Abstract
Several different genetic variants at chromosome 8q24 have been related to prostate, breast and colorectal cancer risk with evidence of region-specific risk differentials for various tumor types. We investigated the association between 15 polymorphisms located in 8q24 regions associated with cancer risk in a pooled analysis of 2587 colorectal adenoma cases, 547 colorectal cancer cases and 2798 controls of European descent from four studies. Logistic regression was used to estimate odds ratios (ORs) and 95% confidence intervals (95% CIs) for the associations. Three polymorphisms (rs10808555, rs6983267 and rs7837328) located between 128.47 and 128.54 Mb were found to be associated with colorectal tumor risk. The association was strongest for the previously reported rs6983267 variant and was similar for both adenoma (ORper allele = 1.16, 95% CI: 1.07–1.25, P = 0.0002) and cancer (OR per allele = 1.17, 95% CI: 1.01–1.35, P = 0.03). The strength of the association of the regional haplotype containing variant alleles at rs10808555, rs6983267 and rs7837328 but not rs10505476 was greater than that of any single variant of both adenoma (OR = 1.27, P = 0.0001) and cancer (OR = 1.26, P = 0.03). The risk associated with rs6983267 was stronger for multiple adenomas (ORper allele = 1.29, P = 5.6 × 10−6) than for single adenoma (ORper allele = 1.10, P = 0.03) with Pheterogeneity = 0.008. This study confirms the association between colorectal neoplasia and the 8q24 polymorphisms located between 128.47 and 128.54 Mb and suggests a role for these variants in the formation of multiple adenomas.
INTRODUCTION
Genetic factors are thought to contribute to colorectal cancer susceptibility as supported by studies showing a 2-fold increased risk of colorectal cancer for individuals with at least one affected first-degree relative and a 4-fold risk for persons with at least two affected relatives (reviewed in 1). Rare, highly penetrant germline mutations have been linked to the development of colorectal cancer syndromes, such as Lynch syndrome, also called hereditary nonpolyposis colorectal cancer, and familial adenomatous polyposis; however, these syndromes account for only a small fraction (3–5%) of all colorectal cancer cases (2). Lower penetrance genetic variants are likely to play a role in the other colorectal cancer cases.
Recently, common polymorphisms located at chromosome 8q24 have been associated with the risk of prostate, breast and colorectal tumors (3–12). Variants in at least three (possibly four) distinct regions have been identified as being independently associated with risk of prostate cancer (3–8). A separate region has been associated with breast cancer risk (9), suggesting a complex contribution of regions in 8q24 to different types of cancer. Simultaneously, three reports identified an association between colorectal cancer and rs6983267, the same variant that had been associated with prostate cancer, as well as a highly correlated polymorphism, rs10505477 (10–12); one of these studies also reported an association with adenoma (10). Initial studies suggested that the association with colorectal cancer may be confined to one region (10,12); however, evidence for regional specificity with colorectal adenoma is sparse, and differences in adenoma subtypes have not been examined. Although the mechanism by which these variants increase risk is not understood, somatic gains in copy number of 8q24 have been observed in 18% of colorectal adenomas and 34% of colorectal cancers (13), suggesting that the region plays an important role in colorectal carcinogenesis.
To determine whether independent loci in multiple 8q24 regions are associated with colorectal neoplasia risk and whether the risk could differ by clinical characteristics of adenomas, we examined a set of single nucleotide polymorphisms (SNPs) from chromosome 8q24 in a large pooled analysis of colorectal adenoma cases and controls and in a smaller series of colorectal cancers. Fifteen SNPs were chosen to interrogate the previously reported regions associated with the risk of prostate, breast or colorectal cancer in 8q24.
RESULTS
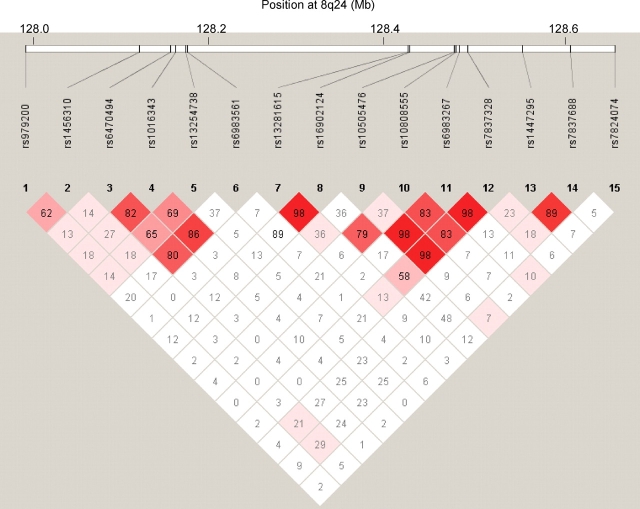
Fifteen SNPs across a 1 Mb region in 8q24 were examined in 547 colorectal cancer cases, 2587 colorectal adenoma cases and 2798 controls of European descent. Three SNPs located between 128.47 and 128.54 Mb (rs10808555, rs6983267 and rs7837328) were associated with colorectal adenoma risk in the pooled analysis (Table 1). Based on the Ptrend, the association was strongest for rs6983267 with an increased risk of 1.18 (95% CI: 1.03–1.35) for heterozygotes and 1.35 (95% CI: 1.16–1.57) for GG homozygotes compared with the TT homozygotes (Ptrend = 0.0002). The three polymorphisms are in strong linkage disequilibrium (Fig. 1) and moderately correlated (r2 range: 0.34–0.66); mutual adjustment attenuated the association for all three polymorphisms. Inclusion of the fourth polymorphism in the region (rs10505476) strengthened the magnitude of the association slightly for rs6983267 (ORper allele = 1.20, 95% CI: 1.08–1.34) and rs7837328 (ORper allele = 1.25, 95% CI: 1.11–1.41) but not rs10808555 (ORper allele = 1.15, 95% CI: 1.05–1.27). Haplotype analysis of the four SNPs genotyped in the 8q24 region between 128.47 and 128.54 Mb revealed that the haplotype containing the variant allele at rs10808555, rs6983267 and rs7837328, but not rs10505476, was associated with an increased risk of adenoma (ORper C-G-G-A haplotype = 1.27, 95% CI: 1.13–1.43, Table 2). Based on the odds ratio (OR) and P-value, the strength of the association for the haplotype was greater than that of any single variant (ORper C-G-G-A haplotype = 1.27, P = 0.0001 versus ORper G allele at rs6983267 = 1.16, P = 0.0002), and if the haplotypes not associated with risk were collapsed into one category, the haplotype analysis was found to provide a better fit for the data than rs6983267 based on the Akaike information criterion (AIChaplotype = 7322.9 versus AICrs6983267 = 7324.3).
