Abstract
Summary: We report a previously uncharacterized CHHC Zn-finger domain identified in spliceosomal U11-48K proteins, tRNA methyl-transferases TRM13 and gametocyte specific factors. We show that this domain behaves as an independent folding unit and that it stoichiometrically binds zinc in a one-to-one ratio. Based on the conserved sequence features we predict that this domain may function as a RNA recognition and binding module.
Contact: tony@mrc-lmb.cam.ac.uk
Supplementary information: Supplementary data are available on Bioinformatics online.
1 INTRODUCTION
Pre-mRNA splicing is an essential step in gene expression. In higher eukaryotes pre-mRNA introns are removed by two distinct spliceosomes. The majority of introns are excised by the U2-dependent (major) spliceosome that consists of five small ribonucleoprotein particles (snRNPs) U1, U2, U4, U5, U6 and a large number of protein splicing factors. A subset of eukaryotic organisms contains a second class of introns, the so-called U12-introns. Although the U12-introns constitute a small fraction of all introns (<1%), they have been identified in various genes implicated in biologically important processes such as DNA replication and repair, transcription, translation, signal transduction and cell cycle control (Burge et al., 1998; Levine et al., 2001). It has been shown that many genes contain both U2- and U12-introns. The U12-introns are recognized by the minor U12-type spliceosome that is formed by the U11, U12 U4atac/U6atac snRNPs and the U5 snRNP, which is common to both spliceosomes.
Both major and minor spliceosomes share similar assembly pathways and catalytic mechanisms. Despite these similarities, the two splicing systems display several fundamental differences. In contrast to U1 and U2 snRNPs, U11 and U12 exist mainly as an 18S U11/U12 di-snRNP complex. The U2-dependent spliceosome assembly is initiated by the interaction of U1 snRNP with the 5′ splice site followed by the association of U2 snRNP with the branch point site of the pre-mRNA. In the U12-dependent assembly, the 18S U11/U12 di-snRNP binds simultaneously to the 5′ splice and branch point site (Frilander et al., 1999).
Both splicing systems share common protein components, but there are proteins that are unique to the minor spliceosome (Will et al., 2004). These include seven proteins with molecular weight 20, 25, 31, 35, 48, 59 and 65 kDa. Four of them (25K, 35K, 48K and 59K) have been shown to associate with the 12S U11 snRNP and probably play a role in the U11/U12 snRNP assembly and/or 5′ splice site recognition.
In contrast to the major spliceosome, the U12-dependent spliceosome is less extensively studied and limited information is available about its protein components and their function. Here, we show that the minor spliceosomal protein U11-48K contains a genuine CHHC Zn-finger domain. We also demonstrate that this domain is similar and probably evolutionarily related to the Zn-finger domains identified in the TRM13 tRNA modifying enzymes and gametocyte specific factors.
2 METHODS
Protein sequence comparisons and database searches were performed with BLAST suite v.2.2.18 (Altschul et al., 1997). Pattern search was carried out with PHI-BLAST (Zhang et al., 1998). Two-step PSI-BLAST searches were performed as described before (Andreeva et al., 2008). TBLASTN was used to scan nucleotide and EST databases at NCBI and ENSEMBL in order to identify additional protein homologs. Initial clustering of protein sequences was performed using BLASTCLUST. Multiple sequence alignments were constructed and analysed using Jalview (Clamp et al., 2004). Analysis of protein disorder and compositionally biased regions were performed with PONDR (Romero et al., 2001) and SEG (Wootton et al., 1996), respectively.
The DNA encoding the second exon of human U11-48K corresponding to residues 53–87 (the CHHC Zn-finger domain) was cloned into a pRSET-derived vector containing an N-terminal fusion of His6-tag, lipoamyl domain and TEV protease cleavage site to generate the plasmid pHLT-ZnF U11-48K. The plasmid was transformed in Escherichiacoli C41 cells (Miroux and Walker, 1996) and the protein was expressed for 12 h at 22○C. The U11-48K Zn-finger domain was first purified using Ni-affinity chromatography followed by TEV protease cleavage. The next steps of purification included a second Ni-column to separate the cleaved fusion partner and a final gel filtration. Protein homogeneity and purity was assessed using mass spectrometry and SDS-PAGE.
