Abstract
Entrez Gene (EG), Online Mendelian Inheritance in Man (OMIM) and the Gene Ontology (GO) are three complementary knowledge resources that can be used to correlate genomic data with disease information. However, bridging between genotype and phenotype through these resources currently requires manual effort or the development of customized software. In this paper, we argue that integrating EG and GO provides a robust and flexible solution to this problem. We demonstrate how the Resource Description Framework (RDF) developed for the Semantic Web can be used to represent and integrate these resources and enable seamless access to them as a unified resource. We illustrate the effectiveness of our approach by answering a real-world biomedical query linking a specific molecular function, glycosyltransferase, to the disorder congenital muscular dystrophy.
Keywords: knowledge integration, Semantic Web, RDF, Entrez Gene, Gene Ontology
Introduction
A common scenario in biomedical research involves the correlation of genomic data with disease information, in other words, associating genotype and phenotype information. In the particular scenario illustrated in this paper, a researcher is interested in glycosylation and its implications for one disorder: congenital muscular dystrophy. The biological process of glycosylation results in the post-translational addition of glycosyl groups (saccharides) to proteins (and lipids). Various enzymes, namely glycosyl-transferases, catalyze glycosylation reactions.
From the functional annotation of gene products with terms from the Gene Ontology (GO), a researcher can identify the genes having the molecular function of catalyzing the transfer of specific glycosyl groups (e.g., hexosyltransferase, for hexosyl groups). Known associations between these genes and diseases can then be mined from resources such as NCBI’s Entrez Gene (EG), where phenotypic information is recorded as pointers to the Online Mendelian Inheritance in Man (OMIM) knowledge base [3]. (See the Materials section for a presentation of GO and EG.)
In order to validate the hypothesis of possible association between the molecular function glycosyltransferase and the disease congenital muscular dystrophy, a researcher could simply search EG for the term glycosyltransferase, and all records containing the string “glycosyltransferase” in GO annotations would be returned. This approach, however, is suboptimal for at least two reasons. First, the term glycosyltransferase might appear as a substring in other GO terms (e.g., in UDP-glycosyltransferase), possibly leading to false positives. Conversely, not all GO terms related to glycosyltransferase actually contain the string “glycosyltransferase” (e.g., acetylglucosaminyltransferase, a kind of glycosyltransferase), possibly leading to false negatives.
To avoid false positives and false negatives, a careful researcher would likely start exploring the Gene Ontology database to create a list of glycosyltransferase-related terms by selecting the term glycosyltransferase itself (GO:0016757) and all its descendants, including specialized types of glycosyltransferase, such as acetylglucosaminyltransferase. This researcher would then look for the genes annotated with any of the glycosyltransferase-related terms. Resources such as the web browser AmiGO [1] support such searches and can retrieve the genes associated with any descendant of a given GO term. Finally, each of the genes found associated with any of the glycosyltransferase-related terms must be searched individually in EG, looking for mentions of the disease congenital muscular dystrophy (as an OMIM phenotype) in the corresponding records.
The procedure described above is evidently inefficient, time consuming and error prone as several web interfaces need to be utilized (AmiGO and Entrez), and as the results of the search in one resource need to be copied and pasted as search terms in the other. The main reason for such inefficiency is that high quality resources such as GO and EG have been designed primarily for consultation by humans, not for automated processing by agents or integration in applications. Moreover, these resources have been developed by different groups, independently of each other and are therefore not interoperable. No system currently supports complex queries such as: Find all the genes annotated with glycosyltransferase-related terms in GO and associated with the disease congenital muscular dystrophy in OMIM. Typically, querying across the different knowledge sources is accomplished manually through meticulous work or requires the development of complex and customized software applications.
In this paper, we propose an integrative approach to querying across knowledge sources. More specifically, we have applied Resource Description Framework (RDF) [4] standard developed by the World Wide Web Consortium (W3C) to integrate knowledge from GO and EG, and used this integrated resource to answer complex queries. We use the scenario presented earlier to illustrate the advantages of this approach. This work is a pilot contribution to the Biomedical Knowledge Repository under development at the U.S National Library of Medicine (NLM) as part of the Advanced Library Services project [8]. This repository integrates knowledge not only from structured resources (database and knowledge bases), but also from the biomedical literature (e.g., MEDLINE), in order to support applications, including knowledge discovery.
