Abstract
Among the myriad of alterations present in cancer cells are an abundance of aberrant mRNA transcripts. Whether abnormal gene transcription is a by-product of cellular transformation or whether it represents an inherent element that contributes to the properties of cancer cells is not yet clear. Here, we present growing evidence that in many cases, aberrant mRNA transcripts contribute to essential phenotypes associated with transformed cells, suggesting that alterations in the splicing machinery are common and functionally important for cancer development. The proteins encoded by these abnormal transcripts are often truncated or missing domains, thereby altering protein function or conferring new functions altogether. Thus, aberrant splicing regulation has genome-wide effects, potentially altering gene expression in many cancer-associated pathways.
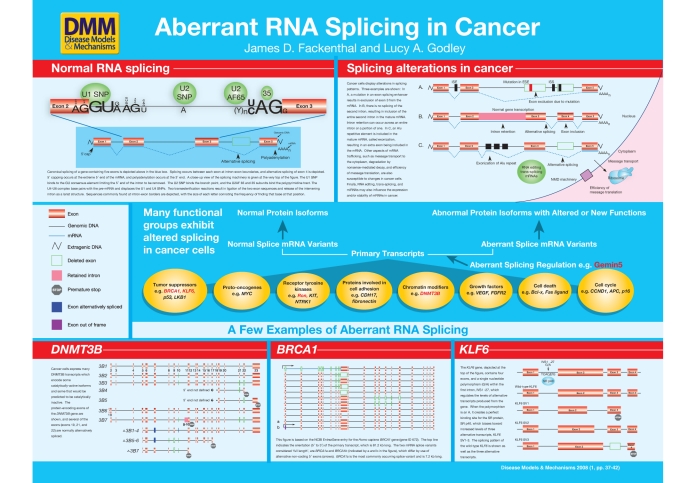
The steps between transcription and translation in eukaryotes are very complicated. In the simplest cases, the primary transcript must acquire a 3′ poly-adenine tail and a 5′ methyl guanine cap in order to be transported to the cytoplasm, undergo translation, and remain stable long enough to produce a functional amount of protein. Additionally, in multicellular eukaryotes, most primary transcripts must undergo splicing reactions, in which exons are joined together and non-coding intervening sequences (introns) are removed (see poster). Genes may produce several protein products by the use of alternative mRNA splicing, in which different exons and different intron/exon junctions may be alternatively used in different splice products from the same primary transcript. Alternative splicing is a highly regulated process, and is recognized increasingly as a player in cancer development.
Human genes challenge the splicing machinery with a seemingly insurmountable problem. The majority of mature mRNAs are 2–4 kb, but primary transcripts can have many introns, each typically 0.5–10 kb, although shorter and longer ones are found frequently. It is common for human primary transcripts to be many times larger than the final mature mRNA after splicing. The challenge, then, is for the splicing machinery to recognize fairly short sections of exonic RNA embedded in very long stretches of intronic RNA (see poster, inset on BRCA1). The higher-order challenge is for the splicing machinery to alter exon usage in alternative splice variants in response to environmental, developmental or tissue-specific cues. It is this regulation of alternative exon usage that can be defective in cancer cells, with potential transcriptome-wide consequences to gene function.
Altered splicing in cancer
In cancer cells, normal mRNA splicing can be misregulated by cancer-specific defects in several splicing mechanisms (Skotheim and Nees, 2007; Srebrow and Kornblihtt, 2006 and references therein). Until recently, it was not clear whether abnormal mRNA splicing was a consequence of transformation, or whether the new RNA and protein isoforms that were produced had actual functional consequences for the cancer cell (Kim et al., 2008). Emerging data suggest that, at least in some cases, the aberrant mRNAs and their encoded proteins have unique properties that confer new properties of growth, differentiation and other cellular characteristics to the cancer cell.
In this review, we will focus on the mechanisms by which splicing has been shown to be abnormal in cancer cells. This review is not meant to provide an exhaustive list of all of the genes abnormally spliced in cancer cells, since the number of such genes is well over 100, and a mere listing of those provides no conceptual framework in which to think of the problem. We have chosen examples in which there is strong evidence that the new proteins encoded by the aberrantly spliced mRNA confer unique function to the expressing cancer cells. The poster provides a schematic view of normal splicing in contrast to the different ways splicing is abnormal in cancer cells.
