Abstract
The synthesis of thymidylate (TMP) occupies a convergence of two critical metabolic pathways: folate metabolism and pyrimidine biosynthesis. Thymidylate is formed from deoxyuridylate (dUMP) using N5, N10 methylene tetrahydrofolate. The metabolic relationship between dUMP, TMP, and folate has been the subject of cancer research from prevention to chemotherapy. Thymidylate stress is induced by nutritional deficiency of folic acid, defects in folate metabolism, and by antifolate and fluoropyrimidine chemotherapeutics. Both classes of chemotherapeutics remain mainstay treatments against solid tumors. Because of the close relationship between dUMP and TMP, thymidylate stress is associated with increased incorporation of uracil into DNA. Genomic uracil is removed by uracil DNA glycosylases of base excision repair (BER). Unfortunately, BER is apparently problematic during thymidylate stress. Because BER requires a DNA resynthesis step, elevated dUTP causes reintroduction of genomic uracil. BER strand break intermediates are clastogenic if not repaired. Thus, BER during thymidylate stress appears to cause genome instability, yet might also contribute to the mechanism of action for antifolates and fluoropyrimidines. However, the precise roles of BER and its components during thymidylate stress remain unclear. In particular, links between BER and downstream events remain poorly defined, including damage signaling pathways and homologous recombination (HR). Evidence is growing that HR responds to persistent BER strand break intermediates and DNA damage signaling pathways mediate cross talk between BER and HR. Examination of crosstalk among BER, HR, and damage signaling may shed light on decades of investigation and provide insight for development of novel chemopreventive and chemotherapeutic approaches.
Keywords: thymidylate deprivation, folate deficiency, base excision repair, homologous recombination
1. Introduction
Thymidylate (TMP) is derived from a unique reaction, representing the convergence of two critical metabolic pathways: the one-carbon donor pathway of folate metabolism and the de novo pyrimidine biosynthetic pathway (Figure 1). The reaction is catalyzed by the enzyme thymidylate synthase (TS), which converts deoxyuridylate (dUMP) to TMP using the folate co-substrate, 5,10-methylenetetrahydrofolate (CH2H4PteGlu), as both carbon donor and reductant. Because TMP synthesis requires a folate derivative and is also required for DNA synthesis, TS is a focal point for research in cancer prevention and treatment. First, nutritional deficiency of folic acid or defects in folate metabolism are potentially pro-carcinogenic [1]. Second, TS is an important target in the treatment of cancer, especially gastrointestinal cancers [2]. TMP is converted through the actions of thymidylate kinase and nucleoside diphosphate kinases to TTP (Figure 2), a substrate for DNA polymerases involved in replication and repair [3]. Thus, reduced TMP synthesis resulting from dietary insufficiency, mutation in genes encoding key biosynthetic enzymes, or chemotherapeutic inhibition causes depletion of TTP, which is cytotoxic in all prokaryotic and eukaryotic organisms examined as opposed to being cytostatic [4]. Although the phenomena of thymineless stress (TLS) and thymineless death (TLD) have been investigated for nearly 50 years, consensus on the critical mechanism(s) underlying the stress response has been difficult to achieve. Moreover, redundancy is observed in TMP synthesis, presumably to reduce the risk of TMP depletion (Figure 2). TMP is formed from thymidine by thymidine kinase (TK) isoforms located in the cytoplasm (TK1) and mitochondria (TK2) [5]. The salvage pathway provides protection from folate deficiency; however, it is presumed to contribute to resistance to chemotherapeutic regimens targeting TS. It is well accepted that TLS and TLD are capable of inducing DNA damage, and that DNA double strand breaks (DSBs), in particular, are associated with cell death. However, the precise source or sources of the DSBs as a result of direct and indirect effects on replication, repair, and damage signaling responses remain obscure and are the focus of this review.
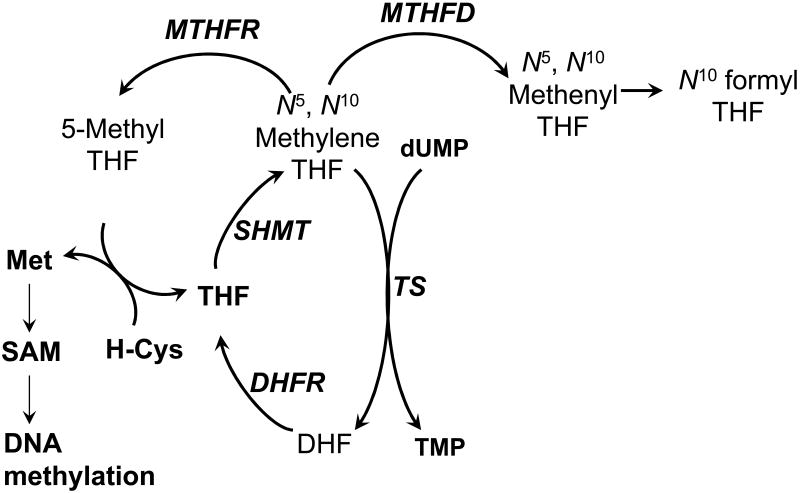
Figure 1. Simplified schematic of folate metabolism focused on the synthesis and utilization of N5,N10 methylene tetrahydrofolate.
N5,N10 Methylene tetrahydrofolate can be utilized as the methyl donor and reductant in TMP synthesis, the methyl donor for the conversion of homocysteine to methionine, and the eventual formyl donor for de novo purine synthesis. Enzymes are in italics. MTHFR, methylene-THF reductase; SHMT, serine hydroxymethyl transferase; TS, thymidylate synthase; MTHFD, methylene-THF dehydrogenase; SAM, S-adenosyl methionine; Met, methionine; H-Cys, homocysteine; THF, tetrahydrofolate; DHF, dihdyrofolate.
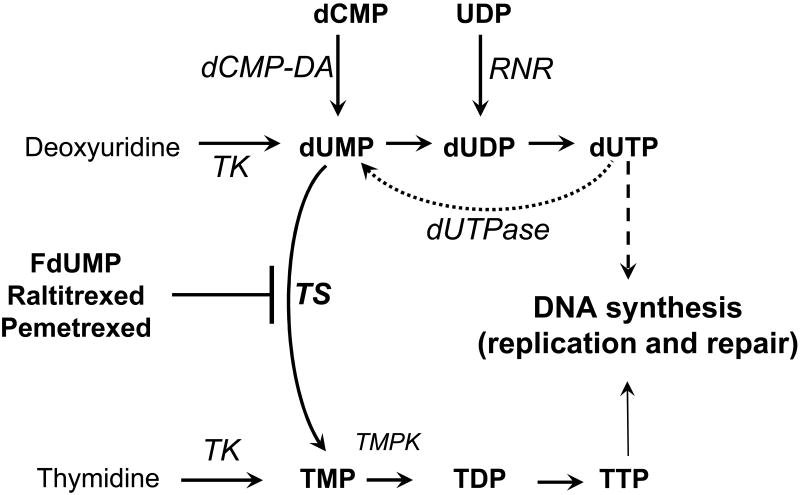
Figure 2. Simplified schematic of dUMP and TMP metabolism leading to DNA synthesis.
Enzymes are in italics. TS, thymidylate synthase; TK, thymidine kinase; TMPK, thymidylate kinase, dUTPase, dUTP nucleotidohydrolase. RNR, ribonucleotide reductase; dCMP-DA, dCMP deaminase. TS is the de novo source of TMP, while TK provides a salvage route to TMP and dUMP. dUTPase prevents dUTP accumulation. Raltitrexed and pemetrexed are folate analogs that interfere with binding of N5,N10 Methylene tetrahydrofolate to TS. Fluorodeoxyuridylate (FdUMP) binds in the nucleotide binding pocket and forms an irreversible ternary complex.
