Abstract
Summary: Many non-synonymous single nucleotide polymor-phisms (nsSNPs) in humans are suspected to impact protein function. Here, we present a publicly available server implementation of the method SNAP (screening for non-acceptable polymorphisms) that predicts the functional effects of single amino acid substitutions. SNAP identifies over 80% of the non-neutral mutations at 77% accuracy and over 76% of the neutral mutations at 80% accuracy at its default threshold. Each prediction is associated with a reliability index that correlates with accuracy and thereby enables experimentalists to zoom into the most promising predictions.
Availability: Web-server: http://www.rostlab.org/services/SNAP; downloadable program available upon request.
Contact: bromberg@rostlab.org
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Non-synonymous SNPs (nsSNPs) are associated with disease: Estimates expect as many as 200 000 nsSNPs in human (Halushka et al., 1999) and about 24 000–60 000 in an individual (Cargill et al., 1999); this implies about 1–2 mutants per protein. While most of these likely do not alter protein function (Ng and Henikoff, 2006), many non-neutral nsSNPs contribute to individual fitness. Disease studies typically face the challenge finding a needle (SNP yielding particular phenotype) in a haystack (all known SNPs). For example, many of the thousands of mutations associated with cancer do not actually lead to the disease. Evaluating functional effects of known nsSNPs is essential for understanding genotype/phenotype relations and for curing diseases. Computational mutagenesis methods can be useful in this endeavor if they can explain the motivation behind assigning a mutant to neutral or non-neutral class or if they can provide a measure for the reliability of a particular prediction.
Screening for non-acceptable polymorphisms is accurate and provides a measure of reliability: here, we present the first web-server implementation of SNAP (screening for non-acceptable polymorphisms), a method that combines many sequence analysis tools in a battery of neural networks to predict the functional effects of nsSNPs (Bromberg and Rost, 2007, 2008). SNAP was developed using annotations extracted from PMD, the Protein Mutant Database (Kawabata et al., 1999; Nishikawa et al., 1994). SNAP needs only sequence as input; it uses sequence-based predictions of solvent accessibility and secondary structure from PROF (Rost, 2000, unpublished data; Rost, 2005; Rost and Sander, 1994), flexibility from PROFbval (Schlessinger et al., 2006), functional effects from SIFT (Ng and Henikoff, 2003), as well as conservation information from PSI-BLAST (Altschul et al., 1997) and PSIC (Sunyaev et al., 1999), and Pfam annotations (Bateman et al., 2004). If available, SNAP can also benefit from SwissProt annotations (Bairoch and Apweiler, 2000). In sustained cross-validation, SNAP correctly identified ∼80% of the non-neutral substitutions at 77% accuracy (often referred to as specificity, i.e. correct non-neutral predictions/all predicted as non-neutral) at its default threshold. When we increase the threshold, accuracy rises at the expense of coverage (fewer of the observed non-neutral nsSNPs are identified). This balance is reflected in a crucial new feature, the reliability index (RI) for each SNAP prediction that ranges from 0 (low) to 9 (high):
| (1) |
where OUTX is the raw value of one of the two SNAP output units.
When given alternative prediction methods, investigators often identify a subset of predictions for which methods agree. This approach may increase accuracy over any single method at the expense of coverage. Well-calibrated method-internal reliability indices can be much more efficient than a combination of different methods (Rost and Eyrich, 2001). Simply put: ‘A basket of rotten fruit does not make for a good fruit salad’ (Chris Sander, CASP1). The SNAP RI has been carefully calibrated.
2 INPUT/OUTPUT
Users submit the wild-type sequence along with their mutants. A comma-separated list gives mutants as: XiY, where X is the wild-type amino acid, Y is the mutant and i is the number of the residue (i=1 for N-terminus). X is not required and a star (⋆) can replace either i or Y. Any combination of characters following these rules is acceptable; e.g. X⋆=replace all residues X in all positions by all other amino acids, ⋆Y=replace all residues in all positions by Y. Users may provide a threshold for the minimal RI [Equation (1)] and/or for the expected accuracy of predictions that will be reported back. These two values correlate; when both are provided, the server chooses the one yielding better predictions. For each mutant, SNAP returns three values (Fig. 1A): the binary prediction (neutral/non-neutral), the RI (range 0–9) and the expected accuracy that estimates accuracy [Equation (1)] on a large dataset at the given RI (i.e. accuracy of test set predictions calculated for each neutral and non-neutral RI; Fig. 1C, Supplementary Online Material Fig. SOM_1).
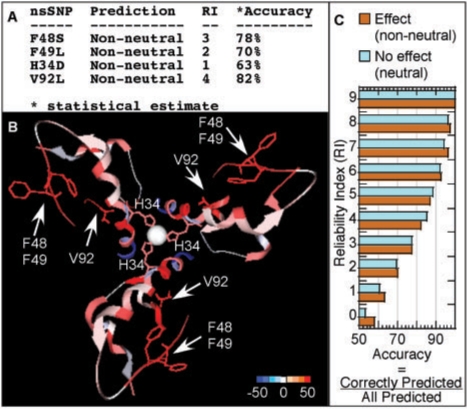
Fig. 1.