Background
Information integration is one of the most challenging areas of research in Computer Science [11]. The use of heterogeneous schemas for data storage, that are designed primarily to ensure optimization of storage space, makes it extremely difficult for users to query data sources in an integrated manner. (The interested reader is referred to [12] for a survey of approaches to information integration.) The Semantic Web provides a common framework that enables the integration, sharing and reuse of data from multiple sources. The use of a representation formalism based on a formal language enables software applications to ‘understand’ and reason over information. Recent research in Semantic Web technologies has delivered promising results to enable information integration across heterogeneous knowledge sources.
The Resource Description Framework (RDF) is a W3C-recommended framework for representing data in a common format that captures the logical structure of the data. This is in contrast to pure storage aspects addressed by traditional relational database schema. The RDF representational model uses a single schema in contrast to multiple heterogeneous schemas or Data Type Definitions (DTD) used to represent data in XML by different sources. Hence in conjunction with a single Uniform Resource Identifier (URI), all data represented in RDF form a single knowledge repository that may be queried as one knowledge resource. An RDF repository consists of a set of assertions or triples. Each triple is constituted of three entities namely, the subject – the triple pertains to this entity, the object – the entity that states something about the object and the predicate – the relationship between the subject and the object. For example, as shown in Figure 1, assertions such as acetylglucosaminyltransferase (GO:0008375) is a kind of hexosyltransferase (GO:0016758) and the gene LARGE (EG:9215) has molecular function acetylglucosaminyltransferase (GO:0008375) can be represented as RDF triples.
Figure 1.
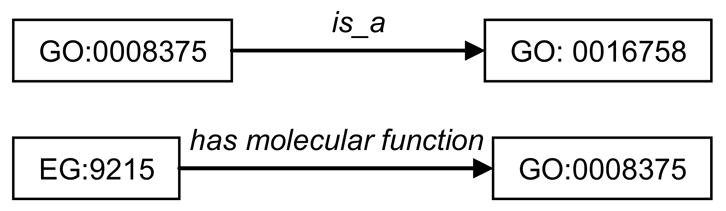
Example of RDF triples
The RDF triples often share nodes, thus forming a graph. For example, the two triples shown in Figure 2 share the node acetylglucosaminyltransferase (GO:0008375). The resulting graph is shown in Figure 2. The graph structure created by RDF is key to information integration in the Semantic Web.
Fiigure 2.
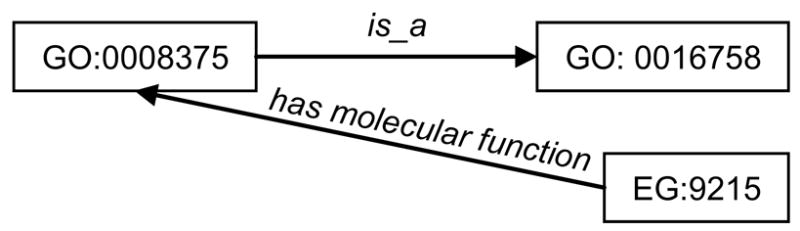
Example of RDF graph
Materials
The Gene Ontology (GO) seeks to provide a consistent description of gene products [13]. GO consists of three controlled vocabularies for biological processes (9,234 terms), molecular functions (7,456 terms) and cellular components (1,804 terms). The GO monthly releases are made available on the GO website in various formats, including RDF. The version of GO used in this study is dated of September 2006.
The Entrez Gene (EG) database records gene-related information from sequenced genomes and of model organisms that are focus of active research [9], totaling about two million genes. EG contains gene information about genomic maps, sequences, homology, and protein expression among others [9]. In contrast to GO, EG is not available in RDF, but in XML (converted from ASN1 by the program gene2xml provided by NCBI), and can be downloaded from the NCBI website. The version of EG used in this study is dated of July 2006.
Methods
Our integration method can be summarized as follows and is illustrated in Figure 3. First, we extract manageable subsets from the two resources to be integrated. We then have to convert the EG subset from XML to RDF. Finally, we load both RDF resources in a common store, apply inference rules, and issue queries against it.
Figure 3.
