Abstract
Start codon selection is a key step in translation initiation as it sets the reading frame for decoding. Two eukaryotic initiation factors, eIF1 and eIF1A, are key actors in this process. Recent work has elucidated many details of the mechanisms these factors use to control start site selection. eIF1 prevents the irreversible GTP hydrolysis that commits the ribosome to initiation at a particular codon. eIF1A both promotes and inhibits commitment through the competing influences of its two unstructured termini. Both factors perform their tasks through a variety of interactions with other components of the initiation machinery, in many cases mediated by the unstructured regions of the two proteins.
Translation initiation begins the final major step in gene expression and is a highly regulated process. Initiation entails the formation of a functional ribosomal complex (80 S in eukaryotes) with an initiator methionyl-tRNA (Met-tRNAi) bound in the P-site, its anticodon base-paired to the start codon of an mRNA, poised to begin peptide synthesis. Formation of the initiation complex is promoted and regulated by a number of non-ribosomal proteins called initiation factors (IFs,2 or for eukaryotes, eIFs).
Many details of eukaryotic translation initiation have been elucidated over the last three decades using genetic, biochemical, and structural techniques. As this review focuses on the roles of eIF1 and eIF1A in start site selection, only a brief overview of the entire process will be given (Fig. 1). For a more complete description, see one of several recent comprehensive reviews (1–3). Met-tRNAi is delivered to the P-site of the small (40 S) ribosomal subunit in the form of a ternary complex (TC) including GTP and the trimeric GTPase eIF2, forming the 43 S pre-initiation complex (PIC) in a process promoted by eIF1, eIF1A, and eIF3. The 43 S PIC then binds the 5′-end of an mRNA with the help of eIF3, eIF4A, eIF4B, and eIF4F, creating the 48 S PIC. The mRNA is thought to be circularized through the interactions of PABP with the 3′-poly(A) tail of the mRNA and the eIF4F 5′-cap-binding complex. The complex is then believed to scan along the mRNA in search of the start codon in an ATP-dependent process. During this search, eIF2 partially hydrolyzes its bound GTP with the help of the GTPase-activating factor eIF5. Prior to start codon recognition, the resultant phosphate ion is not released, producing GTP- and GDP·Pi-bound states of the factor, possibly in equilibrium (4). Upon identification of the start codon, the phosphate is released, making GTP hydrolysis irreversible and committing the complex to proceeding with initiation at that site on the mRNA. After eIF2·GDP release, the 60 S subunit can join the 40 S subunit with the help of a second GTPase, eIF5B. When eIF5B·GDP dissociates after subunit joining, the initiation complex is complete and ready to begin the elongation phase of translation.
FIGURE 1.
Model of eukaryotic translation initiation. The first step of translation initiation is the binding of a TC composed of eIF2, GTP, and Met-tRNAMeti to the 40 S ribosomal subunit forming the 43 S PIC. TC may bind the 40 S subunit as part of the MFC along with eIF1, eIF5, and eIF3. eIF1A also binds and facilitates MFC recruitment. The N- and C-terminal tails of eIF1 and eIF1A are represented in green and pink, respectively. Although their locations in the complex are not known, some proposed interactions are shown here. The PIC then binds mRNA with the help of eIF3, the eIF4F complex, eIF4B, and PABP. For clarity, these factors are not shown. The complex scans the mRNA for the start codon. During this time, GTP bound to eIF2 can be hydrolyzed with the help of eIF5, but this reaction is reversible because phosphate is not released. During the scanning process, the PIC is in an open state, which is in equilibrium with a closed state that is not capable of scanning but is able to investigate the codon in the P-site. In this figure, the closed state is represented by a lock, holding the mRNA in place. When an AUG codon is identified in the P-site, the equilibrium shifts toward the closed state. eIF1 dissociates from the 40 S ribosome, and the phosphate ion bound to eIF2 can now be released, making hydrolysis irreversible and committing the PIC to initiate translation at the codon currently in the P-site. After dissociation from the ribosome, eIF1 may remain bound to the PIC through an interaction with eIF3 (29). After eIF2·GDP and eIF5 dissociate, the 60 S subunit joins the 40 S subunit with the help of the GTPase eIF5B. The 80 S initiation complex is now ready to begin the elongation phase of translation. Factors in this figure are not drawn to scale to make the smaller factors visible.