Zinc binding of U11-48K was determined using the PAR/PMPS assay as previously described (Hunt et al., 1985). For a detailed description of this method and other biophysical methods used in this study, refer to the Supplementary Material.
3 RESULTS
3.1 Domain identification and phyletic distribution
Multiple sequence alignment constructed from U11-48K homologs revealed a highly conserved region containing four invariant Cys and His residues. Using a consensus pattern C-P-X(4)-H-X(9)-H-X(3)-C derived from the alignment and the conserved region of human U11-48K as a query sequence we performed a PHI-BLAST search (E-value threshold of 200) against NR database. The pattern search retrieved similar segments from ∼190 proteins. Some of these proteins produced two distinct matches, suggesting that this region is tandemly repeated. Despite the insignificant PHI-BLAST scores, all identified fragments had a high sequence identity (>30%) with the query sequence.
In order to evaluate the significance of the PHI-BLAST results, we initially clustered the full-length sequences matching the pattern. Multiple alignments generated for each distinct cluster were evaluated for the presence of common sequence motifs. Based on the observed conserved features we further refined and merged the initial clusters. In addition, we performed iterative PSI-BLAST searches (profile inclusion threshold of 0.01) against the NR database using representative sequences from each cluster as queries. These searches yielded the identification of new homologs with similar separation of the Cys- and His-residues (full list of the identified sequences is provided in the Supplementary Material). The profile searches also indicated a number of false positives (7.8% of all PHI-BLAST hits) in some singleton clusters and they were excluded from further consideration.
As a result, we obtained three clusters corresponding to three distinct protein families. Besides the U11-48K family, these include the families of tRNA methyltransferase TRM13 and gametocyte specific factors GTSF (also known as UPF0224), both named after their only experimentally characterized members. The TRM13 family is typified by the yeast protein YOL125w (Genbank identifier gi:74676591) that catalyses the 2′ O-methylation of tRNA, whereas the GTSF family is represented by the D7 protein (gi:118217) and Cue110 (gi:29244024) that are preferentially expressed in germinal tissues. The common region (∼30 residues) containing the conserved pattern was present in a single copy in all identified U11-48K and TRM13 homologs. All GTSF homologs contained two repeats of this motif separated by a short linker.
Cys- and His-residues are usually involved in metal ion coordination. Given the size of the identified fragments, their repetitive nature and strict conservation of Cys- and His-residues, it is likely that this region represents an independent domain folded around a central zinc ion. Therefore, hereafter we refer to it as a CHHC Zn-finger domain.
The CHHC Zn-finger domains were found only in eukaryotes. Multiple alignment and phylogenetic analysis of all identified Zn-finger domains showed that they diverge in four main clusters that to a large extent correspond to the U11-48K, TRM13 and GTSF protein families. Homologs of U11-48K were found in vertebrates, plants and insects that are known to contain U12-type introns. Analysis of the genomic and EST databases revealed previously unreported orthologs in Schistosoma mansoni, Biomphalaria glabrata, Hirudoo medicinalis and Ciona intestinalis. The presence of U11-48K in these organisms is consistent with the proposed early evolution of the minor spliceosome (Russell et al., 2006). U12-type introns have been identified in some protists and fungi, but we were unable to detect U11-48K relatives in these genomes.
In contrast to U11-48K, the TRM13 family is more ubiquitous and present in protozoa, fungi, plants and metazoa. We found that in some amoebozoan genomes, in particular Entamoeba dispar and Entamoeba histolytica, the TRM13 gene is duplicated. The yeast TRM13 is responsible for the 2′-O-methylation of ribose in the position 4 of tRNAPro, His, Gly (Wilkinson et al., 2007). Apparently, this modification is highly conserved in eukaryotes, including humans, and can account for the high conservation and wide distribution of TRM13 enzymes.