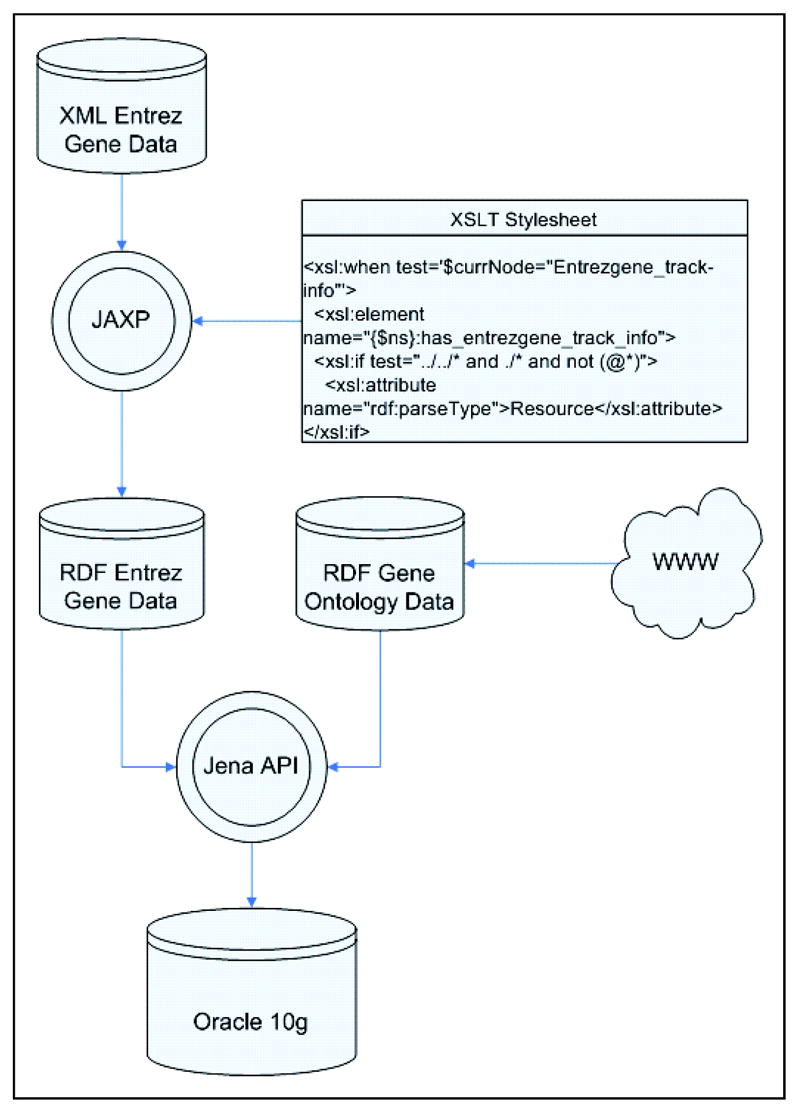
Overview of the integration method
Creating subsets
The entire Entrez Gene data file (in XML format) is very large (50 GB) and unnecessarily difficult to manipulate. In order to obtain a manageable subset from EG, we restricted the gene records to two species: Homo sapiens (human) and Mus musculus (mouse). The resulting EG subset contains a total of 99,861 complete gene records (excluding obsolete records).
Converting XML format Entrez Gene data to RDF
A key element of our integration approach is the conversion of Entrez Gene from XML to RDF. There are many issues involved in the conversion of XML data into RDF format, including modeling the original semantics of the data, filtering redundant XML element tags, linking data entities using meaningful named relationships and identifying entities consistently within and across resources. Unlike traditional XML to XML conversion, XML to RDF conversion should exploit the advantages of the RDF model in representing the logical structure of the information.
We chose not to convert the element tags of the native EG XML representation mechanically into the predicates of the RDF triples. Instead, we manually converted the XML element tags into meaningful and standardized relationship names that convey explicitly the semantics of the connection between the subject and the object. For example, the element <Org-ref_taxname> was mapped to the more meaningful relationship named has_source_organism_taxonomic_name.
We selected the eXtensible Stylesheet Language Transformation (XSLT) [6] for converting the EG XML information into RDF, because this approach allows for a clean separation between the application (using Java API for XML Processing (JAXP)) and the conversion logic (using XSLT stylesheet). Once the stylesheet is created, it can serve as an auxiliary file for existing programs realizing the XML to RDF conversion. In other words, the major interest of this approach is that no specific code is required for the conversion, because the transformation logic resides entirely in the stylesheet.
Loading the two resources into a single data store
Some of the requirements for our RDF store include native support for the RDF graph data model, support for persistence and indexing of the RDF triples, support for extensive collections of triples, and availability of a query language for the RDF graph. After surveying available RDF storage solutions, we decided to use Oracle Spatial 10g [7] as the RDF storage system.
