res. 2004;32:D523–527. doi: 10.1093/nar/gkh013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hendrie PC, Huo Y, Stolitenko RB, Russell DW. A rapid and quantitative assay for measuring neighboring gene activation by vector proviruses. Mol Ther. 2008;16:534–540. doi: 10.1038/sj.mt.6300398. [DOI] [PubMed] [Google Scholar]
- 21.von Kalle C, Fehse B, Layh-Schmitt G, Schmidt M, Kelly P, Baum C. Stem cell clonality and genotoxicity in hematopoietic cells: gene activation side effects should be avoidable. Semin Hematol. 2004;41:303–318. doi: 10.1053/j.seminhematol.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 22.Nienhuis AW, Dunbar CE, Sorrentino BP. Genotoxicity of retroviral integration in hematopoietic cells. Mol Ther. 2006;13:1031–1049. doi: 10.1016/j.ymthe.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 23.Baum C, von Kalle C, Staal FJ, Li Z, Fehse B, Schmidt M, et al. Chance or necessity? Insertional mutagenesis in gene therapy and its consequences. Mol Ther. 2004;9:5–13. doi: 10.1016/j.ymthe.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 24.Modlich U, Bohne J, Schmidt M, von Kalle C, Knoss S, Schambach A, et al. Cell-culture assays reveal the importance of retroviral vector design for insertional genotoxicity. Blood. 2006;108:2545–2553. doi: 10.1182/blood-2005-08-024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hughes SH. The RCAS vector system. Folia Biol (Praha) 2004;50:107–119. [PubMed] [Google Scholar]
- 26.Abel U, Deichmann A, Bartholomae C, Schwarzwaelder K, Glimm H, Howe S, et al. Real-time definition of non-randomness in the distribution of genomic events. PLoS ONE. 2007;2:e570. doi: 10.1371/journal.pone.0000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kustikova O, Fehse B, Modlich U, Yang M, Dullmann J, Kamino K, et al. Clonal dominance of hematopoietic stem cells triggered by retroviral gene marking. Science. 2005;308:1171–1174. doi: 10.1126/science.1105063. [DOI] [PubMed] [Google Scholar]
- 28.Ott MG, Seger R, Stein S, Siler U, Hoelzer D, Grez M. Advances in the treatment of Chronic Granulomatous Disease by gene therapy. Curr Gene Ther. 2007;7:155–161. doi: 10.2174/156652307780859044. [DOI] [PubMed] [Google Scholar]
- 29.Du Y, Spence SE, Jenkins NA, Copeland NG. Cooperating cancer-gene identification through oncogenic-retrovirus-induced insertional mutagenesis. Blood. 2005;106:2498–2505. doi: 10.1182/blood-2004-12-4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du Y, Jenkins NA, Copeland NG. Insertional mutagenesis identifies genes that promote the immortalization of primary bone marrow progenitor cells. Blood. 2005;106:3932–3939. doi: 10.1182/blood-2005-03-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt M, Glimm H, Wissler M, Hoffmann G, Olsson K, Sellers S, et al. Efficient characterization of retro-, lenti-, and foamyvector-transduced cell populations by high-accuracy insertion site sequencing. Ann N Y Acad Sci. 2003;996:112–121. doi: 10.1111/j.1749-6632.2003.tb03239.x. [DOI] [PubMed] [Google Scholar]
- 32.Baum C, Eckert HG, Stockschlader M, Just U, Hegewisch-Becker S, Hildinger M, et al. Improved retroviral vectors for hematopoietic stem cell protection and in vivo selection. J Hematother. 1996;5:323–329. doi: 10.1089/scd.1.1996.5.323. [DOI] [PubMed] [Google Scholar]
- 33.Barry SC, Harder B, Brzezinski M, Flint LY, Seppen J, Osborne WR. Lentivirus vectors encoding both central polypurine tract and posttranscriptional regulatory element provide enhanced transduction and transgene expression. Hum Gene Ther. 2001;12:1103–1108. doi: 10.1089/104303401750214311. [DOI] [PubMed] [Google Scholar]
- 34.Trobridge G, Josephson N, Vassilopoulos G, Mac J, Russell DW. Improved foamy virus vectors with minimal viral sequences. Mol Ther. 2002;6:321–328. doi: 10.1006/mthe.2002.0672. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