Folate deficiency and carcinogenesis
The pro-carcinogenic effects of folic acid deficiency are thought to be due in part to the induction of TLS and associated DNA damage. Low plasma levels of folate lead to a decrease in intracellular levels of tetrahydrofolates, particularly CH2H4PteGlu, which is not only a substrate for TS but also for methylenetetrahydrofolate reductase (MTHFR) and methylenetetrahydrofolate dehydrogenase (MTHFD) (Figure 1). The partitioning of CH2H4PteGlu among three critical pathways leading to TMP, purine, and methionine biosynthesis is likely to limit TMP synthesis under conditions of low dietary intake of folate. Pathways leading to DNA synthesis and methylation are postulated to be perturbed by folate deficiency. DNA strand breakage associated with uracil misincorporation has been reported after exposure of cultivated mammalian tumor cells and human lymphocytes to folate concentrations sub-optimal for growth and in tissues from rodents maintained on low folate diets [1].
Increased uracil in DNA is observed under conditions of TLS (Figure 2). In humans, a significant inverse correlation was reported between uracil misincorporation in lymphocyte DNA and red blood cell folate [6]. Folate supplementation resulted in a decrease in uracil incorporation in DNA, and genomic uracil was reported to serve as a marker of folate status. However, no significant correlations were observed between folate status and DNA strand breakage or global DNA methylation. In studies of genomic alterations associated with folate deficiency, an increase in strand breaks in exons 5-8 of the p53 gene has been reported in the colons of rodents on folate-deficient diets [7]. In recent studies, strand breaks in the p53 gene were detected in human lymphocytes cultivated in vitro in low folate media. Interestingly, p53 protein levels were significantly elevated in cells cultivated in low folate [8]. These studies provide a possible link between folate deficiency and carcinogenesis.
Studies of folate deficiency have also focused on interactions between genotype and pathological processes. An interaction between low dietary folate and the MTHFR gene has been reported in a murine model [9]. Mice heterozygous for a null MTHFR allele and maintained on low dietary folate exhibited an increase in immunostaining for the phosphorylated form of histone H2AX, γ-H2AX. DSBs are known to induce γ-H2AX, although it is less clear during deoxynucleotide deprivation whether stalled replication forks can induce γ-H2AX directly or must first collapse. In addition, MTHFR+/- mice on low dietary folate exhibited decreased expression of polo-like kinase 1 and cell division cycle 25C (CDC25C), genes that are involved in G2/M cell checkpoint control. The activity of MTHFR in heterozygous mice was reported to be similar to that of humans homozygous for the MTHFR C677T allele. MTHFR C677T encodes an enzyme that exhibits reduced catalytic activity (reviewed in [10]). A number of pathological states are associated with MTHFR C677T homozygosity (and even heterozygosity), including neural tube defects, coronary artery disease, and cancer [10]. The relative contributions of reduction in TMP synthesis and other effects associated with folate deficiency, including DNA hypomethylation, to these pathologies remain unclear.
Chemotherapeutic induction of TLS
Two classes of drugs in clinical use are mechanism-based inhibitors of TS. The pro-drugs, 5-fluorouracil (5-FU) and capecitabine, are converted intracellularly to 5-fluorodeoxyuridylate, an analog of dUMP. Pemetrexed and raltitrexed (RTX) are analogs of CH2H4PteGlu. Upon binding to TS, inhibitory complexes are formed that are catalytically inactive, resulting in depletion of TMP.
Loss of TS function leads not only to a decrease in TMP formation but also an increase in substrates, with dUMP accumulating to high levels (Figure 2). Expansion of dUMP pools after exposure to TS inhibitors has been reported in tumors in vivo and in cultured cells [3]. In studies of human and murine tumor cells, steady-state levels of dUMP in unstressed cells were 5-10 μM; however, after exposure of cells to 5-FU, dUMP levels increased approximately 100-fold within 3 hr [11]. The elevation in dUMP occurring after inhibition of TS may be exacerbated by TTP depletion. TTP exerts feedback inhibition on the synthesis of three enzymes involved in dUMP biosynthesis: ribonucleotide reductase, deoxycytidylate deaminase, and TK (Figure 2) [12]. dUMP is also formed from dUTP by the action of dUTP nucleotidohydrolase (dUTPase), which has nuclear and mitochondrial isoforms. In unstressed cells, dUTPase plays an important role in preventing dUTP from incorporation into DNA and in provision of substrate for TS [13]. In concert, the actions of TS and dUTPase are expected to favor high TTP/dUTP. In mutant strains of E. coli with reduced dUTPase activity, DNA fragmentation and an increase in recombination were observed [14]. Under conditions of elevated dUMP (and dUTP) pools associated with folate deficiency or TS inhibition, the activity of dUTPase is likely to be overwhelmed. Since dUTP is an excellent substrate for DNA polymerases, elevation in dUTP in concert with TTP depletion leads to an increase in the incorporation of dUTP into DNA. Elevation in dUTP resulted in DNA fragmentation in mammalian cells even under conditions [deoxyuridine + HAT (hypoxanthine, amethopterin, thymidine)] that generate intracellular ratios of dUTP:TTP of approximately 1 [15]. The data demonstrate that DNA damage occurs upon elevation of dUTP even without TTP depletion and highlight the importance of dUMP accumulation in the genotoxic actions of TLS.
TLS is associated with not only increased uracil in DNA but also an imbalance in nucleotide pools. In studies in murine tumor cells utilizing both genetic (TS-defective mutants that are thymidine auxotrophs) or pharmacological approaches (TS inhibitors) to induce TLS, nucleotide pool analysis indicated that dATP pools increase and dGTP pools decrease in cells undergoing TLS [16]. The TS inhibitor 5-fluoro-2′-deoxyuridine (FdUrd) induced cytotoxicity associated with DSBs and also appeared to activate an endonuclease [16]. The investigators postulated that nucleotide pool imbalance induces DSBs via endonuclease activation, although it was not shown whether the endonuclease activity was repair or apoptotic. A number of investigations have addressed the association of TLD and DNA fragmentation patterns. TLD in either TS-deficient FM3A cells starved of thymidine or FM3A cells exposed to FdUrd led to the formation of DNA fragments 50-200 kb in length [17]. Pulse-labeling studies indicated that strand breaks occurred at newly replicating sites. DNA fragmentation patterns associated with TLD in human colon tumor cell lines were reported to vary [18]. Exposure of three human colon tumor cell lines to FdUrd or CB3717 (folate-based TS inhibitor) resulted in the formation of fragments that are either discrete (50- to 200-kb) or heterogeneous (50 kb to 1-5 Mb) in size. Independent of fragmentation pattern, drug cytotoxicity correlated with extent of DNA fragmentation. More recently, a similar pattern (50-200 kb) of fragmentation was seen in murine embryonic fibroblasts (MEFs) treated with RTX, although caspase induction and PARP cleavage was also seen, suggesting that apoptosis had been induced [19]. In a TS-deficient human colon tumor cell line deprived of thymidine, DNA fragments that are oligonucleosomal in size were observed, a pattern consistent with apoptosis [20]. No accumulation of dUTP was observed after thymidine deprivation and elevation of dATP pools and decline of TTP pools were associated with cytotoxicity. In a subsequent investigation, a dose-dependent and time-dependent accumulation of DNA strand breaks was observed after a brief (2 hr) exposure to the antifolate RTX; no evidence for nucleosomal ladder formation was obtained during the duration of strand break analysis [21]. Clearly, interpretation of the data is hindered by a lack of knowledge concerning the source of the strand breaks leading to DNA fragmentation: activation of DNA repair and/or recombination pathways in response to uracil misincorporation and/or nucleotide pool imbalance or downstream damage response signaling pathways that activate apoptotic pathways.
Studies examining the role of dUTPase in the cytotoxicity of TS inhibitors suggest that dUTP incorporation plays an important role in TLD. TS inhibition after exposure to FdUrd was similar in two human colon tumor cell lines, yet they differed in cytotoxic response [22]. The relative resistance was associated with a > 4-fold higher activity of dUTPase, resulting in a significantly lower accumulation of dUTP. In subsequent studies, transfection of the more sensitive cell line with an expression construct encoding dUTPase from E. coli resulted in protection from FdUrd-mediated DNA fragmentation and cytotoxicity [23]. Down-regulation of dUTPase by siRNA in human cancer cell lines resulted in elevation in dUTP after inhibition of TS by FdUrd. In two of three cell lines studied, elevation in dUTP was associated with an increase in DSBs and hypersensitivity to FdUrd. In the third cell line, dUTP pool elevation had no significant effect on either DNA damage or chemosensitivity [24]. The relevance of dUTP incorporation in TLD has been addressed in short- and long-term cytotoxicity studies. In two cell lines that differed significantly in accumulation of dUTP, cells with higher levels of dUTP were more sensitive to short-term exposure to ZD9331 (folate-based TS inhibitor); however, these cells were less sensitive to ZD9331 as determined by a long-term viability assay [25]. This suggests that dUTP misincorporation causes DNA damage that leads to growth inhibition, but that cell death results from additional events such as dNTP pool perturbations.