The RDF file resulting from the XSLT conversion of the original XML file for EG and the downloaded RDF version of GO are both loaded into a single RDF store. More precisely, the RDF resources are first converted to the NTriple format using the Jena API [10] and loaded into the RDF database using a utility program provided by Oracle.
Applying inference rules
Unlike the Web Ontology language OWL, RDF provides no direct support for inference. However, inference rules can be implemented in the RDF store to make explicit the semantics of some predicates. For example, the relationships is_a and part_of used in GO are partial order relations, thus being reflexive, antisymmetric and transitive. The inference rules we created for implementing the transitivity and combination of these two relationships are shown in Table 1. The inference rules are stored in a rule base created in Oracle 10g.
Table 1.
Inference rules for is_a and part_of in GO
| Relation | is_a | part_of |
|---|---|---|
| is_a | IF <x is_a y>
& <y is_a z> THEN <x is_a z> |
IF <x is_a y>
& <y part_of z> THEN <x part_of z> |
| part_of | IF <x part_of y>
& <y is_a z> THEN <x part_of z> |
IF <x part_of y>
& <y part_of z> THEN <x part_of z> |
Querying the RDF Graph with SPARQL
SPARQL [5] is a query language for RDF graphs, equivalent to SQL, the Structured Query Language, for relational databases. Unlike SQL, SPARQL does not require users to be familiar with the data model (e.g., tables, foreign keys), but simply to indicate how entities of interest relate to each other. For example, the structure of the query: Find all the genes annotated with the GO molecular function glycosyl-transferase (GO:0016757) or any of its descendants and associated with any form of congenital muscular dystrophy is represented in Figure 4.
Figure 4.
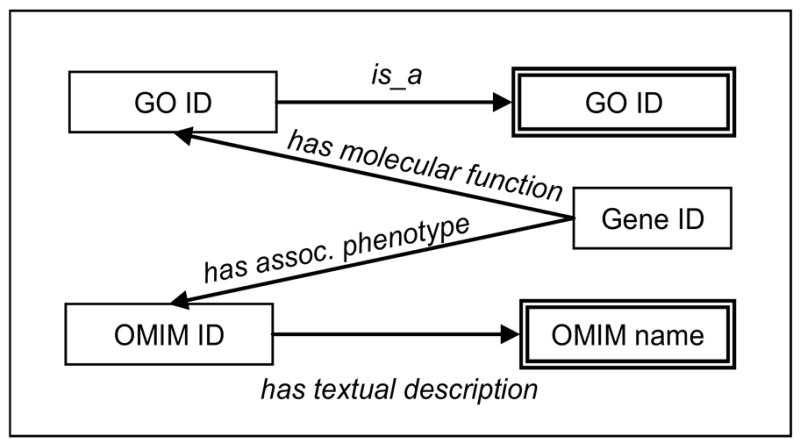
RDF graph corresponding to the query above
The query can be understood as finding a path in the RDF graph using a predetermined set of semantic relationships and would be formulated as follows. Because of the inference rules implementing the transitivity and reflexivity of the is_a relationship, the condition on the GO annotation “glycosyltransferase (GO:0016757) or any of its descendants” is easily expressed by ‘ ?t is_a GO:0016757’. The link between genes and GO terms is expressed by ‘ ?g has_molecular_function ?t’. Similarly, the link between genes and OMIM diseases is expressed by ‘ ?g has_associated_phenotype ?b2’ (OMIM ID) and ‘ ?b2 has_textual_description ?d’ (disease name). Finally, direct constraints are put on the GO term on the one hand (‘?t is_a GO:0016757’, to select glycosyltransferase (GO:0016757)) and on disease names on the other (where a regular expression is used to select disease names containing the strings “congenital”, “muscular” and “dystrophy”). The actual (but simplified) SPARQL query is shown in Figure 5.
Figure 5.