An overview of normal mRNA splicing
The splicing reaction is carried out by a multi-protein/RNA complex called the spliceosome, which consists of five small nuclear ribonucleoproteins (snRNPs), U1, U2, U4, U5 and U6 (Blencowe, 2006 and references therein). The spliceosome recognizes intron-exon boundaries and removes intervening introns via two transesterification reactions that result in ligation of two adjacent exons. The fidelity of this reaction must be exquisite, because if the ligation occurs incorrectly, normal protein-encoding potential may be compromised. For example, in cases where exon-skipping preserves the reading frame of the triplet codons specifying the identity and order of amino acids during translation, the alternatively spliced mRNA may specify a protein that lacks crucial amino acid residues. More commonly, exon-skipping will disrupt the translational reading frame, resulting in premature stop codons. These mRNAs are typically degraded by at least 90% through a process known as nonsense-mediated mRNA degradation (NMD), which reduces the likelihood that such defective messages will accumulate to generate truncated protein products (Lykke-Andersen et al., 2001). If mis-spliced mRNAs escape this pathway, then truncated, mutated or unstable proteins will be produced.
In nonsense-altered splicing (NAS), a process parallel to NMD, cells upregulate the levels of splice variants that skip exons containing frameshift mutations (Maquat, 2002; Valentine, 1998). In these cases, putative null mutations associated with truncation (frameshift, nonsense or splice-site mutations) may promote increased levels of partially functional mRNA species that are potentially associated with non-null phenotypes using NAS.
Alternative splicing is a means of expressing several or many different transcripts from the same genomic DNA and results from the inclusion of a subset of the available exons for a particular protein. By excluding one or more exons, certain protein domains may be lost from the encoded protein, which can result in protein function loss or gain. Several types of alternative splicing have been described: exon skipping; alternative 5′ or 3′ splice sites; mutually exclusive exons; and, much more rarely, intron retention. Kim et al. compared the amount of alternative splicing in cancer versus normal cells using a bioinformatic approach and determined that cancers exhibit lower levels of alternative splicing than normal cells (Kim et al., 2008). Furthermore, the distribution of the types of alternative splicing events differed in cancer versus normal cells. Cancer cells demonstrated less exon skipping, but more alternative 5′ and 3′ splice site selection and intron retention than normal cells (Kim et al., 2008). Interestingly, when Kim et al. examined the phenomenon of exonization (the use of sequences as exons that are used predominantly by other tissues as introns), they found that the genes associated with exonization in cancer cells were preferentially associated with mRNA processing, which indicates a direct link between cancer cells and the generation of aberrant mRNA splice forms.
Exonic and intronic splicing enhancers (ESEs and ISEs, respectively) are purine-rich cis-acting elements that promote splicing of nearby sequences, whereas exonic and intronic splicing suppressors (ESSs and ISSs, respectively) repress splicing of close sequences (Blencowe, 2006). Often enhancer and silencer elements can overlap one another. ESEs are controlled by serine-arginine (SR) proteins that contain an N-terminal RNA binding and C-terminal SR-rich domains. The SR domain is likely to stimulate splicing by promoting spliceosome assembly by recruiting the U1 and U2AF snRNPs, as well as by binding the pre-mRNA branch point directly. The silencing elements, the ESSs and ISSs, are recognized generally by the hnRNPs, which control the likelihood of splicing in combination with the local SR protein composition.
Modern tools for identifying alternative exon usage
Bioinformatic strategies have been used to characterize common sequence features of exons, intron/exon boundaries, and intronic sequences involved in splicing events. These strategies take advantage of large, publicly available databases containing annotated mRNA and corresponding genomic DNA sequences from several species, and have generated several very useful splice site prediction algorithms (Churbanov et al., 2006; Zhang, 1998).
Bioinformatics has also played an important role in identifying transcriptome-wide changes in splicing patterns that vary by developmental stage, tissue, sex, or disease state. Such strategies include alignment of expressed sequence tags (ESTs) (Hui et al., 2004; Xu et al., 2003), high-throughput sequencing, and microarray/tiling array-based strategies, some of which have been combined with analysis of spliced RNAs bound to splicing regulators revealed by co-immunoprecipitation or crosslinking approaches, to quantify use of exons, splice junction sequences and other sequence features (reviewed by Moore and Silver, 2008).