In eukaryotic cells, at least twelve factors, composed of over two dozen polypeptides, are required for initiation complex formation. The same end is achieved by three IFs in bacterial cells. Why is there such a discrepancy between domains for accomplishing virtually the same goal? One important mechanistic difference between initiation in bacteria and eukaryotes is the method of start site selection. Bacterial mRNAs contain a Shine-Dalgarno sequence that base pairs to the 16 S rRNA, holding the mRNA in place so that the start codon is recognized. No such complementary sequences are found in eukaryotic messenger and ribosomal RNAs. Instead, a PIC searches the 5′-untranslated region of the mRNA for the start codon, usually the 5′-most AUG. Consensus sequences have been found in a variety of eukaryotic organisms (5–7) but have not been shown to base pair to rRNA. Rather, the scanning PIC responds to a set of less obvious and less well understood signals that occur upon start codon recognition. The importance of heeding such signals is obvious. Initiation at an incorrect codon will most likely make a completely miscoded protein, wasting the cell's valuable resources and creating a potentially toxic peptide.
Evolution has solved the problem of start codon recognition in eukaryotes with a rather baroque mechanism. Start site recognition is not the job of a single factor but is instead achieved through the cooperative actions of many. This, of course, makes the problem of understanding how the system works that much more challenging. Despite these difficulties, great progress has been made in understanding the roles of individual factors and the mechanisms of their action in start site selection. The picture that has developed over the last few years is one of a constantly changing PIC in which factors interact with a shifting array of different partners as they progress through the steps of initiation. Even the smallest of proteins, 12-kDa eIF1 and 17-kDa eIF1A, interact with a number of other components throughout the process, reaching out their unstructured termini (Fig. 2) to communicate. Between them, these two factors participate in factor recruitment, scanning of the 5′-untranslated region, restraining and promoting GTP hydrolysis and Pi release, controlling key conformational rearrangements of the complex, and subunit joining. Recent work has made it clear that the roles of eIF1 and eIF1A in start codon recognition are intimately linked, and this review will focus on the workings of these proteins in this key step in genetic decoding.
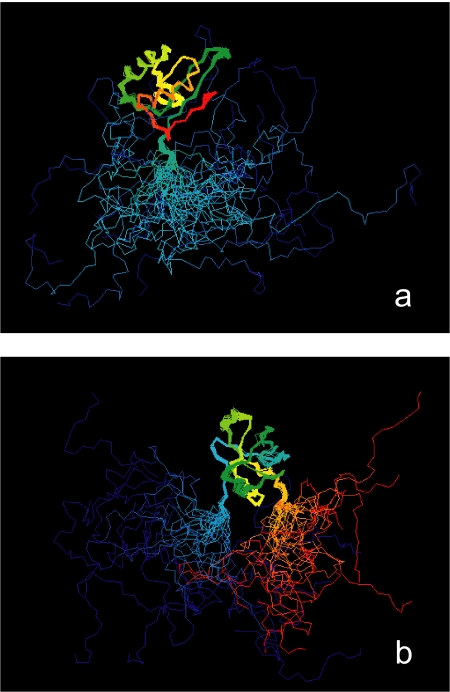
FIGURE 2.
NMR structures of eIF1 and eIF1A. a, the solution structure of human eIF1 (36) is represented in rainbow coloring, from red at the C terminus to blue at the N terminus. The ensemble of structures consistent with the NMR data is overlaid to accentuate the unstructured nature of the N-terminal tail. b, the solution structure of human eIF1A (44) is represented as described for a. Note that both the N- and C-terminal regions of eIF1A are unstructured.