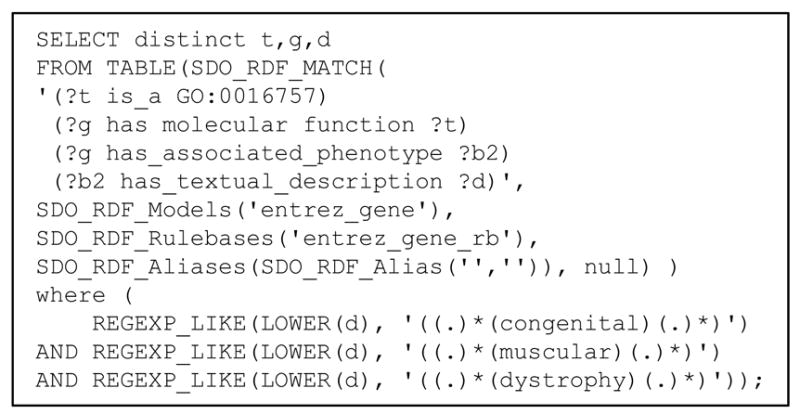
Example of SPARQL query (simplified)
Results
One integrated RDF repository for Entrez Gene and GO
The subset of Entrez Gene restricted to Homo sapiens (human) and Mus musculus (mouse) as biological sources comprises 99,861 gene records. Once converted to RDF, it consists of 772,530 triples. The RDF version of GO contains 293,798 triples. Overall, there are over one million triples in the store created for this experiment, which is relatively small in comparison to the 411 million triples resulting from the conversion of the entire EG to RDF [2].
Biological query result: extended example
The SPARQL query presented above returned one result, corresponding to one path in the graph between the GO term glycosyltransferase (GO:0016757) and OMIM disease names containing (variants of) the string “congenital muscular dystrophy”.
This path involved the human gene LARGE like-glycosyltransferase (EG:9215), annotated with the GO term acetylglucosaminyltransferase (GO:0008375), a descendant of glycosyltransferase (GO:0016757). Also involved in this path is the OMIM disease identified by MIM:608840. The name (textual description) of this disease is Muscular dystrophy, congenital, type 1D and contains the required substrings “congenital”, “muscular” and “dystrophy”. The instantiated RDF graph with path between glycosyltransferase (GO:0016757) and Muscular dystrophy, congenital, type 1D is shown in Figure 6.
Figure 6.
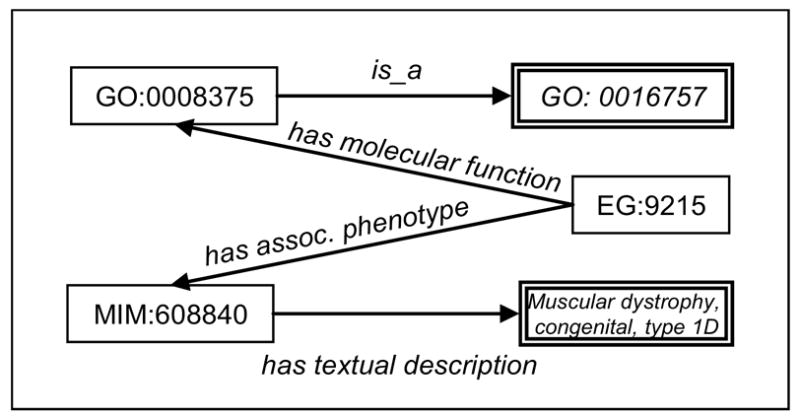
Instantiated RDF graph
This simple SPARQL query provides an easy way of testing the biological hypothesis under investigation, i.e., the existence of a possible link between glycosylation and congenital muscular dystrophy. On manual inspection of the Entrez Gene record, we also note that the given gene may be involved in the development and progression of meningioma through modification of ganglioside composition and other glycosylated molecules in tumor cells.
Discussion
Significance
In this study, we demonstrated the feasibility of integrating two biomedical knowledge resources through RDF. We also provided anecdotal evidence for the benefits of such integration by showing how glycosyltransferase can be linked to congenital muscular dystrophy. The integrated resource is greater than the sum of its parts as it supports complex queries that could typically not be handled otherwise without tedious manual intervention or customized software applications.
Integrated resources based on a graph model are particularly important in an exploratory context where researchers need to “connect the dots” in order to validate an hypothesis. This approach also facilitates intuitive hypothesis formulation and refinement. For example, after verifying that glycosyltransferase is linked to congenital muscular dystrophy, our researchers may narrow the focus of their wet lab experiments to only hexosyltransferase out of the potential seven glycosyltransferases. Analogously, they can focus their research on Muscular dystrophy, congenital, type 1D, out of several other diseases.