Tools like these and others have revealed splicing patterns for numerous genes that differ between tumors and normal tissues. In one study, analysis of EST clusters in normal and tumor cells revealed 383 out of 26,812 splice variants were likely to be tumor associated (Hui et al., 2004), and another showed 89 out of 4100 genes had at least one probable tumor-specific splice variant (Xu et al., 2003). The proportion of alternative splice variants likely to be especially enriched in tumor cells over their normal counterparts is small in these examples. If alternative splicing patterns of crucial genes play an important role in tumor initiation or progression, it may be by a combination of novel splice variants acting in a dominant manner and reduced gene function caused by elimination of potentially functionally distinct products of individual genes. Understanding the role of splicing regulation in cancer will require extensive investigation. However, in the short term, such transcriptome-wide patterns of alternative splicing could conceivably be used to identify tumor subtypes (Baudry et al., 2002; Caballero et al., 2001; Klinck et al., 2008) or provide tumor-specific targets for potential therapies (Volpe et al., 2007).
Abnormal RNA splicing can have functional consequences for cancer cells
As noted above, many groups have documented abnormal or alternative mRNA splicing in cancer cells. An increasing body of evidence suggests that in several cases the novel protein that is produced by the new mRNA alters the characteristics of the expressing cancer cell. Here, we will focus on five examples: DNMT3B, BRCA1, KLF6, Ron, Gemin5, chosen to highlight specific features of aberrant splicing in cancer cells.
DNA methylation is catalyzed by three enzymes in human cells: DNMT1, DNMT3A and DNMT3B, and cancer cells are characterized by abnormal splicing of the DNMT3B gene. There are several splice forms found in non-transformed cells: DNMT3B1 (which contains all of the protein encoding exons 2-23); DNMT3B2 (which lacks exon 10); DNMT3B3 (which lacks exons 10, 21 and 22); and DNMT3B6 (which is expressed from a different promoter that adds 12 amino acids to the N-terminus of the protein). Several aberrant DNMT3B transcripts have been found in cancer cell lines and primary tumors that produce truncated proteins, some of which are catalytically inactive, that may contribute to the abnormal DNA methylation patterns of cancer cells: DNMT3B4 and DNMT3B5; DNMT3B7, a transcript arising from intron retention (in this case, retention of 94 bases of intron 10 sequence); seven transcripts expressed in non-small cell lung cancer (named ΔDNMT3B transcripts) arising from a promoter present within intron 4 (Wang et al., 2006b). Some of these encode proteins that are expected to be catalytically active and some encode catalytically inactive proteins, since they lack crucial portions of the methyltransferase domains; and many other aberrant DNMT3B transcripts present at much lower levels in cancer cells (Ostler et al., 2007). Many of the truncated DNMT3B proteins contain the PWWP domain in the protein’s N-terminus, which may mediate DNA binding.
To test whether truncated DNMT3B isoforms could affect the DNA methylation patterns and phenotypes of cancer cells, two groups independently expressed DNMT3B4 and DNMT3B7 within 293 cells (Saito et al., 2002; Ostler et al., 2007). In both cases, transfected cells expressing the truncated DNMT3B protein altered the DNA methylation pattern relative to control cells. In addition, 293 cells expressing DNMT3B4 grew twice as fast as vector-control cells (Saito et al., 2002). In the context of primary human tumor samples, Mao and colleagues have identified a strong correlation between the expression of ΔDNMT3B4, predicted to encode a catalytically active isoform, and the methylation of the RASSF1A gene (Wang et al., 2007; Wang et al., 2006a). Thus, particular DNMT3B isoforms, derived by alternative splicing, have distinct functions and alter DNA methylation in unique ways.
Interestingly, DNMT3B function is regulated not only by the production of particular protein isoforms that are derived by alternative splicing, but also by a family of micro-RNAs, miR-29a/b/c, which has been shown to regulate DNMT3A and DNMT3B in lung cancer (Fabbri et al., 2007). Forced expression of miR-29s in lung cancer cells restores normal patterns of DNA methylation within the cells as well as re-expression of silenced genes, such as FHIT and WWOX (Fabbri et al., 2007).
