Abstract
Summary: There are thousands of strongly conserved non-coding elements (CNEs) in vertebrate genomes, and their functions remain largely unknown. However, without biologically relevant criteria for prioritizing them, selecting a particular CNE sequences to study can be haphazard. To address this problem, we present cneViewer—a database and webtool that systematizes information on conserved non-coding DNA elements in zebrafish. A key feature here is the ability to search for CNEs that may be relevant to tissue-specific gene regulation, based on known developmental expression patterns of nearby genes. cneViewer provides this and other organizing features that significantly facilitate experimental design and CNE analysis.
Availability: http://cneviewer.zebrafishcne.org
Contact: chuangj@bc.edu
1 INTRODUCTION
There are thousands of conserved non-coding elements (CNEs) in vertebrate genomes. While the functions of most are unknown, their conservation across species suggests they play important biological roles, especially in cis-gene regulation (Visel et al., 2007a; Woolfe et al., 2005). To facilitate the understanding and experimental testing of these sequences, we have built cneViewer—a dynamic webtool and database for studying conserved zebrafish–human non-coding DNA elements.
cneViewer is built on a searchable database of 73 187 strand-specific CNEs each ≥50 bp and with ≥50% identity between the zebrafish Danio rerio and human. This is considerably more conserved than would be expected of non-functional DNA. cneViewer focuses on zebrafish because of its widespread use in developmental studies and because of abundant developmental expression data for many of its genes in the ZFIN database (Sprague et al., 2006).
A novel feature of cneViewer is the ability to search for CNEs near genes expressed exclusively, in particular anatomies or developmental stages. This feature can be used to identify CNEs more likely to influence gene expression in the anatomy of interest, which could then be experimentally validated. This functionality distinguishes cneViewer from tools that focus on one locus at a time (e.g. Ovcharenko et al., 2004). CNEs can be further filtered based on percentage identity, length, motif content, distance or synteny.
cneViewer provides a variety of information for each CNE, including alignments, primers for cloning, distances to nearby genes, sequence identity and links to genome browsers. Such information is ideal for pipeline development for high-throughput studies. All features have been implemented in a simple and user-friendly interface. We believe that these tools significantly improve the selection and procurement of CNE data for both experimental and computational studies of vertebrate CNEs.
2 METHODS
The database is constructed from the perspective of nearby genes, and each CNE is associated with at least one gene. We obtained a list of all gene symbols annotated in Unigene, Ensembl or Genbank from the zebrafish database ZFIN. These genes were mapped to the zv7 genome using the refgene.txt list of locations from UCSC danRer5 annotations. Some ZFIN gene symbols are annotated in refgene.txt under pseudonyms, typically as an EST or deprecated gene name. We identified pseudonyms by parsing the Dr.data Unigene annotation file from NCBI. In cases where a gene has multiple annotated locations, we reported each annotation separately. For example, the gene emx is reported as emx.1 and emx.2.
Genes were associated to aligned sequence blocks in the UCSC axt alignments of zebrafish (danRer5) to human (Hg18). Each block was associated with each gene within 500 kb in either the 5′ or 3′ direction. We removed segments overlapping any RefSeq, GenScan, Ensembl or human TBlastN coding annotations in the zebrafish genome. Tissue-specific and stage-specific expression data for genes were obtained via ZFIN. Syntenic CNEs were defined to be those within 500 kb of at least one orthologous gene in each of the human and zebrafish genomes. The webserver, processing scripts and database were implemented in php, javascript, Perl, Ruby, bash scripts and MySQL. cneViewer is updated three times a year and for new genome builds. A FAQ is available at the site.