Examples of SNAP functionality. (A) SNAP-server predictions for mutations in INS_HUMAN associated with hyperproinsulenemia and diabetes-mellitus type II (Chan et al., 1987; Sakura et al., 1986; Shoelson et al., 1983). (B) SNAP predictions for comprehensive in silico mutagenesis (all-to-alanine). The crystal structure [PDB 2omg (Norrman et al., 2007)] shows an insulin NPH hexamer [insulin co-crystallized with zinc (sphere at the center) in presence of protamine/urea (not highlighted); picture produced by GRASP2 (Petrey and Honig, 2003)]. Red represents mutations predicted as non-neutral and blue represents neutral predictions. Residues in wire depiction are the same as in (A): V92, H34, F48 and F49 of INS_HUMAN (A chain V3, B chain H10, F24 and F25). SNAP predicts all of these to impact function when mutated to alanine. (C) More reliably predicted residues are predicted more accurately: for instance, >90% of the predictions with a reliability index=6 are expected to be right.
At this point, SNAP may take more than an hour to return results (processing status can be tracked on the original submission page). Therefore, most requests will be answered by an email containing a link to the results page. It is also highly recommended to check existing mutant evaluations [available immediately under the ‘known variants’ tab; referenced by RefSeq id (Pruitt et al., 2007) and dbSNP id (Sherry et al., 2001)] prior to submitting sequences for processing. In the near future, PredictProtein (Rost et al., 2004) that provides the framework for SNAP, will store sequences and retrieve predictions for additional mutants in real time. Full sequence analysis (e.g. in silico alanine scans; Fig. 1B) is possible for short proteins (≤150 total mutants/protein) via applicable server query. Analysis of longer sequences and/or local SNAP installation is currently available through the authors.
Supplementary Material
ACKNOWLEDGEMENTS
Thanks to Jinfeng Liu (Genentech) and Andrew Kernytsky (Columbia) for technical assistance; to Chani Weinreb, Marco Punta, Avner Schlessinger (all Columbia) and Dariusz Przybylski (Broad Inst.) for helpful discussions. Particular thanks to Rudolph L. Leibel (Columbia) for crucial support and discussions.
Funding: National Library of Medicine (grant 5-RO1-LM007 329-04).
Conflict of Interest: none declared.
REFERENCES
- Altschul SF, et al. Gapped Blast and PSI-Blast: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000;28:45–48. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, et al. The Pfam Protein Families Database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg Y, Rost B. SNAP: predict effect of non-synonymous poly-morphisms on function. Nucleic Acids Res. 2007;35:3823–3835. doi: 10.1093/nar/gkm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg Y, Rost B. Comprehensive in silico mutagenesis highlights functionally improtant residues in proteins. Bioinformatics. 2008;24:i207–i212. doi: 10.1093/bioinformatics/btn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cargill M, et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat. Genet. 1999;22:231–238. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- Chan SJ, et al. A mutation in the B chain coding region is associated with impaired proinsulin conversion in a family with hyperproinsulinemia. Proc. Natl Acad. Sci. USA. 1987;84:2194–2197. doi: 10.1073/pnas.84.8.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halushka MK, et al. Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nat. Genet. 1999;22:239–247. doi: 10.1038/10297. [DOI] [PubMed] [Google Scholar]
- Kawabata T, et al. The protein mutant database. Nucleic Acids Res. 1999;27:355–357. doi: 10.1093/nar/27.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting the effects of amino acid substitutions on protein function. Annu. Rev. Genomics Hum. Genet. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]
- Nishikawa K, et al. Constructing a protein mutant database. Protein Eng. 1994;7:773. doi: 10.1093/protein/7.5.733. [DOI] [PubMed] [Google Scholar]
- Norrman M, et al. Structural characterization of insulin NPH formulations. Eur. J. Pharm. Sci. 2007;30:414–423. doi: 10.1016/j.ejps.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Petrey D, Honig B. GRASP2: visualization, surface properties, and electrostatics of macromolecular structures and sequences. Methods Enzymol. 2003;374:492–509. doi: 10.1016/S0076-6879(03)74021-X. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, et al. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B. How to use protein 1D structure predicted by PROFphd. In: Walker JE, editor. The Proteomics Protocols Handbook. Humana, Totowa, NJ: 2005. pp. 875–901. [Google Scholar]
- Rost B, Eyrich V. EVA: large-scale analysis of secondary structure prediction. Proteins Struct. Funct. Genet. 2001;45(Suppl. 5):S192–S199. doi: 10.1002/prot.10051. [DOI] [PubMed] [Google Scholar]
- Rost B, Sander C. Conservation and prediction of solvent accessibility in protein families. Proteins Struct. Funct. Genet. 1994;20:216–226. doi: 10.1002/prot.340200303. [DOI] [PubMed] [Google Scholar]
- Rost B, et al. The PredictProtein server. Nucleic Acids Res. 2004;32:W321–W326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakura H, et al. Structurally abnormal insulin in a diabetic patient. Characterization of the mutant insulin A3 (Val----Leu) isolated from the pancreas. J. Clin. Invest. 1986;78:1666–1672. doi: 10.1172/JCI112760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger A, et al. PROFbval: predict flexible and rigid residues in proteins. Bioinformatics. 2006;22:891–893. doi: 10.1093/bioinformatics/btl032. [DOI] [PubMed] [Google Scholar]
- Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoelson S, et al. Identification of a mutant human insulin predicted to contain a serine-for-phenylalanine substitution. Proc. Natl Acad. Sci. USA. 1983;80:7390–7394. doi: 10.1073/pnas.80.24.7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunyaev SR, et al. PSIC: profile extraction from sequence alignments with position-specific counts of independent o, bservations. Protein Eng. 1999;12:387–394. doi: 10.1093/protein/12.5.387. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.