Table 1.
Odds ratio (OR)a of colorectal neoplasia for polymorphisms between 127.9 and 128.7 Mb at chromosome 8q24 among Caucasians
| SNP | Risk allele | MAFb | Colorectal cancer |
Colorectal adenoma |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PLCO 547 cases, 1656 controls | P-value | PLCO I 1174 cases, 1293 controls | PLCO II 364 cases, 363 controls | NHS 544 cases, 542 controls | Minnesota 505 cases, 600 controls | Pooled OR (95% CI) | P-value | |||
| rs979200 G > A | A | 0.35 | 1.01 (0.87–1.18) | 0.89 | 1.04 (0.92–1.17) | 0.99 (0.80–1.22) | 1.16 (0.97–1.38) | 1.12 (0.93–1.34) | 1.06 (0.98–1.15) | 0.13 |
| rs1456310 C > T | T | 0.42 | 1.05 (0.91–1.22) | 0.51 | 0.99 (0.88–1.11) | 0.82 (0.67–1.01) | 1.16 (0.98–1.38) | 1.18 (0.98–1.42) | 1.04 (0.96–1.12) | 0.38 |
| rs6470494 G > A | A | 0.28 | 0.92 (0.78–1.08) | 0.31 | 1.02 (0.90–1.16) | 1.01 (0.80–1.28) | 1.00 (0.83–1.21) | 1.01 (0.83–1.23) | 1.02 (0.94–1.11) | 0.61 |
| rs1016343 G > A | A | 0.20 | 0.97 (0.81–1.15) | 0.72 | 0.96 (0.83–1.10) | 0.99 (0.76–1.29) | 1.02 (0.81–1.27) | 0.97 (0.78–1.21) | 0.98 (0.89–1.08) | 0.70 |
| rs13254738 T > G | G | 0.31 | 0.96 (0.82–1.11) | 0.56 | 1.02 (0.90–1.15) | 0.86 (0.68–1.08) | 1.03 (0.85–1.24) | 1.12 (0.92–1.35) | 1.02 (0.94–1.11) | 0.64 |
| rs6983561 T > G | G | 0.03 | 0.94 (0.61–1.44) | 0.76 | 1.13 (0.81–1.55) | 1.34 (0.76–2.35) | 1.08 (0.67–1.74) | 0.43 (0.24–0.76) | 0.98 (0.78–1.21) | 0.83 |
| rs13281615 A > G | G | 0.41 | 1.08 (0.93–1.24) | 0.31 | 1.02 (0.91–1.14) | 1.01 (0.81–1.24) | 0.98 (0.83–1.17) | 0.89 (0.74–1.07) | 0.98 (0.91–1.06) | 0.62 |
| rs16902124 G > A | A | 0.04 | 1.56 (1.11–2.18) | 0.01 | 1.15 (0.85–1.55) | 0.74 (0.43–1.27) | 0.95 (0.61–1.49) | 0.88 (0.56–1.40) | 0.98 (0.80–1.20) | 0.83 |
| rs10505476c C > T | T | 0.26 | 1.02 (0.86–1.20) | 0.85 | 1.05 (0.92–1.19) | 0.98 (0.77–1.25) | – | 1.20 (0.98–1.47) | 1.07 (0.97–1.18) | 0.18 |
| rs10808555c A > G | G | 0.32 | 1.13 (0.97–1.31) | 0.12 | 1.20 (1.06–1.35) | 1.12 (0.90–1.40) | – | 1.15 (0.96–1.39) | 1.16 (1.06–1.28) | 0.001 |
| rs6983267 T > G | G | 0.49 | 1.17 (1.01–1.35) | 0.03 | 1.24 (1.11–1.39) | 0.93 (0.75–1.16) | 1.10 (0.93–1.30) | 1.21 (1.01–1.44) | 1.16 (1.07–1.25) | 0.0002 |
| rs7837328 G > A | A | 0.40 | 1.12 (0.97–1.30) | 0.11 | 1.21 (1.08–1.36) | 0.98 (0.79–1.20) | 1.04 (0.87–1.23) | 1.24 (1.04–1.49) | 1.14 (1.05–1.23) | 0.001 |
| rs1447295 C > A | A | 0.10 | 0.87 (0.68–1.10) | 0.24 | 1.08 (0.90–1.31) | 0.85 (0.61–1.20) | 1.06 (0.81–1.39) | 0.99 (0.74–1.31) | 1.02 (0.90–1.16) | 0.74 |
| rs7837688 G > T | T | 0.10 | 0.93 (0.73–1.19) | 0.57 | 1.04 (0.86–1.26) | 0.88 (0.63–1.24) | 0.98 (0.75–1.28) | 1.05 (0.78–1.40) | 1.00 (0.88–1.13) | 0.97 |
| rs7824074 G > T | T | 0.29 | 0.98 (0.84–1.15) | 0.81 | 1.06 (0.94–1.20) | 0.93 (0.73–1.18) | 0.95 (0.78–1.15) | 0.95 (0.78–1.16) | 1.00 (0.92–1.09) | 0.97 |
aOR per risk allele assuming a log-additive model, adjusted for age, sex, and study (where appropriate).