3 RESULTS
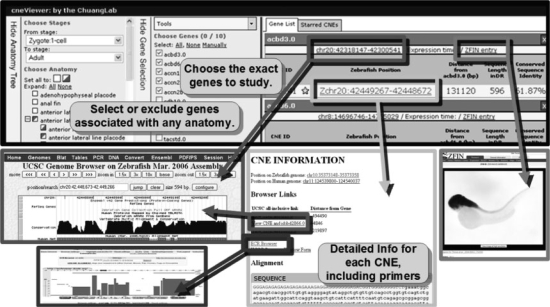
cneViewer implements a number of tools for identifying and describing CNEs (Fig. 1). The first is a tool to search for CNEs by the expression patterns of genes within 500 kb. An Anatomy Tree pane on the left-hand side of the page allows one to search for genes with expression in particular tissues. Tissues can be specified with Boolean logic using checkboxes. These tissue annotations are hierarchical, in accordance with the ZFIN framework. Genes can also be restricted by expression timing.
Fig. 1.
cneViewer allows one to select CNEs by the expression of nearby genes, with practical data about each CNE.
All genes matching the anatomical criteria are loaded into a Gene Selection pane. This contains a Tools menu to add or remove specific genes. When a checkbox next to a gene is selected, the CNEs within 500 kb appear in the CNE pane at the right of the window. CNE sequences are reported on the DNA strand of the gene.
The CNE pane provides essential information on each CNE, including location in the zebrafish genome, distance to the nearby gene (either 5′ or 3′), length and sequence identity with human. CNEs can be sorted by clicking on the feature name. Each gene has links to its location in the UCSC genome browser and to its ZFIN gene entry. Individual CNEs can be marked with a star for storage in a secondary tab.
Clicking on an individual CNE opens a CNE information page. This page contains links to the CNE locus on the UCSC, ECR and Ancora genome browsers. A list of genes within 500 kb is provided with links to the UCSC genome browser displaying each gene and the CNE together. The information page also contains the zebrafish and human CNE sequences, the sequence alignment and the 50 bp adjacent to each end of the CNE, which can be used as primer sequence to clone the CNE experimentally.
The Tools menu in the Gene pane contains features including the ability to: search for CNEs containing a desired subsequence, specify a list of CNEs to view in the CNE pane or download CNE data to a text file. Users may also restrict to CNEs meeting specific length, distance or conservation cutoffs, or with conserved synteny between human and zebrafish (similar to Engstrom et al., 2008).
In summary, while a few hundred CNEs have been experimentally studied in mouse (Visel et al., 2007b) and fish (Woolfe et al., 2007), most CNEs remain uncharacterized. Understanding the remaining CNEs requires not just a description of their characteristics, but also their organization into biologically relevant groups. This is analogous to the importance of the Gene Ontology for protein-coding genes. cneViewer organizes CNEs by considering the expression of nearby zebrafish genes, which may make it useful for identifying sequence features relevant to tissue-specific or timing-specific expression. cneViewer implements such organization and other features in a user-friendly tool, which can help researchers develop and test new hypotheses about the functions of CNEs.
Funding
National Institutes of Health (grant R21-HD051835).
Conflict of Interest: none declared.
REFERENCES
- Engstrom P, et al. Ancora: a web resource for exploring highly conserved noncoding elements and their association with developmental regulatory genes. Genome Biol. 2008;9:R34. doi: 10.1186/gb-2008-9-2-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovcharenko I, et al. ECR browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 2004;32(Suppl. 2):W280–W286. doi: 10.1093/nar/gkh355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague J, et al. The Zebrafish information network: the zebrafish model organism database. Nucleic Acids Res. 2006;34(Suppl. 1):D581–D585. doi: 10.1093/nar/gkj086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, et al. Enhancer identification through comparative genomics. Semin. Cell Dev. Biol. 2007a;18:140. doi: 10.1016/j.semcdb.2006.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, et al. VISTA enhancer browser—a database of tissue-specific human enhancers. Nucleic Acids Res. 2007b;35(Suppl. 1):D88–D92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfe A, et al. CONDOR: a database resource of developmentally associated conserved non-coding elements. BMC Dev. Biol. 2007;7:100. doi: 10.1186/1471-213X-7-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfe A, et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]