Finally, Elela and colleagues have used large scale reverse transcription-PCR and deep sequencing to analyze the transcriptome of primary ovarian cancers, borderline ovarian tumors, and normal ovarian tissue (Klinck et al., 2008). The group found that normal and tumor tissue can be distinguished based on the differing splicing patterns of several genes. One of the highest scoring genes in their system was DNMT3B. Notably, ovarian cancers expressed both transcripts encoding full-length catalytically active DNMT3B as well as mRNAs encoding truncated, catalytically inactive isoforms. The ability of this group to distinguish normal from cancer cells merely based on splicing patterns underscores the prevalence of aberrant splicing in primary human tumors.
Splicing patterns in BRCA1, a gene associated with tumor initiation
The breast cancer susceptibility gene BRCA1 generates several splice variants that may play an important role in tumor development. Many BRCA1 splice variants result from mutations in consensus intronic splice donor and acceptor sites, and others result from mutations in degenerate splicing consensus sites in sequences near intron-exon boundaries (Bonatti et al., 2006; Chen et al., 2006; Claes et al., 2003; Tesoriero et al., 2005). Mutations that result in exon skipping without maintaining the original translation reading frame inevitably generate premature stop codons, rendering mis-spliced messages subject to NMD.
There are at least two known examples of BRCA1 mutations falling within exonic splicing enhancers (ESEs). One, E1694X, would be classified as a nonsense mutation by conceptual translation, but the G>T transversion that generates the E169X mutation within exon 18 of BRCA1 disrupts a consensus ESE sequence, which likely explains the observed exon skipping (Liu et al., 2001). Additionally, the BRCA1 C64G mutation, often cited as a deleterious missense mutation, has been shown to be associated with skipping of BRCA1 exon 5, and also disrupts a consensus ESE motif (Yang et al., 2003).
A variety of BRCA1 splice variants have been found to accumulate normally in several tissues. Among those that result from exon skipping events and retain the translational reading frame are variants that skip exon 5, exon 11 (all of it or most of it), exons 2–10, exons 9–11, exons 14–17 and exons 14–18 (Orban et al., 2003). Of these, the variant lacking exon 11 is detected in other mammalian species. Additionally, a BRCA1 mRNA variant (BRCA1-IRIS) that lacks exons 12–24 but contains a short segment of coding sequence from intron 11 has been reported (ElShamy et al., 2004). It has been suggested that different BRCA1 splice forms are associated with breast and ovarian cancer, based on the observations that different relative levels of splice variants involving exons 9, 10 and 11 are seen in breast and ovarian cancer cell lines compared with normal or leukemia cell lines (Orban et al., 2001; Wilson et al., 1997).
Alternative splicing of KLF6 is affected by an intronic SNP, producing a novel transcript that changes cancer cell behavior
The Kruppel-like factor 6 (KLF6) gene is a zinc-finger-containing transcription factor and tumor suppressor that is inactivated by deletion or mutation in several cancer types, including prostate, colon, hepatocellular carcinoma, gastric, nasopharyngeal, ovarian and gliomas (DiFeo et al., 2008 and references therein). An intronic single nucleotide polymorphism (SNP), IVS –27 G>A/IVSΔA, generates a novel SRp40-binding site and obliterates two other overlapping SR-protein-binding sites, resulting in the increased production of three alternatively spliced transcripts, KLF6 SV1-3 (Narla et al., 2005a). The presence of this SNP confers an increased risk of prostate cancer. KLF6 SV1 and SV2 protein products are mislocalized to the cytoplasm, and the KLF6 SV1 protein product opposes the function of wild-type KLF6 (Narla et al., 2005b). Expression of KLF6 SV1 is increased in hormone-refractory metastatic prostate cancer and is associated with decreased survival in patients with lung adenocarcinoma (DiFeo et al., 2008; Narla et al., 2005b)
Changes in splicing factor levels within cancer cells alters gene splicing
Why are splice patterns different in normal and tumor cells? As splice patterns normally vary during embryogenesis and between tissue types, it is clear that splicing is regulated by responses to developmental cues. It is therefore easy to imagine that such signaling pathways may become altered in cancer cells. Indeed, there is a fascinating body of literature showing that some kinases and phosphatases known to play important roles in tumor development also play regulatory roles in the phosphorylation status of SR proteins, the serine-arginine-rich proteins that bind to exonic splicing enhancers (ESEs) to signal exon identity to the spliceosome. The phosphorylation status of SR proteins affects their function by regulating their subcellular localization and their protein-protein interactions. The kinases that regulate the phosphorylation status of SR proteins such as SRp40 include Akt, an environmentally regulated kinase that participates in numerous tumor types. Likewise, the phosphatases that affect the phosphorylation status of SR proteins such as SRp70, SRp55, SRp40 and ASF/SF2, include protein phosphatases PP1 and PP2A, and affect pathways that regulate apoptosis (reviewed by Blaustein et al., 2007). The SR proteins serve many functions, so effects on their phosphorylation states could have effects on gene splicing, mRNA export and/or mRNA translation.