bMAF, minor allele frequency among controls.
cThe assay for this SNP failed within the NHS samples, reducing the sample size for the pooled analysis of adenoma risk to 2043 cases and 2256 controls.
Figure 1.
Pairwise linkage disequilibrium (D′) among the genotyped 8q24 polymorphisms in Caucasian controls from all studies. Diamond boxes represent the pairwise D′ estimates between polymorphisms.
Table 2.
Odds ratio of colorectal neoplasia for haplotypes between 128.47 and 128.54 Mb at chromosome 8q24 among Caucasians
| Haplotypea | Freqb | Colorectal cancer |
Colorectal adenoma |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PLCO 547 cases, 1656 controls | P-value | PLCO I 1174 cases, 1293 controls | PLCO II 364 cases, 363 controls | Minnesota 505 cases, 600 controls | Pooled OR w/o NHS (95% CI) | P-value | Pooled OR w/ NHSc (95% CI) | P-value | ||
| C-A-T-G | 0.48 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | – | 1.0 | ||
| C-A-G-G | 0.09 | 1.18 (0.92–1.52) | 0.20 | 1.20 (0.98–1.48) | 0.87 (0.59–1.29) | 1.05 (0.75–1.48) | 1.12 (0.96–1.32) | 0.16 | 1.15 (1.00–1.32) | 0.06 |
| C-G-T-G | 0.03 | 1.01 (0.61–1.67) | 0.98 | 1.01 (0.70–1.47) | 4.07 (1.63–10.18) | 1.19 (0.60–2.34) | 1.23 (0.92–1.65) | 0.17 | 1.22 (0.91–1.64) | 0.18 |
| C-G-G-A | 0.14 | 1.26 (1.02–1.55) | 0.03 | 1.42 (1.20–1.68) | 1.04 (0.77–1.41) | 1.25 (0.96–1.63) | 1.29 (1.14–1.47) | 0.00008 | 1.27 (1.13–1.43) | 0.0001 |
| T-A-G-A | 0.11 | 1.07 (0.83–1.38) | 0.60 | 1.14 (0.94–1.39) | 1.07 (0.74–1.54) | 1.29 (0.96–1.74) | 1.17 (1.00–1.35) | 0.05 | 1.14 (0.99–1.32) | 0.07 |
| T-G-G-A | 0.15 | 1.12 (0.91–1.39) | 0.28 | 1.18 (1.00–1.39) | 0.95 (0.68–1.31) | 1.26 (0.96–1.64) | 1.15 (1.01–1.31) | 0.03 | 1.13 (1.00–1.27) | 0.05 |
aHaplotypes are comprised of rs10505476, rs10808555, rs6983267 and rs7837328, respectively.
bHaplotype frequency among controls.
cAs genotype data was not available for all SNPs in the NHS, haplotype frequencies for NHS estimated using the genotype data from NHS as well as the other studies.
Table 3 shows the risks associated with the four polymorphisms in the region between 128.47 and 128.54 Mb by adenoma tumor multiplicity. The association with rs6983267 was stronger for individuals with multiple adenomas (ORper allele = 1.29, 95% CI: 1.16–1.45) than those with single adenomas (ORper allele = 1.10, 95% CI: 1.01–1.20) with Pheterogeneity = 0.008. Similarly, the association with the haplotype containing the variant allele at rs10808555, rs6983267 and rs7837328, but not rs10505476, was stronger for multiple adenomas (ORper C-G-G-A haplotype = 1.38, 95% CI: 1.16–1.63) than for single adenomas (ORper C-G-G-A haplotype = 1.22, 95% CI: 1.06–1.40). No statistically significant heterogeneity was observed with rs6983267 or the haplotypes for large (≥1 cm) versus small (<1 cm), advanced versus non-advanced, or proximal versus distal adenoma. With regard to colorectal adenoma risk, no statistically significant interactions were found with rs6983267 or the haplotypes and smoking, family history, age (<60 versus ≥60), or sex.
Table 3.
Odds ratio (OR)a of colorectal adenoma of four SNPs between 128.47 and 128.54 Mb at chromosome 8q24 among Caucasians stratified by adenoma multiplicity
| SNP | Risk allele | MAFb | Single adenoma |
Multiple adenomas |
Pheterogeneity | ||
|---|---|---|---|---|---|---|---|
| Pooled OR (95% CI) | P-value | Pooled OR (95% CI) | P-value | ||||
| rs10505476 C > T | T | 0.26 | 1.03 (0.92–1.15) | 0.65 | 1.14 (1.00–1.30) | 0.05 | 0.16 |
| rs10808555 A > G | G | 0.32 | 1.15 (1.04–1.28) | 0.008 | 1.19 (1.05–1.34) | 0.005 | 0.64 |
| rs6983267 T > G | G | 0.49 | 1.10 (1.01–1.20) | 0.03 | 1.29 (1.16–1.45) | 5.6 × 10−6 | 0.008 |
| rs7837328 G > A | A | 0.40 | 1.10 (1.00–1.20) | 0.04 | 1.24 (1.11–1.38) | 0.0002 | 0.05 |
aOR per risk allele assuming a log-additive model, adjusted for age, sex, and study (where appropriate).
bMAF, minor allele frequency among controls.