GTSF homologs were found in two protozoan, Tetrahymena thermophila and Paramecium tetraurelia, and a vast number of metazoan genomes. No orthologs were identified in fungi and plants. In most of the genomes, the predicted GTSF gene is present in multiple copies, but some lineages have a single copy of this gene (Supplementary Material). The P. tetraurelia, for instance, possesses three predicted GTSF homologs, whereas T. thermophila has only one. A recent analysis of the P. tetraurelia genome suggests that Paramecium lineage probably underwent at least three whole-genome duplications (Aury et al., 2006). Thus the presence of two or more GTSF copies in the genomes can account for a gene duplication event that had happened early in the eukaryotic evolution. This also suggests a plausible evolutionary scenario that includes an early duplication and fusion of the Zn-finger domain followed by multiple gene duplications. The metazoan GTSF-like proteins formed two paralogous subfamilies (named GTSF-FamA and GTSF-FamB) with distinct sequence motifs and predicted gene structure. The duplicated CHHC Zn-finger motif in GTSF-FamA corresponds to a single exon, whereas each Zn-finger domain in GTSF-FamB is encoded by an individual exon. Similar domain-exon correlation was observed in all U11-48K homologs. These findings suggests that the Zn-finger domain might be a minimal unit of evolution and events such as exon shuffling may have contributed to the architectural and functional diversity observed in the CHHC Zn-finger containing proteins.
3.2 Domain features and architectures
The CHHC Zn-finger does not show sequence similarity to any known or previously characterized Zn-finger domain. Multiple alignment of all identified CHHC Zn-finger domains indicated several conserved residues, some of which may contribute to the fold stability (Fig. 1 and Supplementary Material, Fig. 1). Besides the strictly conserved Cys- and His-residues, residues with hydrophobic side-chains tend to occupy positions 1, 2, 6, 14, 19 and 23. Highly conserved are a Pro residue at position 5 and Asp/Asn (Asx) at position 7. Asx and Pro are usually found in turns. The high conservation of these residues in the CHHC Zn-finger domains may reflect a requirement for a specific backbone conformation. The multiple alignment also revealed several clusters of positively charged residues that may have a functional role. In Znf1-GTSF, all vertebrate U11-48K and nearly all TRM13 Zn-finger domains these residues were located near the C-termini. The amoebozoan TRM13 and the Znf2-GTSF Zn-finger domains had very distinct patterns of basic residues that could account for different function.
Fig. 1.
(A) Representative multiple alignment of the CHHC Zn-finger domains (a complete alignment is provided in the Supplementary Materials). Amino acid residues are coloured according to Clustal colour scheme. Sequences derived from ESTs are marked with *. (B) Sequence logo of all identified CHHC Zn-finger domains, generated with WebLogo tool (Crooks et al., 2004).
With the exception of TRM13, the other two protein families had very simple domain architectures with one or two Zn-finger domains as their only globular domains (Supplementary Material, Fig. 2). We found that the amino acid composition of U11-48K proteins significantly differs from the average amino acid composition observed in globular domains (data not shown). These proteins are enriched in disorder promoting residues (Q, R, S, P, E, K) and have a low content of order promoting residues (Dunker et al., 2001). For instance, human U11-48K is enriched with E, R, K and S as much as 2.0-, 2.2-, 1.5- and 1.4-fold, respectively. This analysis was supported by the prediction of nearly 55% protein disorder with PONDR and the identification of two long low-complexity regions at the N- and C-terminus with SEG. Taken together, this suggests that the central part containing the CHHC Zn-finger is probably the only region of folded globular structure.
In addition to the CHHC Zn-finger domain, the TRM13 proteins contain a putative CCCH-type Zn-finger and methyltransferase domain (Pfam domain PF05206, Finn et al., 2006). Using a transitive sequence database search we were able to detect similarity between the TRM13 methyltransferase domain and classical methyltransferases. This result was confirmed by two-step iterative PSI-BLAST search that retrieved with significant E-values a number of methyltransferase domains classified in SCOP (e.g. d1p91a_with E=1e-34, d1kpga_with E=1e-31, d1sqga2 with E=4e-14; SCOP Superfamily sunid=53335). A combination of Zn-finger and methyltransferase domains is not uncommon. It is found, for instance, in another family of RNA modification enzymes, RlmAI/RlmAII, that catalyzes the methylation of the 23S rRNA nucleotides G745/G748 in g–/g+bacteria, respectively. The Zn-finger domain in these enzymes is thought to be responsible for the specific recognition and binding to the rRNA substrate (Das et al., 2004).