Additionally, altering the levels of different splicing factors within a cell could change the ratio of particular splicing events. For example, SR proteins and hnRNPs are known to antagonize one another in a concentration-dependent manner. The end result of altered splicing ratios within a gene could be to produce proteins of altered function and to confer a growth or other advantage to the cell expressing them. Several examples of this type of dysregulated splicing occur in cancer cells, and recent evidence suggests that at least some of these events result in changes in cancer cell motility.
Ron encodes a tyrosine kinase receptor for macrophage-stimulating protein, which regulates cell dissociation, motility and invasive properties. Exclusion of exon 11 results in a transcript that encodes a constitutively active receptor, ΔRon (Collesi et al., 1996). Exclusion or inclusion of exon 11 within Ron transcripts is controlled through an ESE and an ESS, respectively, that are located in exon 12. The accumulation of ΔRon transcripts within breast and colon tumors correlates with the levels of SF2/ASF, a highly conserved SR protein (Ghigna et al., 2005). By regulating the amount of ΔRon produced within a cell, SF2/ASF controls the epithelial to mesenchymal transition leading to cell locomotion (Ghigna et al., 2005).
Interestingly, cell motility in breast cancer cells is also controlled by the levels of Gemin 5, a component of the spliceosomal complex. Lee and colleagues used isotope capture affinity tag proteomic analysis to compare the global protein expression patterns in MDA-MB-435 breast cancer cells versus the Nm23-H1 metastasis suppressor cells (Lee et al., 2008). Ingenuity pathway analysis revealed that the most prominent class of altered proteins were those involved in RNA post-transcriptional modification. Levels of Gemin 5, ACIN1, PABPC1, HNRPA2B1 and Bop1 were all higher in the less metastatic cells. Furthermore, expression of gemin5 was sufficient to alter the splicing pattern of the more metastatic MDA-MB-435 cells to that of the less metastatic cells. In addition, gemin5-expressing transfectants were less mobile than vector-expressing cells.
Altered splicing regulation and the tumor microenvironment
Signals from tumor microenvironments often play important roles in promoting tumor development and metastasis. Alternative splice variants of several genes are known to have different accumulation patterns in tumors and surrounding stromal tissues, and soluble signals from stromal cells and the extracellular matrix can influence epithelial cell transformation by altering splicing regulation. For example, the CD44 gene, which encodes a cell hyaluronate receptor, undergoes extensive alternative splicing both as part of its normal function and also in response to tumor-specific splicing regulation. Whereas the full-length CD44 mRNA protein product is detected at high levels in both colorectal cancer cells and surrounding stromal tissues, at least one splice variant (v6) is present in 77% of tumors but only in 17% of stromal cells (Furuta et al., 1998). Additionally, Tenascin-C (TN), an extracellular matrix glycoproteinprotein, exists as several splice variant isoforms, two of which are especially associated with an invasive phenotype. As these isoforms are also seen associated with a subset of ductal carcinomas in situ (DCIS), it is possible they could be used as predictors of aggressive tumor behavior (Adams et al., 2002). Fibronectin, another extracellular matrix protein that plays roles in cell adhesion and migration, also exists as multiple isoforms generated by alternative splicing. Growth factors produced by mammary mesenchymal cells can stimulate a mammary epithelial cell line to produce specific fibronectin mRNA splice variants especially associated with proliferation, migration and tissue remodeling (Blaustein et al., 2004). Similarly, MMP-3, a stromal protein that promotes malignant transformation in cultured mammary epithelial cells, has been found to promote the expression of an alternative splice form of the small GTPase rac1. This in turn causes an increase in cellular reactive oxygen species, resulting in DNA damage and genomic instability (Radisky et al., 2005). Collectively, these examples show that splicing of cancer-associated mRNAs may be quite different in epithelial and surrounding stromal cells, and that stromal signaling may cause changes in splicing regulation that result in aggressive tumor behavior.