The rs6983267 variant was also associated with an increased risk of colorectal cancer (Table 1) with ORs of 1.25 (95% CI: 0.97–1.62) and 1.37 (95% CI: 1.03–1.83) for the heterozygotes and GG homozygotes, respectively, compared with the TT homozygotes (Ptrend = 0.03). The ORs associated with rs10808555 and rs7837328 for colorectal cancer were similar in magnitude to those observed for adenoma (Table 1); however, the sample size for colorectal cancer was smaller and the corresponding P-values did not reach statistical significance. Haplotype analysis of the polymorphisms in the region showed that the haplotype containing the variant allele at rs10808555, rs6983267 and rs7837328, but not rs10505476, was associated with an increased risk of cancer (Table 2) that was stronger than the association for any single variant in the region (ORper C-G-G-A haplotype = 1.26 versus ORper G allele at rs6983267 = 1.17). The association with rs6983267 and colorectal cancer did not differ significantly by anatomical location (proximal versus distal), and no statistically significant interactions were found with smoking, family history, age, or gender.
The A variant at rs16902124 (located in region spanning 128.14 to 128.28 Mb) was also found to be associated with an increased risk of colorectal cancer (ORper allele = 1.56, 95% CI: 1.11–2.18). Adjustment for the polymorphisms in the neighboring region (rs10505476, rs10808555, rs6983267 and rs7837328) strengthened the association slightly (ORper allele = 1.65, 95% CI: 1.15–2.36); however, the rs16902124 variant was relatively infrequent in this population (3.8% among controls), and no association was observed with adenoma (ORper allele = 0.98, 95% CI: 0.80–1.20).
DISCUSSION
This study confirms the previously observed association between rs6983267 and the risk of colorectal neoplasia and explores additional genetic variation in the 8q24 region for colorectal cancer and adenoma. Several studies have replicated the increased risk of colorectal cancer observed in the initial reports with the G variant at rs6983267 and/or the T allele at rs10505477, a nearby SNP highly correlated (r2 = 0.94 among Centre d'Etude du Polymorphisme Humain individuals in HapMap) with rs6983267 (14–17); however, this is the first study to confirm the association with colorectal adenoma. Tomlinson et al. (10) found that the G allele at rs6983267 was associated with an increased risk of colorectal adenoma with a comparable OR (OR = 1.22, 95% CI: 1.10–1.34) with that observed in our study (OR = 1.16, 95% CI: 1.07–1.25). Similar to previous studies (10,12), we did not find an association with variants outside the 8q24 region spanning 128.47 and 128.54 Mb. Although smoking is a major risk factor for adenoma (18), we observed no statistically significant differences in the risk associated with rs6983267 by smoking status. Similarly, we did not find any statistically significant interactions with age, sex, or family history, and risk did not differ appreciably by tumor location.
When examining haplotypes within the region surrounding rs6983267 from 128.47 to 128.54 Mb, we found that the haplotype containing the variant allele at rs10808555, rs6983267 and rs7837328, but not rs10505476, was associated with a greater increased risk of both colorectal adenoma and cancer than the G variant at rs6983267 alone, suggesting that perhaps the haplotype captures the risk associated with this 8q24 region better than the single variant rs6983267. However, other haplotypes were also associated with a borderline increased risk of adenoma. Similarly, Tomlinson et al. (10) reported that inclusion of rs10505477 improved the model fit for rs6983267.
The association for both the rs6983267 variant and haplotype was stronger for persons with multiple adenomas, who may be at higher risk of colorectal cancer (19), than those with single adenoma. Although this subgroup finding needs to be replicated in other studies, it suggests that the 8q24 variant may play a role in adenoma initiation similar to, but not nearly as strong as, mutations in APC, which accelerate adenoma formation through the accumulation of β-catenin and increased transcription of MYC as a part of the Wnt signaling pathway (20). MYC is amplified in ∼32% of colorectal cancers (21). Although MYC is >300 kb away from rs6983267, in light of the central role of MYC in colorectal carcinogenesis, it is plausible that common genetic variants could alter distant regulatory elements of MYC expression.
The rs6983267 polymorphism is located ∼15kb upstream of the processed pseudogene, POU5F1P1, which is a retrotransposed copy of the POU-domain transcription factor gene, POU5F1, with 97.5% shared identity (22). POU5F1 (otherwise known as OCT4) plays a critical role in maintaining stem cell pluripotency, self-renewal and chromatin structure (23), and has been shown to promote tumor growth in a dose-dependent manner (24). A conserved POU5F1-binding site in the 5′ promoter region of the WNT-signaling gene, FZD5, has been reported (25). Transcription of OCT4 pseudogenes, including POU5F1P1, has been observed in colorectal cancer tissues and cell lines (10,22), and at least one mouse OCT4 pseudogene has been shown to mediate stem cell regulatory function (26), suggesting that OCT4 pseudogenes may play a role in regulating stem cell proliferation and/or OCT4 activity. Tomlinson et al. (10) did not find evidence that POU5F1P1 expression levels in a panel of colorectal cancer cell lines and tumors varied with rs6983267 genotype; however, as somatic gains of 8q24 are present in approximately one-third of colorectal cancers (13), the expression levels observed may have been more reflective of somatic changes than of any effect of the genotypes.