Collectively, DNA damage in the form of DSBs is commonly seen following TLS caused by folic acid deficiency or by chemotherapeutic inhibition of TS, and in the latter case is associated with cell death. Among the potential sources of DNA damage, uracil misincorporation into DNA and its removal by base excision repair (BER) has been the most commonly presumed source. It is interesting to note that under folic acid deficiency, BER would exert a pro-carcinogenic influence, while BER activity during chemotherapeutic TS inhibition would be antiproliferative to cancer cells. BER is discussed in greater detail in the next section.
2. Base excision repair and uracil DNA glycosylases
Genomic Uracil and UDG-initiated BER
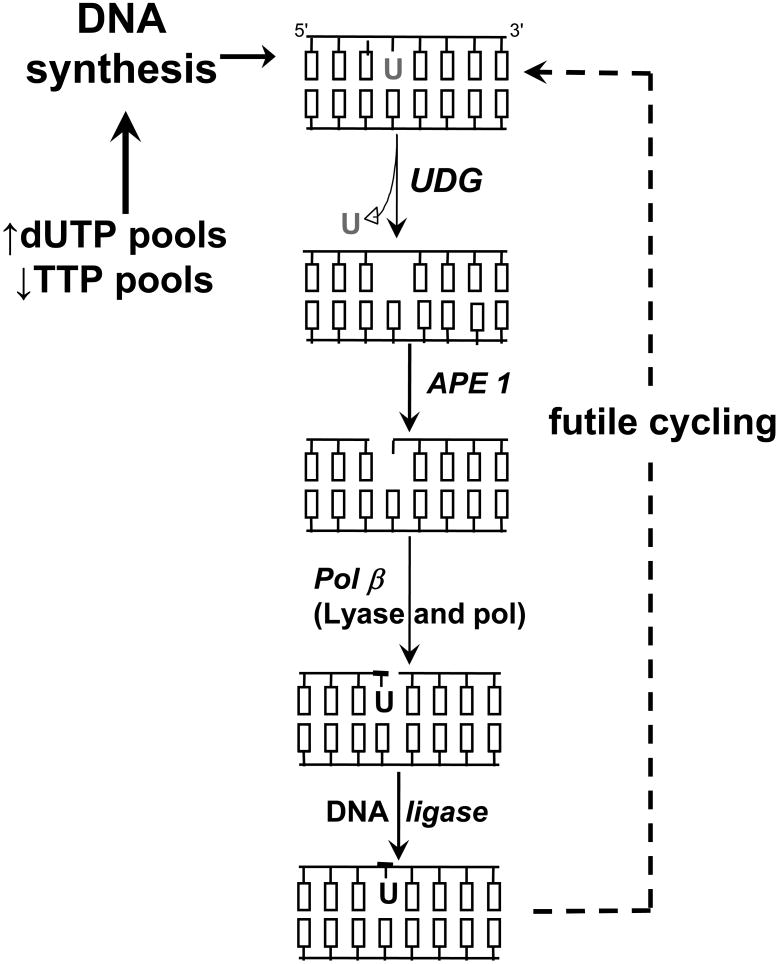
There are two sources of genomic uracil, namely hydrolytic deamination of cytosine and incorporation of dUMP during replication [26]. Base excision repair (BER) is the only pathway known to remove genomic uracil in mammalian cells [27]. BER is initiated by DNA glycosylases that cleave the glycosidic bond to release incorrect or damaged bases. Briefly, the subsequent enzymatic steps of BER are: 1) abasic site (apurinic/apyrimidinic, AP) endonuclease activity that produces a strand break with a 3′-OH and 5′-deoxyribosephosphate (5′-dRP) group; 2) dRP lyase to remove the 5′-dRP group and produce a free 5′-phosphate; 3) polymerase activity for DNA resynthesis; and 4) ligase to seal the nick (Figure 3).
Figure 3. Simplified schematic of base excision repair during TLS.
In the first step of BER, UDG activity catalyzes uracil release and produces an abasic site in DNA. AP endonuclease produces a nick 5′ to the abasic site. The dRP lyase activity of pol β creates a 5′-phosphate and polymerase activity fills the one nucleotide gap in short-patch BER. DNA ligase seals the nick. During TLS, insufficient TS activity decreases TTP and increases dUTP, overwhelming dUTPase and driving genomic uracil incorporation.
There are four known genetic loci in humans that encode for proteins capable of removing uracil [26]. Note that the abbreviation UDG (uracil DNA glycosylase) refers to the biochemical activity of one or more enzymes, whereas the abbreviations below refer to specific loci and polypeptides. Biochemical characterization of the four human enzymes suggests specialized roles [27]. The UNG genetic locus encodes mitochondrial (UNG1) and nuclear (UNG2) isoforms [27]. UNG2 appears to account for the bulk of cellular UDG activity [27]. The primary role of UNG2 seems to be counteracting uracil misincorporation during replication [28, 29]. Ung-/- mice are viable but develop later onset B-lymphomas [30]. Interestingly, a model of somatic hypermutation promoted by enzymatic deamination of cytosines in DNA via activation-induced deaminase (AID) involves UNG–induced BER [30]. A second UDG named SMUG1 excises a broader range of damaged pyrimidines not excised by UNG [27]. SMUG1 deficient mice have not yet been reported. UNG and SMUG1 do not show an opposing base preference and can remove U from single stranded DNA. In contrast to UNG and SMUG1, MBD4 (Methyl Binding Domain protein 4, also known as MED1) and thymine DNA glycosylase (TDG) specifically recognize substrates mispaired with guanine in double stranded DNA [27]. MBD4 has a preference for removing uracil or thymine in the CpG sequence context, thus suggesting that MBD4 prevents mutagenesis at sites of promoter methylation [31-33]. Mbd4-/- mice are viable, but show a 2-3 fold increased mutation frequency at CpG sites, and the absence of MBD4 accelerates the intestinal tumor predisposition of the ApcMin/+ mice [32, 33]. Mutations in the MBD4 gene have also been reported in human colorectal cancers with high microsatellite instability (MSI-H) [34, 35], cancers most commonly associated with defects in mismatch repair (MMR). TDG removes other damaged pyrimidines in addition to uracil, most notably 3,N4-ethenocytosine [36]. Interestingly, Tdg-/- mice are informally reported to be embryonic lethal and deficient cell lines have not yet been described [36]. Collectively, these observations suggest that UNG2 is the primary UDG that removes U incorporated during replication and MBD4 specializes in preventing mutagenesis at CpG sequences, while TDG and SMUG1 can remove other types of damaged pyrimidines in addition to genomic uracil. The precise biological roles of the four DNA glycosylases and their contribution towards or against mammalian carcinogenesis have yet to be fully elucidated.
UDG-initiated BER during TLS
Uracil excision and the DNA resynthesis step of BER become problematic during folate deficiency or chemotherapeutic inhibition of TS (Figure 3). The lack of TTP and elevation in dUTP cause the reintroduction of genomic uracil. Thus, continued UDG activity and uracil incorporation during repair synthesis would create a ‘futile cycling’ of BER (Figure 3). In contrast to base damage that predominantly causes point mutations, BER strand break intermediates are clastogenic and in most cases are more toxic than the initial damaged base [37]. A simple prediction from the futile cycling model is that limiting UDG activity would reduce BER intermediates from accumulating and reduce clastogenic events. The biological implications from this model are that UDG activity would be pro-carcinogenic to normal cells during folate deficiency, but exhibit anti-tumor activity during treatment with TS inhibitors. What is the evidence for BER as a procarcinogenic process during folate deficiency? Although a few studies have determined genomic uracil and strand breaks during folate deficiency, there is surprisingly little evidence directly implicating specific UDGs in mammalian cell models. Ames and coworkers found that genomic uracil incorporation and DNA strand breaks appeared to directly associate with folate deficiency [38, 39]. Other studies noted an association between genome instability as measured by the comet assay and folate deficiency [40, 41]. Collectively, there is reasonable evidence that folate deficiency is associated with genomic uracil and genome instability, but the mechanistic details and precise links between BER and clastogenesis are not well understood.