3.3 Biochemical characterization
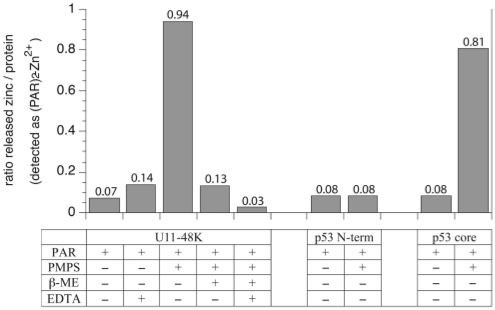
We have cloned and expressed the CHHC Zn-finger of the human U11-48K protein. The CD spectrum indicates that the purified domain is partly structured. The 1D-NMR spectrum shows several resonances beyond 8.5 p.p.m. (up to 9.3 p.p.m.), which clearly demonstrates that the domain is folded and thus provides evidence that the CHHC Zn-finger can exist as an independent folding unit (Supplementary Material, Fig. 3). By using colorimetric zinc-binding assay, we confirm that the predicted CHHC Zn-finger domain indeed binds zinc. Our results show that it stoichiometrically binds zinc ions in a one-to-one ratio (Fig. 2).
Fig. 2.
Molecular ratio of released zinc per mole protein. Zinc release was measured by following absorbance changes at 500 nm (using 100μM PAR as reference) for 15 μM U11-48K CHHC Zn-finger domain, p53 N-terminus (negative control) and p53 core domain (positive control), respectively, at 25○C. Upon addition of PMPS, the Zn2+-complexing cysteines are modified by PMPS and one Zn2+ is released per U11-48K and forms an orange Zn2+-(PAR)2 complex. The zinc release can be almost completely reversed by addition of excess β-mercaptoethanol, which competes off PMPS from cysteines.
3.4 Probable biological function
The CHHC Zn-finger motif may possess a RNA binding function. This prediction arises from the fact that this domain is present in two proteins known to bind RNA (U11-48K and TRM13) and that it contains the sequence determinants that can mediate this function.
Yeast TRM13 catalyses the methylation of a ribose 2′ OH in position 4 of tRNAPro, tRNAHis and tRNAGly. This is one of the few modifications found in the middle of duplex RNA and one of only four modifications that occur in the acceptor stem of the tRNA. Binding to tRNA is crucial for the enzyme selectivity and substrate recognition. TRM13 binds with high affinity to its cognate substrates but exhibits no detectable binding to other tRNA species (Wilkinson et al., 2007). It is conceivable that the CHHC Zn-finger domain may mediate this selective binding by recognizing a specific shape or particular structure of the cognate tRNAs. It may also contribute to the correct positioning of the substrate tRNA in the enzyme active site.
During the preparation of this article, Turunen et al. (2008) reported that U11-48K protein contacts the 5′ splice site in the early spliceosomal complex and that this interaction requires U11-5′ ss base-pairing as a prerequisite. Besides the conserved Zn-finger domain, U11-48K contains a strictly conserved, probably non-globular region rich in basic residues that can potentially bind to RNA. Having in mind that the 5′ splice site is highly conserved in the U12-type introns, two hypotheses can account for the interaction of U11-48K with the 5′ splice site. One possibility is that the non-globular region of U11-48K mediates this interaction. Alternatively, the CHHC Zn-finger domain may directly contact the 5′ splice site and thereby contribute to the U12-intron recognition.
The molecular function of the GTSF family is unknown, but the presence of two copies of a CHHC Zn-finger motif allows function prediction. Two members of this protein family, the xenopus protein D7 and mouse Cue110, are preferentially expressed in gametocytes and are localized in the cytoplasm (Smith et al., 1988; Yoshimura et al., 2007). We propose that these proteins may play a role in the storage of maternal and paternal RNA that is synthesized in large amounts during early gametogenesis.