In conclusion, this large pooled study confirms the previously reported association between colorectal neoplasia risk and polymorphisms found in the 8q24 region located between 128.47 and 128.54 Mb. In contrast to the multiple 8q24 regions observed to be associated with prostate cancer risk, but consistent with current evidence suggesting only a primary 8q24 region for breast cancer, our study also indicates only a single locus for colorectal neoplasia. This study also suggest that the variants in the 8q24 region between 128.47 and 128.54 Mb are more strongly associated with the risk of multiple adenomas than that of single adenoma, potentially indicating a role in accelerating adenoma formation, which may have implications for screening. Additional studies are needed to locate the specific causal variant(s) in this region and to determine the biological mechanism by which colorectal neoplasia risk is increased.
MATERIALS AND METHODS
Study subjects
PLCO Trial
The prostate, lung, colorectal, and ovarian (PLCO) cancer screening trial is a randomized trial of ∼155,000 men and women, aged 55–74, enrolled during 1993–2001 from 10 US centers, to determine whether screening reduces the mortality from PLCO cancers (27). Participants randomized to the screening arm underwent a 60 cm flexible sigmoidoscopy examination at study entry (T0) and at year 3 or year 5 of the study (T3/T5; protocol for the second sigmoidoscopy changed from year 3 to year 5 in 1999). Subjects with screen-detected abnormalities were referred to their personal physician for diagnostic follow-up, and PLCO staff subsequently abstracted medical records pertaining to the diagnostic work-ups, identifying pathologically verified cases of colorectal adenoma and cancer (28). Subjects in both arms of the trial were sent annual questionnaires asking about recent cancer diagnoses. Medical records were obtained for all colorectal cancers reported on the questionnaire or through death certificates, and all colorectal cancers were pathologically verified. The trial was approved by the institutional review boards of the National Cancer Institute and the 10 screening centers.
Participants were eligible for this study if they: (i) consented to participate in PLCO ancillary epidemiologic studies of cancer and related diseases; (ii) completed a risk factor questionnaire; (iii) provided a biologic sample (blood sample in the screened arm or buccal cell sample in the control arm) (29); and (iv) reported a negative history of selected diseases related to colorectal tumors (i.e. inflammatory bowel disease, colorectal polyps or polyposis syndrome, or cancer – except basal cell skin cancer). For the first study (PLCO I), we identified 1463 participants from the screening arm with advanced colorectal adenoma (≥1 cm in size, containing villous/tubulovillous characteristics, high-grade dysplasia or carcinoma in situ) of the distal colon or rectum from the T0 exam. For the second study (PLCO II), we identified 407 participants who screened negative on the T0 exam but were identified with colorectal adenoma of the distal colon or rectum (≥0.5 cm in size, displaying villous/tubulovillous characteristics high-grade dysplasia or carcinoma in situ) on the T3/T5 exam. From those eligible, 651 colorectal cancer cases (355 cases from the screening arm and 296 from the control arm of the trial) were also identified. Controls for the first case–control study (PLCO I) were 1631 participants who had a successful baseline screening exam that was negative for polyps in the distal colon and rectum, and controls for the second study (PLCO II) were 407 participants who had a successful T0 (baseline) and T3/T5 (follow-up) sigmoidoscopic exam, both negative for adenomatous polyps of the distal colon and rectum. Controls were frequency-matched to the respective case series on ethnicity, gender, and for a subset, on age (55–59, 60–64, 65–69, 70–74 years). All controls were used for analyses involving the colorectal cancer cases.
Minnesota study
Details of this case–control study have been described elsewhere (30). In brief, cases with adenomatous or hyperplastic polyps and polyp-free controls were recruited through a large, multi-clinic gastroenterology practice in metropolitan Minneapolis. All patients aged 30–74 years, who were scheduled for colonoscopy from April 1991 to April 1994, were screened for eligibility and recruited prior to colonoscopy. Indications for colonoscopy included bleeding, diagnostic follow-up, screening and family history. Eligibility criteria included English-speaking, residence in the Twin Cities metropolitan area, no known inherited syndrome associated with increased risk of colorectal neoplasia, no prior diagnosis of cancer (except non-melanoma skin cancer) or colorectal polyps, and no history of inflammatory bowel disease. Patients whose colonoscopy did not reach the cecum were ineligible; removed polyps were examined histologically using standard diagnostic criteria (31). Cases were subjects found to have adenoma at the time of colonoscopy (N = 530); controls were subjects without adenomatous or hyperplastic polyps at colonoscopy (N = 649). This study was approved by the institution review boards at the clinical centers and the University of Minnesota, and written informed consent was obtained from all participants.
Nurses' Health Study
The Nurses' Health Study (NHS) is a longitudinal cohort that began in 1976 when 121 700 female registered nurses between the ages of 30 and 55 years in the US completed a self-administered questionnaire on baseline health-related exposures and medical history. Subsequently, participants completed mailed, self-administered questionnaires biennially, which asked for updated medical history and other risk factor information. In 1989 and 1990, 32 826 women, between the ages of 43 and 69 years, provided blood samples for use in etiologic studies.