Conclusions
Alternative splicing is one of the most important ways cells regulate the different protein products encoded by individual genes. Yet the complexity of the splicing regulatory mechanisms provides numerous targets for cancer-associated aberrations. In some cases, cancer-specific misregulation of one or a few genes acting early in tumorigenesis can be sufficient to initiate cancer development. In other cases, cancer-specific splicing alterations in genes that regulate expression of numerous downstream genes can contribute to a tumorigenic environment traditionally associated with high frequency mutation. As with gene expression patterns, tumor-associated alternative splicing patterns may now be examined with high-density microarrays and/or high-throughput DNA sequencing, which could be used to define tumor subtypes, provide high-resolution diagnostic tools, and even suggest targets for novel therapies. Such therapies could be directed at the splicing machinery itself, as it is subject to regulation by cell signaling pathways. Hypotheses generated by transcriptome-wide analyses of splicing events are now being tested in vivo to reveal the functional consequences of aberrant splice products of crucial tumor-associated genes.
ACKNOWLEDGEMENTS
We regret that, owing to space constraints, we were not able to describe other colleagues’ work that has defined the functional consequences of abnormal splicing of many more genes in cancer cells. We thank Jonathan Staley for his critical review and insightful comments on this manuscript.
Footnotes
COMPETING INTERESTS:
L.G. serves as a consultant for MGI Pharma, Inc.
References
- Adams M., Jones J. L., Walker R. A., Pringle J. H., Bell S. C. (2002). Changes in tenascin-C isoform expression in invasive and preinvasive breast disease. Cancer Res. 62, 3289–3297 [PubMed] [Google Scholar]
- Baudry D., Faussillon M., Cabanis M. O., Rigolet M., Zucker J. M., Patte C., Sarnacki S., Boccon-Gibod L., Junien C., Jeanpierre C. (2002). Changes in WT1 splicing are associated with a specific gene expression profile in Wilms’ tumour. Oncogene 21, 5566–5573 [DOI] [PubMed] [Google Scholar]
- Blaustein M., Pelisch F., Coso O. A., Bissell M. J., Kornblihtt A. R., Srebrow A. (2004). Mammary epithelial-mesenchymal interaction regulates fibronectin alternative splicing via phosphatidylinositol 3-kinase. J. Biol. Chem. 279, 21029–21037 [DOI] [PubMed] [Google Scholar]
- Blaustein M., Pelisch F., Srebrow A. (2007). Signals, pathways and splicing regulation. Int. J. Biochem. Cell Biol. 39, 2031–2048 [DOI] [PubMed] [Google Scholar]
- Blencowe B. J. (2006). Alternative splicing: new insights from global analyses. Cell 126, 37–47 [DOI] [PubMed] [Google Scholar]
- Bonatti F., Pepe C., Tancredi M., Lombardi G., Aretini P., Sensi E., Falaschi E., Cipollini G., Bevilacqua G., Caligo M. A. (2006). RNA-based analysis of BRCA1 and BRCA2 gene alterations. Cancer Genet. Cytogenet. 170, 93–101 [DOI] [PubMed] [Google Scholar]
- Caballero O. L., de Souza S. J., Brentani R. R., Simpson A. J. (2001). Alternative spliced transcripts as cancer markers. Dis. Markers 17, 67–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Truong T. T., Weaver J., Bove B. A., Cattie K., Armstrong B. A., Daly M. B., Godwin A. K. (2006). Intronic alterations in BRCA1 and BRCA2: effect on mRNA splicing fidelity and expression. Hum. Mutat. 27, 427–435 [DOI] [PubMed] [Google Scholar]
- Churbanov A., Rogozin I. B., Deogun J. S., Ali H. (2006). Method of predicting splice sites based on signal interactions. Biol. Direct 1, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claes K., Poppe B., Machackova E., Coene I., Foretova L., De Paepe A., Messiaen L.Center for Medical Genetics GUHBKCrab (2003). Differentiating pathogenic mutations from polymorphic alterations in the splice sites of BRCA1 and BRCA2. Genes Chromosomes Cancer 37(3), 314–320 [DOI] [PubMed] [Google Scholar]