Arguably, the graph data model of RDF resources is more intuitive than the database schemas. In fact, the RDF data model enables us to model the inherent logical relations between entities that mirror the human cognitive model of the real world. Additionally, the RDF data model offers more flexibility than database schemas for accommodating changes to the underlying model.
Generalization
The integration approach demonstrated in this study can be generalized to more complex queries and to additional information sources. For example, many additional constraints can be easily added to the query presented earlier by exploiting other properties represented in GO or EG. Examples of such constraints include restricting the annotations to specific evidence codes (e.g., TAS) and narrowing the query to a specific model organism.
Only two resources are currently integrated in our RDF store. However, this approach can be generalized to other resources including pathway databases, microarray resources, disease ontologies and virtually all the structured knowledge bases currently under the umbrella of the Entrez system, including UniGene and HomoloGene. Knowledge extracted from unstructured sources such as the biomedical literature can also be integrated. Creating such an extensive repository of biomedical knowledge is one of the goals of the Advanced Library Services project under development at NLM.
Unresolved issues and challenges
In addition to scalability issues, which can be addressed by mature software and the next generation of hardware, challenges include the identification and organization of entities and relationships. Heterogeneous resources can interoperate in a RDF graph only if the entities shared by these resources are identified consistently. The namespace provided by the UMLS is expected to play an important role for the permanent identification of biomedical entities. In contrast to entities for which organizational schemes currently exist (terminologies and ontologies), the named relationships used to connect data entities during the conversion of EG from XML to RDF are currently not formalized in an ontology of relationships. As a consequence, only limited reasoning can be supported by the RDF graph. As sizeable ontologies of relationships become available, they too will be used for normalizing knowledge in our repository. RDF schemas and OWL will also be investigated.
Acknowledgments
This research was supported in part by the Intramural Research Program of the National Institutes of Health (NIH), National Library of Medicine (NLM) and by the Integrated Technology Resource for Biomedical Glycomics (5 P41 RR18502), funded by the National Institutes of Health National Center for Research Resources.
References
- 1.AmiGO: Gene Ontology browser [cited 11/29/06; Available from: http://www.godatabase.org/]
- 2.BioRDF subgroup. Health Care and Life Sciences interest group. [cited 11/29/06; Available from: http://esw.w3.org/topic/HCLSIG_BioRDF_Subgroup]
- 3.Online Mendelian Inheritance in Man, OMIM (TM) McKusick-Nathans Institute for Genetic Medicine, Johns Hopkins University (Baltimore, MD) and National Center for Biotechnology Information, National Library of Medicine. [cited 11/29/06; Available from: http://www.ncbi.nlm.nih.gov/omim/]
- 4.Resource Description Framework (RDF) [cited 11/29/06; Available from: http://www.w3.org/TR/2004/REC-rdf-primer-20040210/]
- 5.SPARQL Query Language for RDF. W3C Working Draft 2006. [cited 11/29/06; Available from: http://www.w3.org/TR/rdf-sparql-query]
- 6.XML Schema Language Transformation (XSLT) [cited 11/29/06; Available from: http://www.w3.org/TR/xslt]
- 7.Alexander N, Ravada S. “RDF Object Type and Reification in Oracle”. Technical White Paper. [cited 11/29/06; Available from: http://download-east.oracle.com/otndocs/tech/semantic_web/pdf/rdf_reification.pdf]
- 8.Bodenreider O, Rindflesch TC. Advanced library services: Developing a biomedical knowledge repository to support advanced information management applications. Bethesda, Maryland: Lister Hill National Center for Biomedical Communications, National Library of Medicine; September 14, 2006. Technical report. [Google Scholar]
- 9.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005 January 1;33(Database Issue):D54–D58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McBride B. IEEE Internet Computing. 6. Vol. 6. 2002. Nov, 2002, Jena: A Semantic Web Toolkit; pp. 55–59. [Google Scholar]
- 11.Sheth AP. Changing Focus on Interoperability in Information Systems: From System, Syntax, Structure to Semantics. In: Goodchild MF, Egenhofer MJ, Fegeas R, Kottman CA, editors. Interoperating Geographic Information Systems. Kluwer, Academic Publishers; 1999. pp. 5–30. [Google Scholar]
- 12.Shvaiko P, Euzenat J. A survey of schema-based matching approaches. Journal on Data Semantics. 2005;4:146–71. [Google Scholar]
- 13.The Gene Ontology Consortium. Gene Ontology: tool for the unification of biology. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]