Does BER futile cycling contribute to cell death in cancer cells treated with TS inhibitors? The indirect inference is yes, although most of these anti-metabolites have multiple mechanisms of action. The extent to which UDG-initiated BER directly contributes to cancer cell death caused by chemotherapeutic antifolates and fluoropyrimidines remains controversial despite decades of clinical use [42, 43]. 5-FU metabolites have additional mechanisms of action, notably incorporation into nucleic acids. Incorporation into DNA has received attention more recently as a mechanism of cell killing for 5-FU. 5-FU in DNA can be recognized by the SMUG1, TDG, and MBD4 DNA glycosylases in addition to mismatch repair [42, 44-46], but will not be discussed in detail here. Increasing dUTP via manipulation of dUTPase activity pools during TS inhibition sensitized some, but not all, cell types examined, which indirectly suggests that uracil incorporation and BER futile cycling occurs [23-25, 47, 48]. In all of the above-cited studies, the amount of uracil incorporated into DNA was not directly measured and BER status was not evaluated. A number of studies directly examining BER proficient and deficient cells have not provided consistent evidence that BER futile cycling occurs or contributes to [19, 42, 43, 49-52]. For example, there was no difference in RTX, FdUrd, or 5-FU toxicity in cells inhibited for UNG activity compared to UNG proficient cells, despite an accumulation of genomic uracil in the UNG inhibited cells [51]. RTX and FdUrd treatment induced γ-H2AX independent of UNG activity, which suggests that collapsed replication forks might be the source of DSBs [51]. This and other studies suggest that UNG activity does not play a role [49, 52], yet it has also been proposed that FdUrd can cause the premature degradation of UNG2 in some cancer cell lines [43]. In short, the role of UDGs in contributing to cell killing caused by TS inhibitors remains enigmatic.
Downstream BER events
DNA polymerase β (Pol β) is a central component of BER in that it performs two enzymatic functions, namely DNA polymerase and 5′-dRP lyase (Figure 3). The 5′-dRP group results from AP endonuclease activity and must be removed to create ligatable 3′-OH and 5′-phosphate ends. Importantly, the 5′-dRP lyase activity of Pol β, not polymerase activity, is required to protect against toxicity caused by DNA damaging agents [37]. Initiating BER in the absence of Pol β leads to negative consequences including sister chromatid exchanges (SCEs) and apoptosis [37]. Folate deficiency has been examined in Pol β+/- mice [53]. The conclusion was that combining folate deficiency and Pol β haploinsufficiency increased unrepaired BER intermediates [53]. The authors were surprised to find that Pol β was not upregulated as a result of folate deficiency, when the prediction was that increased genomic uracil might upregulate BER in response to DNA damage. Interestingly, upregulation of UNG activity in liver tissue of folate deficient animals was reported while downstream BER activity (APE1 and Pol β) remained unchanged [53]. The upregulation of UDG activity would create a BER imbalance in the absence of APE1 and Pol β upregulation, thus resulting in clastogenic events. The mechanism by which UDG activity was increased in this model was not examined nor were genomic uracil and markers of chromosomal instability measured. Several studies have examined the influence of BER components in S. cerevisiae in response to antifolates or 5-FU [54-56]. Eliminating UDG activity conferred resistance to short but not prolonged TLS in S. cerevisiae [54, 56]. S. cerevisiae lacks SMUG1, MBD4, and TDG homologues [26], which potentially limit the applicability of the observations to mammalian cells. The absence of APN1, the major AP endonuclease in S. cerevisiae, leads to heightened sensitivity to 5-FU or antifolates [54, 55]. In a variation from what occurs in mammalian cells, S. cerevisiae lack a paralog of Pol β, which removes the 5′-dRP group caused by AP endonuclease activity. Instead, S. cerevisiae seem to rely on RAD27 to remove the 5′-dRP as part of the displaced strand, analogous to FEN1-dependent long-patch BER in mammalian cells [26]. It is intriguing to note that a rad27 null strain was markedly resistant to 5-FU [55], which is analogous to the finding that Pol β deficient MEFs were resistant to TS inhibitors [50].
There is another paradox of the futile cycling hypothesis during TLS. Although it is known that dUTPase activity can vary greatly among cell lines and in tumor biopsies, no studies have directly demonstrated that increased UDG-initiated BER or increased genomic uracil occurs as a consequence of altered dUTPase activity. There is something more complex and frankly not well understood about having too much uracil in DNA. A minimum amount of dUTPase activity is required for viability of all living cells. The nonviability of dUTPase deficient strains of E. coli and S. cerevisiae even in the absence of UDG activity suggest that excessive genomic uracil cannot be tolerated [57, 58]. To our knowledge, there have been no systematic examinations of whether the tolerance of genomic uracil occurs as part of an adaptation in any mammalian cell culture model during tumorigenesis. In summary, the direct role of UDG-initiated BER remains vague. The myriad cell types such as cancer cells and MEFs used in the above mentioned studies invoke the usual caveats, yet the lack of consensus also emphasizes several issues that require further exploration. First, other sources of damage such as collapsed replication forks can potentially be contributing to cell death independent of BER activity. Second, the connections among BER, DNA damage signaling pathways, homologous recombination (HR), and cell death pathways in mammalian cells remain poorly understood. Epistatic analyses in E. coli and S. cerevisiae have demonstrated that HR responds to unresolved BER intermediates created by DNA glycosylase-mediated removal of alkylated bases [37]. If UDG-initiated BER is invoked under conditions of TLS, do BER strand break intermediates invoke HR? Third, it is known that altered S-phase progression occurs in cancer cells, and that defects in S-phase kinase signaling affects sensitivity to TS inhibitors [59-61]. Defects in DNA damage signaling checkpoint and HR status may be an underlying modulator of the contribution of BER, and these processes are discussed in the next section.
3. Homologous Recombination and DNA damage signaling responses
HR and DSBs formed during TLS
It has been suggested that DSBs and chromosome rearrangements are likely the main cause of TLD [4]. Although the mechanistic details that connect TLS, genomic instability, and cell death are poorly understood, there is suggestive evidence that HR is playing a role (Figure 4). Sister chromatid exchanges (SCEs) are thought to represent homologous recombination events because they are reciprocal exchanges of DNA between sister chromatids that occur during replication. Importantly, mammalian cells treated with the antifolate methotrexate had increased SCEs [62]. The antifolate RTX, which is specific for TS, also induced SCEs [19]. Suggestive evidence supporting a role for recombinational repair in TLS was also reported in FM3A mouse cells [63]. DSBs are repaired by two primary pathways: non-homologous end joining (NHEJ), an error prone pathway, or homologous recombination (HR), an error free pathway. In mammalian cells, NHEJ is generally thought to be responsible for resolving DSBs during the G1 phase of the cell cycle, while HR is responsible for resolving DSBs during S and G2 phases [64]. Investigations examining the role of NHEJ in TLS have not been reported, although the evidence that TLS occurs in S-phase and that SCEs occur suggests that HR is more likely involved. To repair a broken DNA molecule by HR, the site of the break is processed into 3′ overhanging ends, and a complex of proteins polymerizes along the single stranded DNA forming a nucleoprotein filament. This complex directs strand invasion of a homologous stretch of DNA within the sister chromatid that serves as a replication template, and an endonuclease resolves the Holliday junction. Key proteins involved in HR are RecA/RAD51, RAD51-related proteins and members of the RAD52 epistasis group including MRE11, RAD50, and NBS1 (Xrs2 in S. cerevisiae) [65].