- Collesi C., Santoro M. M., Gaudino G., Comoglio P. M. (1996). A splicing variant of the RON transcript induces constitutive tyrosine kinase activity and an invasive phenotype. Mol. Cell. Biol. 16, 5518–5526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFeo A., Feld L., Rodriguez E., Wang C., Beer D, G., Martignetti J. A., Narla G. (2008). A functional role for KLF6-SV1 in lung adenocarcinoma prognosis and chemotherapy response. Cancer Res. 68, 965–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ElShamy W. M., Livingston D. M. (2004). Identification of BRCA1-IRIS, a BRCA1 locus product. Nat. Cell Biol. 6, 954–967 [DOI] [PubMed] [Google Scholar]
- Fabbri M., Garzon R., Cimmino A., Liu Z., Zanesi N., Callegari E., Liu S., Alder H., Costinean S., Fernandez-Cymering C., et al. (2007). MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl. Acad. Sci. USA 104, 15805–15810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta K., Zahurak M., Goodman S. N., Hamilton S. R., August J. T. (1998). CD44 expression in the stromal matrix of colorectal cancer: association with prognosis. Clin. Cancer Res. 4, 21–29 [PubMed] [Google Scholar]
- Ghigna C., Giordano S., Shen H., Benvenuto F., Castiglioni F., Comoglio P. M., Green M. R., Riva S., Biamonti G. (2005). Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol. Cell 20, 881–890 [DOI] [PubMed] [Google Scholar]
- Hui L., Zhang X., Wu X., Lin Z., Wang Q., Li Y., Hu G. (2004). Identification of alternatively spliced mRNA variants related to cancers by genome-wide ESTs alignment. Oncogene 23, 3013–3023 [DOI] [PubMed] [Google Scholar]
- Kim E., Goren A., Ast G. (2008). Insights into the connection between cancer and alternative splicing. Trends Genet. 24, 7–10 [DOI] [PubMed] [Google Scholar]
- Klinck R., Bramard A., Inkel L., Dufresne-Martin G., Gervais-Bird J., Madden R., Paquet E. R., Koh C., Venables J. P., Prinos P., et al. (2008). Multiple alternative splicing markers for ovarian cancer. Cancer Res. 68, 657–663 [DOI] [PubMed] [Google Scholar]
- Lee J. H., Horak C. E., Khanna C., Meng Z., Yu L. R., Veenstra T. D., Steeg P. S. (2008). Alterations in Gemin5 expression contribute to alternative mRNA splicing patterns and tumor cell motility. Cancer Res. 68, 639–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. X., Cartegni L., Zhang M. Q., Krainer A. R. (2001). A mechanism for exon skipping caused by nonsense or missense mutations in BRCA1 and other genes. [see comments]. Nat. Genet. 27, 55–58 [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J.Department of Molecular, B. and Biochemistry, Y. U. S. o. M. C. A. N. H. C. T. U. S. A. j. e. m. y. e (2001). mRNA quality control: Marking the message for life or death. Curr. Biol. 11(3), R88–R91 [DOI] [PubMed] [Google Scholar]
- Maquat L. E. (2002). NASty effects on fibrillin pre-mRNA splicing: another case of ESE does it, but proposals for translation-dependent splice site choice live on. Genes Dev. 16, 1743–1753 [DOI] [PubMed] [Google Scholar]
- Moore M. J., Silver P. A. (2008). Global analysis of mRNA splicing. RNA 14, 197–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narla G., Difeo A., Reeves H. L., Schaid DJ., Hirshfeld J., Hod E., Katz A., Isaacs W. B., Hebbring S., Komiya A., et al. (2005a). A germline DNA polymorphism enhances alternative splicing of the KLF6 tumor suppressor gene and is associated with increased prostate cancer risk. Cancer Res. 65, 1213–1222 [DOI] [PubMed] [Google Scholar]
- Narla G., DiFeo A., Yao S., Banno A., Hod E., Reeves H. L., Qiao R. F., Camacho-Vanegas O., Levine A., Kirschenbaum A., et al. (2005b). Targeted inhibition of the KLF6 splice variant, KLF6 SV1, suppresses prostate cancer cell growth and spread. Cancer Res 65, 5761–5768 [DOI] [PubMed] [Google Scholar]
- Orban T. I., Olah E. (2001). Expression profiles of BRCA1 splice variants in asynchronous and in G1/S synchronized tumor cell lines. Biochem. Biophys. Res. Commun. 280, 32–38 [DOI] [PubMed] [Google Scholar]