4 CONCLUSIONS
The CHHC Zn-finger is a novel domain present in spliceosomal U11-48K proteins, TRM13 enzymes and gametocyte specific factors. Our experimental analysis confirms that this domain is a genuine Zn-binding module that in isolation behaves as an independent folding unit. Based on the conserved sequence features, we predict that this domain may function as a RNA recognition and binding module. Future analysis will aim to structurally characterize this domain and to elucidate its molecular function.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs Alexey Murzin and Kiyoshi Nagai for the careful reading of the article and helpful suggestions, Dr Maria Garcia-Alai for advice on the Zn-binding assay and Dr Trevor Rutherford for help with the NMR experiment. A.A. thanks Prof. Sir Alan Fersht for the continuing financial support.
Funding: Trinity College Junior Research Fellowship to H.T.
Conflict of Interest: none declared.
REFERENCES
- Altschul S, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva A, et al. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aury J, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- Burge C, et al. Evolutionary fates and origins of U12-type introns. Mol. Cell. 1998;2:773–785. doi: 10.1016/s1097-2765(00)80292-0. [DOI] [PubMed] [Google Scholar]
- Clamp M, et al. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- Crooks G, et al. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das K, et al. Crystal structure of RlmAI: implications for understanding the 23S rRNA G745/G748-methylation at the macrolide antibiotic-binding site. Proc. Natl Acad. Sci. USA. 2004;101:4041–4046. doi: 10.1073/pnas.0400189101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker A, et al. Intrinsically disordered proteins. J. Mol. Graph. Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- Finn R, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frilander M, Steitz J. Initial recognition of U12-dependent introns requires both U11/5' splice-site and U12/branchpoint interactions. Genes Dev. 1999;13:851–863. doi: 10.1101/gad.13.7.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt J, et al. The use of 4-(2-pyridylazo)resorcinol in studies of zinc release from Escherichia coli aspartate transcarbamoylase. Anal. Biochem. 1985;146:150–157. doi: 10.1016/0003-2697(85)90409-9. [DOI] [PubMed] [Google Scholar]
- Levine A, Durbin R. A computational scan for U12-dependent introns in the human genome sequence. Nucleic Acids Res. 2001;29:4006–4013. doi: 10.1093/nar/29.19.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miroux B, Walker J. Over-production of proteins in Escherichia.coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- Romero P, et al. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Russell A, et al. An early evolutionary origin for the minor spliceosome. Nature. 2006;443:863–866. doi: 10.1038/nature05228. [DOI] [PubMed] [Google Scholar]
- Smith R, et al. Destruction of a translationally controlled mRNA in Xenopus oocytes delays progesterone-induced maturation. Genes Dev. 1988;2:1296–1306. doi: 10.1101/gad.2.10.1296. [DOI] [PubMed] [Google Scholar]
- Turunen J, et al. The U11-48K protein contacts the 5′ splice site of U12-type introns and the U11-59K protein. Mol. Cell. Biol. 2008;28:3548–3560. doi: 10.1128/MCB.01928-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson M, et al. The 2′-O-methyltransferase responsible for modification of yeast tRNA at position 4. RNA. 2007;13:404–413. doi: 10.1261/rna.399607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will C, et al. The human 18S U11/U12 snRNP contains a set of novel proteins not found in the U2-dependent spliceosome. RNA. 2004;10:929–941. doi: 10.1261/rna.7320604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton J, Federhen S. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 1996;266:554–571. doi: 10.1016/s0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]
- Yoshimura T, et al. Gene expression pattern of Cue110: a member of the uncharacterized UPF0224 gene family preferentially expressed in germ cells. Gene~Expr. Patterns. 2007;8:27–35. doi: 10.1016/j.modgep.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Zhang Z, et al. Protein sequence similarity searches using patterns as seeds. Nucleic Acids Res. 1998;26:3986–3990. doi: 10.1093/nar/26.17.3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.