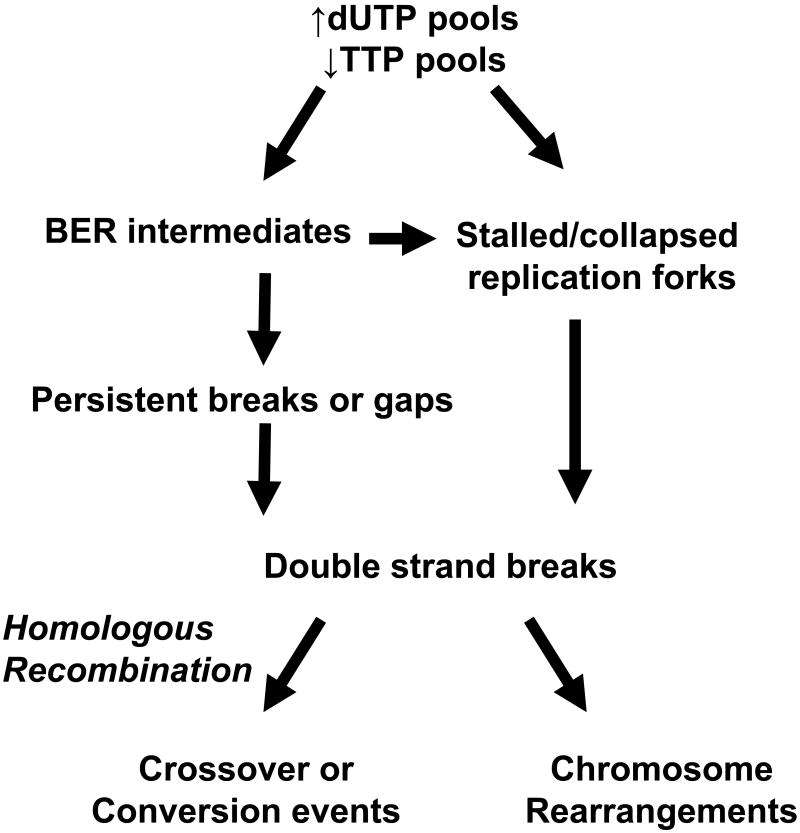
Figure 4. Simplified schematic of the links between TLS and homologous recombination repair.
DNA double strand breaks generated from persistent BER strand break intermediates and/or stalled/collapsed replication forks are thought to be processed by HR. The simplified biochemical steps necessary for HR include search for homology, strand invasion, DNA synthesis, and an endonuclease to resolve the Holliday junction. The outcome of HR could potentially be detectable as a crossover event resulting in sister chromatid exchanges or gene conversion events. If left unrepaired, double strand breaks lead to non-viable cells or chromosomal rearrangements detectable as translocations in a population of surviving cells.
It has been known for half a century that E. coli cells are sensitive to TLS and later investigations determined that the cells accumulate single-strand and double-strand DNA breaks. However, structures of the breaks resulting from TLS and how recombination proteins repair them remain unresolved. Nakayama et al. investigated break formation in E. coli DNA using pulsed field gel electrophoresis [66]. Large restriction fragments were observed in thymine-starved cells, which were primarily associated with replication origins. Electron microscopy analysis showed that these structures contain single-stranded tails, gaps and branchings consistent with single- and double-strand break accumulation. Furthermore, the break formation was dependent upon the E. coli RecA protein consistent with HR pathways being involved in the processing of resulting breaks [66]. Further investigations are required to elucidate whether futile repair cycling occurs with regards to leading versus lagging strand synthesis and whether breaks induced by TLS differ from DNA replication breaks caused by other DNA damaging agents.
In baker's yeast, S. cerevisiae, RAD50, RAD55 and RAD57 are involved in the HR repair pathway in addition to RAD51 and RAD52 [65]. Like RecA, the RAD51 protein forms nucleoprotein filaments along DNA and mediates strand exchange between homologous DNA molecules. In yeast, starvation for thymine nucleotides was lethal and recombinogenic [67]. Absence of RAD51 resulted in increased sensitivity to TLD and further increased genetic recombination frequencies, which correlated with DNA strand breaks as measured by sucrose gradient analysis. The increased sensitivity to antifolate-induced TLD of yeast strains deficient in RAD52, RAD54, and RAD57 provide further support for involvement of HR. Additionally, RAD50, which acts upstream of the RAD51/RAD52 pathway, was necessary for normal levels of recombination and resistance to TLS. In contrast, mutants defective in nucleotide excision repair (RAD3) or error-prone repair (RAD6, 18) were more resistant to antifolate-induced TLD [67]. The authors conclude that DSBs are among the lethal lesions induced by TLS.
As mentioned above, RAD51/RecA is a key HR protein involved in the strand invasion step. Vertebrates possess seven members of the RAD51/RecA family of HR proteins (RAD51, RAD51B, RAD51C, RAD51D, DMC1, XRCC2, and XRCC3), inferring more complex coordination of HR than in yeast and E. coli. Interaction of the RAD51 protein with the human breast cancer susceptibility proteins BRCA1 and BRCA2 was also a key finding suggesting a link between DSB repair defects with human cancer formation and progression [68, 69]. RAD51 protein complexes are visualized by immunofluorescence as distinct nuclear foci during replication, and the frequency and intensity of foci substantially increase upon exposure to DNA damaging agents. RAD51 foci fail to form in cell lines deficient for each of the RAD51 paralogs, suggesting they are involved in recruiting RAD51 to the damaged DNA. It is interesting to note that damage foci appearing in response to S-phase stress include known HR proteins as well as prominent S-phase checkpoint proteins, discussed in the next section.
DNA damage signaling during TLS
There are intriguing links among HR proteins and S-phase checkpoint signaling pathways that respond to DNA damage and stalled replication forks [70]. The ATM and ATR kinases are central players that provide an early response to DNA damage and replication stress. Evidence suggests that ATR is activated in response to multiple types of replication stress, whereas the ATM response is specific for double strand breaks [70]. Mutations in the ATM and ATR genes are responsible for ataxia telangiectasia (AT) and seckel syndrome, respectively. The downstream cascade includes the CHK1 and CHK2 signaling kinases among targets that number in the hundreds [71]. The MRN complex consists of the HR-associated proteins MRE11, RAD50, and NBS1. Mutations in MRE11 and NBS1 are responsible for an AT-like disorder and Nijmegan breakage syndrome, respectively. One classic phenotype associated with the above syndromes is elevated spontaneous clastogenesis. The MRN complex appears to act both up and down stream of ATR signaling via interactions with replication protein A (RPA) [72]. Recruitment of the MRN complex then stimulates RAD51 loading onto DNA facilitated by RPA, RAD52, and BRCA2 to initiate homology searching [73].
CHK1 has received attention as a chemotherapeutic target of UCN-01 [74]. It has been shown that CHK1 is required for HR [75] and that deficiency in CHK1 (or inhibition with UCN-01) leads to increased initiation of replication, ATR activation, and strand breaks [76]. Thus, it is hypothesized that CHK1 serves to regulate S-phase progression and HR initiation in response to collapsed replication forks. UCN-01 can heighten cellular sensitivity to 5-FU [77]. Also, TS inhibitors induce CHK1 phosphorylation [59] and CHK1 deficiency sensitizes cells to 5-FU [60, 61]. CHK1 is thought to be phosphorylated by ATR in response to replication stress, and ATR hypomorphic cells are also sensitive to 5-FU [78]. Collectively, the results suggest that ATR and CHK1-dependent S-phase checkpoint responses influence cell survival following TLS induced by chemotherapy.