As previously described (32), colorectal adenoma cases and controls were selected from among women who provided a blood sample, had a sigmoidoscopy or colonoscopy between 1989 and 1998, and had no previous diagnosis of ulcerative colitis, adenoma, or cancer (except non-melanoma skin cancer) prior to the reported endoscopy. Cases included participants who reported a diagnosis of a polyp (after the date of blood draw) on any of the biennial questionnaires through 1998 that was confirmed to be adenomatous by review of pathology reports (N = 557). One case was subsequently found to be hyperplastic and excluded from the study, leaving 556 cases. Controls were subjects who reported having a negative sigmoidoscopy or colonoscopy (N = 557). One control was selected for each case, matched on year of birth, year of blood draw, time period of endoscopy, routine screening, gastrointestinal symptoms, and indication for endoscopy and time period of first or most recent endoscopy. This study was approved by the institutional review board at the Brigham and Women's Hospital.
Adenoma and cancer phenotypes
Colorectal adenoma and cancer subtypes were based on pathology reports. Both adenoma and cancer cases were classified by location (proximal colon, distal colon, rectum or multiple locations). Adenomas were also classified by size (<1 cm or ≥1 cm), histology (tubular, tubulovillous, villous, or not specified), and number (single versus multiple). Advanced adenoma was defined as adenoma ≥1 cm in size or tubulovillous/villous characteristics or displaying high-grade dysplasia or carcinoma in situ.
Genotyping
Fifteen common SNPs from regions in chromosome 8q24 were selected for genotyping based on suggestive or strong associations with prostate, breast, or colorectal cancer in one or more genome-wide association studies or high correlation (r2 > 0.8) with a SNP previously reported to be associated. The following SNPs were associated with disease: for breast cancer, rs13281615 (9) and an additional SNP in linkage disequilibrium, rs16902124; for colorectal and prostate cancer, rs6983267 (5,7,10–12) and three additional SNPs in linkage disequilibrium, rs10505476, rs10808555 and rs7837328; and for prostate cancer, rs1447295 (3) and an additional SNP in linkage disequilibrium, rs7837688. Two SNPs (rs13254738 and rs6983561) were chosen on the basis of the association observed in non-Caucasians populations for prostate cancer risk (7). Five additional SNPs were selected because of observed significance in the Cancer Genetic Markers of Susceptibility breast or prostate cancer genome-wide scans: rs979200 (breast), rs1456310 (prostate), rs6470494 (prostate), rs1016343 (breast and prostate), and rs7824074 (prostate).
Previously extracted germline DNA was available for 621 colorectal cancer cases, 1666 adenoma cases, and 1821 controls in PLCO, 554 adenoma cases and 553 controls in NHS, and 518 adenoma cases and 618 controls in the Minnesota study. All SNPs were genotyped using the TaqMan assay system (ABI, Foster City, CA, USA) (see Supplementary Material, Table S1 for details). Genotyping was successfully completed for 15 SNPs of the PLCO and Minnesota samples with average completion rates of 98.7 and 99.3%, respectively, and for 13 SNPs of the NHS samples with an average completion rate of 97.4%. The assays for two SNPs (rs10505476 and rs10808555) failed within the NHS samples. Replicate quality control samples included in the studies yielded >99% concordance for all SNPs. All genotype frequencies for controls by race were consistent with Hardy–Weinberg proportions (P > 0.05), except rs6983561 for whites in the Minnesota study (P = 0.05) and rs10808555 for blacks in PLCO (P = 0.03).
Statistical analysis
Analyses were restricted to Caucasians (n = 5,932, 93% of subjects), as the other ethnic groups (African-American, n = 195; Asian, n = 127; Other races, n = 97) were too small to draw meaningful conclusions from their results. Logistic regression was used to estimate the ORs and 95% confidence intervals (95% CIs) for the association between each SNP and colorectal neoplasm, adjusting for age, sex, and study. Although multiple genetic models were explored, the log-additive model proved to be a reasonable model for most SNPs and was tested by including a single variable coded as the number of variant alleles in the regression model.
Polytomous logistic regression was used to evaluate the association between the SNPs and adenoma subtypes, and the Wald test was used to assess heterogeneity among subtypes. Heterogeneity between studies and interactions between the SNPs and other covariates were assessed by including the cross-product terms as well as the main effect terms in logistic regression models, and the statistical significance of the interaction was evaluated by comparing nested models with and without the cross-product terms using a likelihood ratio test.
Pairwise linkage disequilibrium measures (D′ and r2) were estimated using the program, Haploview (http://www.broad.mit.edu/personal/jcbarret/haploview/). Haplotypes were estimated using an expectation-maximization algorithm (33) for SNPs within the same region, and risks for individual haplotypes were calculated assuming a log-additive model for each haplotype and using the generalized linear model implemented in HaploStats (34,35), adjusting for age, sex, and study. The most common haplotype was used as the referent.
All statistical analyses, except the linkage disequilibrium and haplotype analyses, were conducted using STATA 7.0 (College Station, TX, USA).
SUPPLEMENTARY MATERIAL
FUNDING
This study was supported by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health; National Cancer Institute, National Institutes of Health under contract N01-CO-12400; and National Institutes of Health research grants CA87969, CA55075 and CA059045.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Drs Christine Berg and Philip Prorok, Division of Cancer Prevention, NCI, the screening center investigators and staff of the PLCO Cancer Screening Trial, Mr Thomas Riley and staff at Information Management Services, Inc., and Ms Barbara O'Brien and staff at Westat, Inc. for their contributions to the PLCO Cancer Screening Trial. The authors also thank Mr John Whitton, Ms Lisa Fosdick, and Dr Roberd M. Bostick for their work on the Minnesota case–control study. In addition, we wish to acknowledge the study participants for spending their time and making this study possible.