- Orban T. I., Olah E. (2003). Emerging roles of BRCA1 alternative splicing. MP, Mol. Pathol. 56, 191–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostler K. R., Davis E. M., Payne S. L., Gosalia B. B., Exposito-Cespedes J., Le Beau M. M., Godley L. A. (2007). Cancer cells express aberrant DNMT3B transcripts encoding truncated proteins. Oncogene 26, 5553–5563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radisky D. C., Levy D. D., Littlepage L. E., Liu H., Nelson C. M., Fata J. E., Leake D., Godden E. L., Albertson D. G., Nieto M. A., et al. (2005). Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature 436, 123–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y., Kanai Y., Sakamoto M., Saito H., Ishii H., Hirohashi S. (2002). Overexpression of a splice variant of DNA methyltransferase 3b, DNMT3b4, associated with DNA hypomethylation on pericentromeric satellite regions during human hepatocarcinogenesis. Proc. Natl. Acad. Sci. USA 99, 10060–10065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skotheim R. I., Nees M. (2007). Alternative splicing in cancer: noise, functional, or systematic. Int. J. Biochem. Cell Biol. 39, 1432–1449 [DOI] [PubMed] [Google Scholar]
- Srebrow A., Kornblihtt A. R. (2006). The connection between splicing and cancer. J. Cell. Sci. 119, 2635–2641 [DOI] [PubMed] [Google Scholar]
- Tesoriero A. A., Wong E. M., Jenkins M. A., Hopper J. L., Brown M. A., Chenevix-Trench G., Spurdle A. B., Southey M. C. (2005). Molecular characterization and cancer risk associated with BRCA1 and BRCA2 splice site variants identified in multiple-case breast cancer families. Hum. Mutat. 26, 495. [DOI] [PubMed] [Google Scholar]
- Valentine C. R. (1998). The association of nonsense codons with exon skipping. Mutat. Res. 411, 87–117 [DOI] [PubMed] [Google Scholar]
- Volpe G., Cignetti A., Panuzzo C., Kuka M., Vitaggio K., Brancaccio M., Perrone G., Rinaldi M., Prato G., Fava M., et al. (2007). Alternative BCR/ABL splice variants in Philadelphia chromosome-positive leukemias result in novel tumor-specific fusion proteins that may represent potential targets for immunotherapy approaches. Cancer Res. 67, 5300–5307 [DOI] [PubMed] [Google Scholar]
- Wang J., Bhutani M., Pathak A. K., Lang W., Ren H., Jelinek J., He R., Shen L., Issa J. P., Mao L. (2007). Delta DNMT3B variants regulate DNA methylation in a promoter-specific manner. Cancer Res. 67, 10647–10652 [DOI] [PubMed] [Google Scholar]
- Wang J., Walsh G., Liu D. D., Lee J. J., Mao L. (2006a). Expression of Delta DNMT3B variants and its association with promoter methylation of p16 and RASSF1A in primary non-small cell lung cancer. Cancer Res. 66, 8361–8366 [DOI] [PubMed] [Google Scholar]
- Wang L., Wang J., Sun S., Rodriguez M., Yue P., Jang S. J., Mao L. (2006b). A novel DNMT3B subfamily, DeltaDNMT3B, is the predominant form of DNMT3B in non-small cell lung cancer. Int. J. Oncol. 29, 201–207 [PubMed] [Google Scholar]
- Wilson C. A., Payton M. N., Elliott G. S., Buaas F. W., Cajulis E. E., Grosshans D., Ramos L., Reese D. M., Slamon D. J., Calzone F. J. (1997). Differential subcellular localization, expression and biological toxicity of BRCA1 and the splice variant BRCA1-delta11b. Oncogene 14, 1–16 [DOI] [PubMed] [Google Scholar]
- Xu Q., Lee C. (2003). Discovery of novel splice forms and functional analysis of cancer-specific alternative splicing in human expressed sequences. Nucleic Acids Res. 31, 5635–5643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Swaminathan S., Martin B. K., Sharan S. K. (2003). Aberrant splicing induced by missense mutations in BRCA1: clues from a humanized mouse model. Hum. Mol. Genet. 12, 2121–2131 [DOI] [PubMed] [Google Scholar]
- Zhang M. Q. (1998). Statistical features of human exons and their flanking regions. Hum. Mol. Genet. 7, 919–932 [DOI] [PubMed] [Google Scholar]