4. Conclusions and future directions
It is well accepted that TLS induces DNA damage, but elucidating the precise causes of DNA damage is hindered by a lack of knowledge concerning the source of the strand breaks observed. UDG-initiated BER, stalled/collapsed replication forks, and initiation of HR in response to BER intermediates or collapsed replication forks are each plausible and non-exclusive mechanisms of DSB formation during TLS. When directly examined across a number of models, UDG-initiated BER only appears to contribute to cell death transiently or during TLS of shorter duration. One possibility is that BER's role is overshadowed by more profound or prolonged effects associated with TTP deprivation and dNTP pool imbalance. Yet the clinical relevance might remain when conditions of dietary insufficiency, polymorphic difference in metabolism, or heterogeneity of tumor cell exposure to chemotherapeutics in vivo provide varying degrees of TLS. It is interesting to note that with TLS induced by dietary or metabolic insufficiency of folate, BER would seemingly be pro-carcinogenic, while BER activity during chemotherapeutic TS inhibition would be antiproliferative to cancer cells. In a similar vein, HR could act as a pro-survival or pro-apoptotic process depending on the context. For example, HR protects against exogenous DNA damaging agents that are known to induce DSBs. However, one plausible hypothesis implicating HR as deleterious during TLS would be the demand for deoxynucleotides that the resynthesis step of HR impose, which can stretch up to thousands of nucleotides in comparison to the 1-5 nucleotides required to complete BER. Invoking HR during nucleotide pool imbalance would appear to be deleterious, yet may be the only means by which stalled and/or collapsed replication forks might recover. In either scenario, the S-phase checkpoint signaling processes that regulate S-phase arrest and invocation of HR certainly would be expected to influence cell death decisions during TLS.
BER and HR do not act in isolation, but are intricately orchestrated in the crowded confines of the nucleus during replication. The precise links between BER and HR in mammalian cells remain largely unexplained. S-phase checkpoint signaling pathways can influence cell survival in response to TLS, thus implicating these responses in facilitating the coordination of BER and HR. Defects in DNA damage signaling checkpoints and HR status are likely an underlying modulator of the contribution of BER during TLS. Of course, cancer cells have multiple and as yet incompletely understood defects in signaling pathways. Further studies in this regard should shed better light on the downstream consequences for carcinogenesis and chemotherapy associated with TLS.
Acknowledgments
Supported by grants from the NIH (1 R01 CA100450) and ACS (RSG-030158-01-GMC).
Abbreviations
- TS
thymidylate synthase
- TMP
thymidylate
- TTP
thymidine triphosphate
- dUMP
deoxyuridylate
- dUTP
deoxyuridine triphosphate
- CH2H4PteGlu
N5,N10-methylenetetrahydrofolate
- 5-FU
5-fluorouracil
- FdUrd
5-fluoro-2′-deoxyuridine
- RTX
raltitrexed (Tomudex)
- TLS
thymineless stress
- TLD
thymineless death
- DSB
double strand breaks
- MTHFR
methylenetetrahydrofolate reductase
- MTHFD
methylenetetrahydrofolate dehydrogenase
- TK
thymidine kinase
- DHFR
dihydrofolate reductase
- dUTPase
deoxyuridine triphosphate nucleotidohydrolase
- UDG
uracil DNA glycosylase
- BER
base excision repair
- PARP
poly (ADP-ribose) polymerase
- HR
homologous recombination
- SCE
sister chromatid exchange
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Powers HJ. Interaction among folate, riboflavin, genotype, and cancer, with reference to colorectal and cervical cancer. J Nutr. 2005;135:2960S–6S. doi: 10.1093/jn/135.12.2960S. [DOI] [PubMed] [Google Scholar]
- 2.Costi MP, Ferrari S, Venturelli A, Calo S, Tondi D, Barlocco D. Thymidylate synthase structure, function and implication in drug discovery. Curr Med Chem. 2005;12:2241–58. doi: 10.2174/0929867054864868. [DOI] [PubMed] [Google Scholar]
- 3.Parker WB, Cheng YC. Metabolism and mechanism of action of 5-fluorouracil. Pharmacol Ther. 1990;48:381–95. doi: 10.1016/0163-7258(90)90056-8. [DOI] [PubMed] [Google Scholar]
- 4.Ahmad SI, Kirk SH, Eisenstark A. Thymine metabolism and thymineless death in prokaryotes and eukaryotes. Ann Rev Microbiol. 1998;52:591–625. doi: 10.1146/annurev.micro.52.1.591. [DOI] [PubMed] [Google Scholar]
- 5.Eriksson S, Arner E, Spasokoukotskaja T, Wang L, Karlsson A, Brosjo O, et al. Properties and levels of deoxynucleoside kinases in normal and tumor cells; implications for chemotherapy. Adv Enzyme Regul. 1994;34:13–25. doi: 10.1016/0065-2571(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 6.Basten GP, Duthie SJ, Pirie L, Vaughan N, Hill MH, Powers HJ. Sensitivity of markers of DNA stability and DNA repair activity to folate supplementation in healthy volunteers. Br J Cancer. 2006;94:1942–7. doi: 10.1038/sj.bjc.6603197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim YI, Shirwadkar S, Choi SW, Puchyr M, Wang Y, Mason JB. Effects of dietary folate on DNA strand breaks within mutation-prone exons of the p53 gene in rat colon. Gastroenterology. 2000;119:151–61. doi: 10.1053/gast.2000.8518. [DOI] [PubMed] [Google Scholar]
- 8.Crott JW, Liu Z, Choi SW, Mason JB. Folate depletion in human lymphocytes up-regulates p53 expression despite marked induction of strand breaks in exons 5-8 of the gene. Mutat Res. 2007;626:171–9. doi: 10.1016/j.mrgentox.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Knock E, Deng L, Wu Q, Leclerc D, Wang XL, Rozen R. Low dietary folate initiates intestinal tumors in mice, with altered expression of G2-M checkpoint regulators polo-like kinase 1 and cell division cycle 25c. Cancer Res. 2006;66:10349–56. doi: 10.1158/0008-5472.CAN-06-2477. [DOI] [PubMed] [Google Scholar]
- 10.Friso S, Choi SW. Gene-nutrient interactions in one-carbon metabolism. Curr Drug Metab. 2005;6:37–46. doi: 10.2174/1389200052997339. [DOI] [PubMed] [Google Scholar]
- 11.Berger SH, Hakala MT. Relationship of dUMP and free FdUMP pools to inhibition of thymidylate synthase by 5-fluorouracil. Mol Pharmacol. 1984;25:303–9. [PubMed] [Google Scholar]
- 12.Jackson RC. The regulation of thymidylate biosynthesis in Novikoff hepatoma cells and the effects of amethopterin, 5-fluorodeoxyuridine, and 3-deazauridine. J Biol Chem. 1978;253:7440–6. [PubMed] [Google Scholar]
- 13.Ladner RD. The role of dUTPase and uracil-DNA repair in cancer chemotherapy. Curr Protein Pept Sci. 2001;2:361–70. doi: 10.2174/1389203013380991. [DOI] [PubMed] [Google Scholar]
- 14.Tye BK, Lehman IR. Excision repair of uracil incorporated in DNA as a result of a defect in dUTPase. J Mol Biol. 1977;117:293–306. doi: 10.1016/0022-2836(77)90128-0. [DOI] [PubMed] [Google Scholar]
- 15.Ingraham HA, Dickey L, Goulian M. DNA fragmentation and cytotoxicity from increased cellular deoxyuridylate. Biochemistry. 1986;25:3225–30. doi: 10.1021/bi00359a022. [DOI] [PubMed] [Google Scholar]
- 16.Yoshioka A, Tanaka S, Hiraoka O, Koyama Y, Hirota Y, Ayusawa D, et al. Deoxyribonucleoside triphosphate imbalance. 5-Fluorodeoxyuridine-induced DNA double strand breaks in mouse FM3A cells and the mechanism of cell death. J Biol Chem. 1987;262:8235–41. [PubMed] [Google Scholar]