Conflict of Interest statement. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services or does mention trade names, commercial products, or organizations indicate endorsement by the US Government.
REFERENCES
- 1.Butterworth A.S., Higgins J.P., Pharoah P. Relative and absolute risk of colorectal cancer for individuals with a family history: a meta-analysis. Eur. J. Cancer. 2006;42:216–227. doi: 10.1016/j.ejca.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 2.Kinzler K.W., Vogelstein B. In: The Genetic Basis of Human Cancer. Vogelstein B., Kinzler K.W., editors. New York: McGraw-Hill Companies, Inc.; 2002. pp. 583–612. [Google Scholar]
- 3.Amundadottir L.T., Sulem P., Gudmundsson J., Helgason A., Baker A., Agnarsson B.A., Sigurdsson A., Benediktsdottir K.R., Cazier J.B., Sainz J., et al. A common variant associated with prostate cancer in European and African populations. Nat. Genet. 2006;38:652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 4.Freedman M.L., Haiman C.A., Patterson N., McDonald G.J., Tandon A., Waliszewska A., Penney K., Steen R.G., Ardlie K., John E.M., et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc. Natl Acad. Sci. USA. 2006;103:14068–14073. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yeager M., Orr N., Hayes R.B., Jacobs K.B., Kraft P., Wacholder S., Minichiello M.J., Fearnhead P., Yu K., Chatterjee N., et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 6.Gudmundsson J., Sulem P., Manolescu A., Amundadottir L.T., Gudbjartsson D., Helgason A., Rafnar T., Bergthorsson J.T., Agnarsson B.A., Baker A., et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat. Genet. 2007;39:631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 7.Haiman C.A., Patterson N., Freedman M.L., Myers S.R., Pike M.C., Waliszewska A., Neubauer J., Tandon A., Schirmer C., McDonald G.J., et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat. Genet. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng S.L., Sun J., Cheng Y., Li G., Hsu F.C., Zhu Y., Chang B.L., Liu W., Kim J.W., Turner A.R., et al. Association between two unlinked loci at 8q24 and prostate cancer risk among European Americans. J. Natl Cancer Inst. 2007;99:1525–1533. doi: 10.1093/jnci/djm169. [DOI] [PubMed] [Google Scholar]
- 9.Easton D.F., Pooley K.A., Dunning A.M., Pharoah P.D., Thompson D., Ballinger D.G., Struewing J.P., Morrison J., Field H., Luben R., et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomlinson I., Webb E., Carvajal-Carmona L., Broderick P., Kemp Z., Spain S., Penegar S., Chandler I., Gorman M., Wood W., et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat. Genet. 2007;39:984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 11.Zanke B.W., Greenwood C.M., Rangrej J., Kustra R., Tenesa A., Farrington S.M., Prendergast J., Olschwang S., Chiang T., Crowdy E., et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat. Genet. 2007;39:989–994. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 12.Haiman C.A., Le M.L., Yamamato J., Stram D.O., Sheng X., Kolonel L.N., Wu A.H., Reich D., Henderson B.E. A common genetic risk factor for colorectal and prostate cancer. Nat. Genet. 2007;39:954–956. doi: 10.1038/ng2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes S., Williams R.D., Webb E., Houlston R.S. Meta-analysis and pooled re-analysis of copy number changes in colorectal cancer detected by comparative genomic hybridization. Anticancer Res. 2006;26:3439–3444. [PubMed] [Google Scholar]
- 14.Gruber S.B., Moreno V., Rozek L.S., Rennert H.S., Lejbkowicz F., Bonner J.D., Greenson J.K., Giordano T.J., Fearon E.R., Rennert G. Genetic variation in 8q24 associated with risk of colorectal cancer. Cancer Biol. Ther. 2007;6:1143–1147. doi: 10.4161/cbt.6.7.4704. [DOI] [PubMed] [Google Scholar]
- 15.Poynter J.N., Figueiredo J.C., Conti D.V., Kennedy K., Gallinger S., Siegmund K.D., Casey G., Thibodeau S.N., Jenkins M.A., Hopper J.L., et al. Variants on 9p24 and 8q24 are associated with risk of colorectal cancer: results from the Colon Cancer Family Registry. Cancer Res. 2007;67:11128–11132. doi: 10.1158/0008-5472.CAN-07-3239. [DOI] [PubMed] [Google Scholar]
- 16.Tuupanen S., Niittymaki I., Nousiainen K., Vanharanta S., Mecklin J.P., Nuorva K., Jarvinen H., Hautaniemi S., Karhu A., Aaltonen L.A. Allelic imbalance at rs6983267 suggests selection of the risk allele in somatic colorectal tumor evolution. Cancer Res. 2008;68:14–17. doi: 10.1158/0008-5472.CAN-07-5766. [DOI] [PubMed] [Google Scholar]
- 17.Li L., Plummer S.J., Thompson C.L., Merkulova A., Acheson L.S., Tucker T.C., Casey G. A common 8q24 variant and the risk of colon cancer: a population-based case–control study. Cancer Epidemiol. Biomarkers Prev. 2008;17:339–342. doi: 10.1158/1055-9965.EPI-07-0713. [DOI] [PubMed] [Google Scholar]