- 17.Ayusawa D, Arai H, Wataya Y, Seno T. A specialized form of chromosomal DNA degradation induced by thymidylate stress in mouse FM3A cells. Mutat Res. 1988;200:221–30. doi: 10.1016/0027-5107(88)90086-3. [DOI] [PubMed] [Google Scholar]
- 18.Canman CE, Tang HY, Normolle DP, Lawrence TS, Maybaum J. Variations in patterns of DNA damage induced in human colorectal tumor cells by 5-fluorodeoxyuridine: implications for mechanisms of resistance and cytotoxicity. Proc Natl Acad Sci USA. 1992;89:10474–8. doi: 10.1073/pnas.89.21.10474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L, Connor EE, Berger SH, Wyatt MD. Determination of apoptosis, uracil incorporation, DNA strand breaks, and sister chromatid exchanges under conditions of thymidylate deprivation in a model of BER deficiency. Biochem Pharmacol. 2005;70:1458–68. doi: 10.1016/j.bcp.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 20.Houghton JA, Tillman DM, Harwood FG. Ratio of 2′-deoxyadenosine-5′-triphosphate/thymidine-5′-triphosphate influences the commitment of human colon carcinoma cells to thymineless death. Clin Cancer Res. 1995;1:723–30. [PubMed] [Google Scholar]
- 21.Matsui SI, Arredondo MA, Wrzosek C, Rustum YM. DNA damage and p53 induction do not cause ZD1694-induced cell cycle arrest in human colon carcinoma cells. Cancer Res. 1996;56:4715–23. [PubMed] [Google Scholar]
- 22.Canman CE, Lawrence TS, Shewach DS, Tang HY, Maybaum J. Resistance to fluorodeoxyuridine-induced DNA damage and cytotoxicity correlates with an elevation of deoxyuridine triphosphatase activity and failure to accumulate deoxyuridine triphosphate. Cancer Res. 1993;53:5219–24. [PubMed] [Google Scholar]
- 23.Canman CE, Radany EH, Parsels LA, Davis MA, Lawrence TS, Maybaum J. Induction of resistance to fluorodeoxyuridine cytotoxicity and DNA damage in human tumor cells by expression of Escherichia coli deoxyuridinetriphosphatase. Cancer Res. 1994;54:2296–8. [PubMed] [Google Scholar]
- 24.Koehler SE, Ladner RD. Small interfering RNA-mediated suppression of dUTPase sensitizes cancer cell lines to thymidylate synthase inhibition. Mol Pharmacol. 2004;66:620–6. doi: 10.1124/mol.66.3.. [DOI] [PubMed] [Google Scholar]
- 25.Webley SD, Welsh SJ, Jackman AL, Aherne GW. The ability to accumulate deoxyuridine triphosphate and cellular response to thymidylate synthase (TS) inhibition. Br J Cancer. 2001;85:446–52. doi: 10.1054/bjoc.2001.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. Washington, D.C.: ASM Press; 2006. [Google Scholar]
- 27.Krokan HE, Drablos F, Slupphaug G. Uracil in DNA--occurrence, consequences and repair. Oncogene. 2002;21:8935–48. doi: 10.1038/sj.onc.1205996. [DOI] [PubMed] [Google Scholar]
- 28.Nilsen H, Rosewell I, Robins P, Skjelbred CF, Andersen S, Slupphaug G, et al. Uracil-DNA glycosylase (UNG)-deficient mice reveal a primary role of the enzyme during DNA replication. Mol Cell. 2000;5:1059–65. doi: 10.1016/s1097-2765(00)80271-3. [DOI] [PubMed] [Google Scholar]
- 29.Otterlei M, Warbrick E, Nagelhus TA, Haug T, Slupphaug G, Akbari M, et al. Post-replicative base excision repair in replication foci. EMBO J. 1999;18:3834–44. doi: 10.1093/emboj/18.13.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nilsen H, Stamp G, Andersen S, Hrivnak G, Krokan HE, Lindahl T, et al. Gene-targeted mice lacking the Ung uracil-DNA glycosylase develop B-cell lymphomas. Oncogene. 2003;22:5381–6. doi: 10.1038/sj.onc.1206860. [DOI] [PubMed] [Google Scholar]
- 31.Hendrich B, Hardeland U, Ng HH, Jiricny J, Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;401:301–4. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- 32.Millar CB, Guy J, Sansom OJ, Selfridge J, MacDougall E, Hendrich B, et al. Enhanced CpG mutability and tumorigenesis in MBD4-deficient mice. Science. 2002;297:403–5. doi: 10.1126/science.1073354. [DOI] [PubMed] [Google Scholar]
- 33.Wong E, Yang K, Kuraguchi M, Werling U, Avdievich E, Fan K, et al. Mbd4 inactivation increases C to T transition mutations and promotes gastrointestinal tumor formation. Proc Natl Acad Sci U S A. 2002;99:14937–42. doi: 10.1073/pnas.232579299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bader S, Walker M, Hendrich B, Bird A, Bird C, Hooper M, et al. Somatic frameshift mutations in the MBD4 gene of sporadic colon cancers with mismatch repair deficiency. Oncogene. 1999;18:8044–7. doi: 10.1038/sj.onc.1203229. [DOI] [PubMed] [Google Scholar]
- 35.Riccio A, Aaltonen LA, Godwin AK, Loukola A, Percesepe A, Salovaara R, et al. The DNA repair gene MBD4 (MED1) is mutated in human carcinomas with microsatellite instability. Nat Genet. 1999;23:266–8. doi: 10.1038/15443. [DOI] [PubMed] [Google Scholar]
- 36.Cortazar D, Kunz C, Saito Y, Steinacher R, Schar P. The enigmatic thymine DNA glycosylase. DNA Repair (Amst) 2007;6:489–504. doi: 10.1016/j.dnarep.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 37.Wyatt MD, Pittman DL. Methylating agents and DNA repair responses: methylated bases and sources of strand breaks. Chem Res Toxicol. 2006;19:1580–94. doi: 10.1021/tx060164e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blount BC, Mack MM, Wehr CM, MacGregor JT, Hiatt RA, Wang G, et al. Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: implications for cancer and neuronal damage. Proc Natl Acad Sci U S A. 1997;94:3290–5. doi: 10.1073/pnas.94.7.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mashiyama ST, Courtemanche C, Elson-Schwab I, Crott J, Lee BL, Ong CN, et al. Uracil in DNA, determined by an improved assay, is increased when deoxynucleosides are added to folate-deficient cultured human lymphocytes. Anal Biochem. 2004;330:58–69. doi: 10.1016/j.ab.2004.03.065. [DOI] [PubMed] [Google Scholar]
- 40.Duthie SJ, Hawdon A. DNA instability (strand breakage, uracil misincorporation, and defective repair) is increased by folic acid depletion in human lymphocytes in vitro. Faseb J. 1998;12:1491–7. [PubMed] [Google Scholar]
- 41.Duthie SJ, Narayanan S, Blum S, Pirie L, Brand GM. Folate deficiency in vitro induces uracil misincorporation and DNA hypomethylation and inhibits DNA excision repair in immortalized normal human colon epithelial cells. Nutr Cancer. 2000;37:245–51. doi: 10.1207/S15327914NC372_18. [DOI] [PubMed] [Google Scholar]
- 42.An Q, Robins P, Lindahl T, Barnes DE. 5-Fluorouracil incorporated into DNA is excised by the smug1 DNA glycosylase to reduce drug cytotoxicity. Cancer Res. 2007;67:940–5. doi: 10.1158/0008-5472.CAN-06-2960. [DOI] [PubMed] [Google Scholar]
- 43.Fischer JA, Muller-Weeks S, Caradonna SJ. Fluorodeoxyuridine Modulates Cellular Expression of the DNA Base Excision Repair Enzyme Uracil-DNA Glycosylase. Cancer Res. 2006;66:8829–37. doi: 10.1158/0008-5472.CAN-06-0540. [DOI] [PubMed] [Google Scholar]
- 44.Cortellino S, Turner D, Masciullo V, Schepis F, Albino D, Daniel R, et al. The base excision repair enzyme MED1 mediates DNA damage response to antitumor drugs and is associated with mismatch repair system integrity. Proc Natl Acad Sci U S A. 2003;100:15071–6. doi: 10.1073/pnas.2334585100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fischer F, Baerenfaller K, Jiricny J. 5-Fluorouracil is efficiently removed from DNA by the base excision and mismatch repair systems. Gastroenterology. 2007;133:1858–68. doi: 10.1053/j.gastro.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 46.Meyers M, Wagner MW, Mazurek A, Schmutte C, Fishel R, Boothman DA. DNA mismatch repair-dependent response to fluoropyrimidine-generated damage. J Biol Chem. 2005;280:5516–26. doi: 10.1074/jbc.M412105200. [DOI] [PubMed] [Google Scholar]