- 18.Botteri E., Iodice S., Raimondi S., Maisonneuve P., Lowenfels A.B. Cigarette smoking and adenomatous polyps: a meta-analysis. Gastroenterology. 2008;134:388–395. doi: 10.1053/j.gastro.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 19.Atkin W.S., Morson B.C., Cuzick J. Long-term risk of colorectal cancer after excision of rectosigmoid adenomas. N. Engl. J. Med. 1992;326:658–662. doi: 10.1056/NEJM199203053261002. [DOI] [PubMed] [Google Scholar]
- 20.He T.C., Sparks A.B., Rago C., Hermeking H., Zawel L., da Costa L.T., Morin P.J., Vogelstein B., Kinzler K.W. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 21.Augenlicht L.H., Wadler S., Corner G., Richards C., Ryan L., Multani A.S., Pathak S., Benson A., Haller D., Heerdt B.G. Low-level c-myc amplification in human colonic carcinoma cell lines and tumors: a frequent, p53-independent mutation associated with improved outcome in a randomized multi-institutional trial. Cancer Res. 1997;57:1769–1775. [PubMed] [Google Scholar]
- 22.Suo G., Han J., Wang X., Zhang J., Zhao Y., Dai J. Oct4 pseudogenes are transcribed in cancers. Biochem. Biophys. Res. Commun. 2005;337:1047–1051. doi: 10.1016/j.bbrc.2005.09.157. [DOI] [PubMed] [Google Scholar]
- 23.Campbell P.A., Perez-Iratxeta C., ndrade-Navarro M.A., Rudnicki M.A. Oct4 targets regulatory nodes to modulate stem cell function. PLoS ONE. 2007;2:e553. doi: 10.1371/journal.pone.0000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gidekel S., Pizov G., Bergman Y., Pikarsky E. Oct-3/4 is a dose-dependent oncogenic fate determinant. Cancer Cell. 2003;4:361–370. doi: 10.1016/s1535-6108(03)00270-8. [DOI] [PubMed] [Google Scholar]
- 25.Katoh Y., Katoh M. Conserved POU-binding site linked to SP1-binding site within FZD5 promoter: transcriptional mechanisms of FZD5 in undifferentiated human ES cells, fetal liver/spleen, adult colon, pancreatic islet, and diffuse-type gastric cancer. Int. J. Oncol. 2007;30:751–755. [PubMed] [Google Scholar]
- 26.Lin H., Shabbir A., Molnar M., Lee T. Stem cell regulatory function mediated by expression of a novel mouse Oct4 pseudogene. Biochem. Biophys. Res. Commun. 2007;355:111–116. doi: 10.1016/j.bbrc.2007.01.106. [DOI] [PubMed] [Google Scholar]
- 27.Prorok P.C., Andriole G.L., Bresalier R.S., Buys S.S., Chia D., Crawford E.D., Fogel R., Gelmann E.P., Gilbert F., Hasson M.A., et al. Design of the prostate, lung, colorectal and ovarian (PLCO) cancer screening trial. Control Clin. Trials. 2000;21:273S–309S. doi: 10.1016/s0197-2456(00)00098-2. [DOI] [PubMed] [Google Scholar]
- 28.Schoen R.E., Weissfeld J.L., Pinsky P.F., Riley T. Yield of advanced adenoma and cancer based on polyp size detected at screening flexible sigmoidoscopy. Gastroenterology. 2006;131:1683–1689. doi: 10.1053/j.gastro.2006.08.025. [DOI] [PubMed] [Google Scholar]
- 29.Hayes R.B., Sigurdson A., Moore L., Peters U., Huang W.Y., Pinsky P., Reding D., Gelmann E.P., Rothman N., Pfeiffer R.M., et al. Methods for etiologic and early marker investigations in the PLCO trial. Mutat. Res. 2005;592:147–154. doi: 10.1016/j.mrfmmm.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 30.Potter J.D., Bostick R.M., Grandits G.A., Fosdick L., Elmer P., Wood J., Grambsch P., Louis T.A. Hormone replacement therapy is associated with lower risk of adenomatous polyps of the large bowel: the Minnesota Cancer Prevention Research Unit Case–Control Study. Cancer Epidemiol. Biomarkers Prev. 1996;5:779–784. [PubMed] [Google Scholar]
- 31.O'Brien M.J., Winawer S.J., Zauber A.G., Gottlieb L.S., Sternberg S.S., Diaz B., Dickersin G.R., Ewing S., Geller S., Kasimian D. The National Polyp Study. Patient and polyp characteristics associated with high-grade dysplasia in colorectal adenomas. Gastroenterology. 1990;98:371–379. [PubMed] [Google Scholar]
- 32.Tranah G.J., Giovannucci E., Ma J., Fuchs C., Hunter D.J. APC Asp1822Val and Gly2502Ser polymorphisms and risk of colorectal cancer and adenoma. Cancer Epidemiol. Biomarkers Prev. 2005;14:863–870. doi: 10.1158/1055-9965.EPI-04-0687. [DOI] [PubMed] [Google Scholar]
- 33.Excoffier L., Slatkin M. Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol. Biol. Evol. 1995;12:921–927. doi: 10.1093/oxfordjournals.molbev.a040269. [DOI] [PubMed] [Google Scholar]
- 34.Schaid D.J., Rowland C.M., Tines D.E., Jacobson R.M., Poland G.A. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am. J. Hum. Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lake S.L., Lyon H., Tantisira K., Silverman E.K., Weiss S.T., Laird N.M., Schaid D.J. Estimation and tests of haplotype-environment interaction when linkage phase is ambiguous. Hum. Hered. 2003;55:56–65. doi: 10.1159/000071811. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.