- 47.Parsels LA, Parsels JD, Wagner LM, Loney TL, Radany EH, Maybaum J. Mechanism and pharmacological specificity of dUTPase-mediated protection from DNA damage and cytotoxicity in human tumor cells. Cancer Chemother Pharmacol. 1998;42:357–62. doi: 10.1007/s002800050829. [DOI] [PubMed] [Google Scholar]
- 48.Webley SD, Hardcastle A, Ladner RD, Jackman AL, Aherne GW. Deoxyuridine triphosphatase (dUTPase) expression and sensitivity to the thymidylate synthase (TS) inhibitor ZD9331. Br J Cancer. 2000;83:792–9. doi: 10.1054/bjoc.2000.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andersen S, Heine T, Sneve R, König I, Krokan HE, Epe B, et al. Incorporation of dUMP into DNA is a major source of spontaneous DNA damage, while excision of uracil is not required for cytotoxicity of fluoropyrimidines in mouse embryonic fibroblasts. Carcinogenesis. 2005;26:547–55. doi: 10.1093/carcin/bgh347. [DOI] [PubMed] [Google Scholar]
- 50.Li L, Berger SH, Wyatt MD. Involvement of base excision repair in response to therapy targeted at thymidylate synthase. Mol Cancer Ther. 2004;3:747–53. [PubMed] [Google Scholar]
- 51.Luo Y, Walla M, Wyatt MD. Uracil incorporation into genomic DNA does not predict toxicity caused by chemotherapeutic inhibition of thymidylate synthase. DNA Repair. 2008;8:162–9. doi: 10.1016/j.dnarep.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Welsh SJ, Hobbs S, Aherne GW. Expression of uracil DNA glycosylase (UDG) does not affect cellular sensitivity to thymidylate synthase (TS) inhibition. Eur J Cancer. 2003;39:378–87. doi: 10.1016/s0959-8049(02)00610-x. [DOI] [PubMed] [Google Scholar]
- 53.Cabelof DC, Raffoul JJ, Nakamura J, Kapoor D, Abdalla H, Heydari AR. Imbalanced base excision repair in response to folate deficiency is accelerated by polymerase beta haploinsufficiency. J Biol Chem. 2004;279:36504–13. doi: 10.1074/jbc.M405185200. [DOI] [PubMed] [Google Scholar]
- 54.Dornfeld K, Johnson M. AP endonuclease deficiency results in extreme sensitivity to thymidine deprivation. Nucleic Acids Res. 2005;33:6644–53. doi: 10.1093/nar/gki975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seiple L, Jaruga P, Dizdaroglu M, Stivers JT. Linking uracil base excision repair and 5-fluorouracil toxicity in yeast. Nucleic Acids Res. 2006;34:140–51. doi: 10.1093/nar/gkj430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tinkelenberg BA, Hansbury MJ, Ladner RD. dUTPase and uracil-DNA glycosylase are central modulators of antifolate toxicity in Saccharomyces cerevisiae. Cancer Res. 2002;62:4909–15. [PubMed] [Google Scholar]
- 57.el-Hajj HH, Wang L, Weiss B. Multiple mutant of Escherichia coli synthesizing virtually thymineless DNA during limited growth. J Bacteriol. 1992;174:4450–6. doi: 10.1128/jb.174.13.4450-4456.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gadsden MH, McIntosh EM, Game JC, Wilson PJ, Haynes RH. dUTP pyrophosphatase is an essential enzyme in Saccharomyces cerevisiae. Embo J. 1993;12:4425–31. doi: 10.1002/j.1460-2075.1993.tb06127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parsels LA, Parsels JD, Tai DC, Coughlin DJ, Maybaum J. 5-fluoro-2′-deoxyuridine-induced cdc25A accumulation correlates with premature mitotic entry and clonogenic death in human colon cancer cells. Cancer Res. 2004;64:6588–94. doi: 10.1158/0008-5472.CAN-03-3040. [DOI] [PubMed] [Google Scholar]
- 60.Robinson HM, Jones R, Walker M, Zachos G, Brown R, Cassidy J, et al. Chk1-dependent slowing of S-phase progression protects DT40 B-lymphoma cells against killing by the nucleoside analogue 5-fluorouracil. Oncogene. 2006;25:5359–69. doi: 10.1038/sj.onc.1209532. [DOI] [PubMed] [Google Scholar]
- 61.Xiao Z, Xue J, Sowin TJ, Zhang H. Differential roles of checkpoint kinase 1, checkpoint kinase 2, and mitogen-activated protein kinase-activated protein kinase 2 in mediating DNA damage-induced cell cycle arrest: implications for cancer therapy. Mol Cancer Ther. 2006;5:1935–43. doi: 10.1158/1535-7163.MCT-06-0077. [DOI] [PubMed] [Google Scholar]
- 62.Banerjee A, Benedict WF. Production of sister chromatid exchanges by various cancer chemotherapeutic agents. Cancer Res. 1979;39:797–9. [PubMed] [Google Scholar]
- 63.Ayusawa D, Koyama H, Shimizu K, Kaneda S, Takeishi K, Seno T. Induction, by thymidylate stress, of genetic recombination as evidenced by deletion of a transferred genetic marker in mouse FM3A cells. Mol Cell Biol. 1986;6:3463–9. doi: 10.1128/mcb.6.10.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–74. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 65.Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol Mol Biol Rev. 2002;66:630–70. doi: 10.1128/MMBR.66.4.630-670.2002. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nakayama K, Kusano K, Irino N, Nakayama H. Thymine Starvation-Induced Structural-Changes in Escherichia-Coli DNA - Detection by Pulsed-Field Gel-Electrophoresis and Evidence for Involvement of Homologous Recombination. J Mol Biol. 1994;243:611–20. doi: 10.1016/0022-2836(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 67.Kunz BA, Haynes RH. DNA repair and the genetic effects of thymidylate stress in yeast. Mutat Res. 1982;93:353–75. [Google Scholar]
- 68.Scully R, Chen J, Ochs RL, Keegan K, Hoekstra M, Feunteun J, et al. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell. 1997;90:425–35. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- 69.Sharan SK, Morimatsu M, Albrecht U, Lim DS, Regel E, Dinh C, et al. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature. 1997;386:804–10. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- 70.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–23. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 71.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–6. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 72.Olson E, Nievera CJ, Lee AY, Chen L, Wu X. The Mre11-Rad50-Nbs1 complex acts both upstream and downstream of ataxia telangiectasia mutated and Rad3-related protein (ATR) to regulate the S-phase checkpoint following UV treatment. J Biol Chem. 2007;282:22939–52. doi: 10.1074/jbc.M702162200. [DOI] [PubMed] [Google Scholar]
- 73.Thorslund T, West SC. BRCA2: a universal recombinase regulator. Oncogene. 2007;26:7720–30. doi: 10.1038/sj.onc.1210870. [DOI] [PubMed] [Google Scholar]
- 74.Tse AN, Carvajal R, Schwartz GK. Targeting checkpoint kinase 1 in cancer therapeutics. Clin Cancer Res. 2007;13:1955–60. doi: 10.1158/1078-0432.CCR-06-2793. [DOI] [PubMed] [Google Scholar]
- 75.Sorensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, Bartek J, et al. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat Cell Biol. 2005;7:195–201. doi: 10.1038/ncb1212. [DOI] [PubMed] [Google Scholar]
- 76.Syljuasen RG, Sorensen CS, Hansen LT, Fugger K, Lundin C, Johansson F, et al. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol Cell Biol. 2005;25:3553–62. doi: 10.1128/MCB.25.9.3553-3562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hsueh CT, Kelsen D, Schwartz GK. UCN-01 suppresses thymidylate synthase gene expression and enhances 5-fluorouracil-induced apoptosis in a sequence-dependent manner. Clin Cancer Res. 1998;4:2201–6. [PubMed] [Google Scholar]
- 78.Wilsker D, Bunz F. Loss of ataxia telangiectasia mutated- and Rad3-related function potentiates the effects of chemotherapeutic drugs on cancer cell survival. Mol Cancer Ther. 2007;6:1406–13. doi: 10.1158/1535-7163.MCT-06-0679. [DOI] [PubMed] [Google Scholar]