Searching for the Start Codon
One of the first functions attributed to eIF1 and eIF1A was facilitating TC binding to the 40 S ribosome (8–13). In addition to assisting TC recruitment, eIF1 and eIF1A facilitate recruitment of one another by binding cooperatively to the 40 S subunit (14). eIF1A was also shown to have other functions that promote the formation of PICs, including dissociating 40 S dimers (15) and recruiting mRNA (8).
Early genetic work identified eIF1 as an essential player in start codon recognition (16, 17). A screen for suppressors of a mutation in the start codon of the HIS4 gene identified mutations in eIF1 that produced a Sui– (suppressor of initiation codon mutations) phenotype (17). These mutations allow initiation to occur at an in-frame UUG near the correct start site. The ability of eIF1 to ensure correct identification of the start codon was further demonstrated by its ability to suppress a Sui– mutation in eIF5 when overexpressed (18). More recently, genetic studies have identified Sui– mutants of eIF1A, as well as mutants that read through a start codon, a phenotype called leaky scanning (19). The leaky scanning phenotype of the eIF1A mutants and the ability of eIF1 to suppress initiation at non-AUG codons suggested that eIF1 and eIF1A might act in antagonistic ways, with eIF1 being responsible for preventing premature engagement with putative start codons and eIF1A facilitating pausing at the correct start codon long enough to proceed with downstream initiation events. Biochemical data have supported this good cop/bad cop model for the actions of these factors but also revealed a surprising amount of complexity in their mechanisms.
The inhibitory action of eIF1 targets GTP hydrolysis by eIF2. In the absence of eIF1, the rate of eIF5-stimulated GTP hydrolysis is the same, regardless of the presence of mRNA on the PIC (20). The addition of eIF1 decreases the amount of GTP hydrolysis observed in the absence of mRNA. In the presence of cap-bound eIF4F and eIF1, eIF1A, and eIF3, hydrolysis in the absence of a start codon is reduced by 80% (21). Phosphate release was later identified as the point at which eIF1 inhibits overall GTP hydrolysis prior to start codon recognition (4).
The importance of both factors in start codon selection was indicated by primer extension (“toeprinting”) studies, which allow the positions of PICs on mRNAs to be determined. These experiments revealed that in the absence of both eIF1 and eIF1A, the 40 S subunit is located very close to the 5′-cap of the mRNA and is unable to locate the start codon. The addition of eIF1 removes these complexes, but eIF1A is required for the proper start codon to be found (22). This result is consistent with eIF1 acting as a negative regulator of incorrectly located PICs and with eIF1A acting to stabilize properly formed complexes. Primer extension studies also showed that complexes can form at the near-cognate AUU codon in the absence of eIF1 (23), further indicating that eIF1 is involved in distinguishing between cognate and near-cognate start codons. Both eIF1 and eIF1A also help differentiate between start codons in a “good” context, i.e. the Kozak consensus sequence surrounding many start codons in mammals (5), and a “bad” context. In the absence of eIF1A, PICs are able to scan through good context AUG codons, and in the absence of eIF1, a PIC is more likely to stop at an AUG codon in a bad context, consistent with roles as positive and negative regulators of start codon recognition, respectively (23). The authors of this work speculated that the two factors might influence the equilibrium of the PIC between two states. In one state, which they called the “open” state, the complex is able to scan the mRNA, and base pairing with the tRNA cannot occur. In the other conformation, the “closed” state, the complex can no longer scan the mRNA, and the tRNA is able to interact with the P-site codon. The existence of such an equilibrium has been supported by a growing body of genetic, biochemical, and structural data.
Kinetic studies of the stability of eIF1A in PICs, which probe the strength of interactions between the factor and other components in the complex, provided the first evidence that two types of complexes exist and that eIF1A plays a key role in determining the balance between the two (24). In the presence of both eIF5 and an AUG codon, the equilibrium between these two states is shifted toward the one in which eIF1A makes more favorable interactions. Mutations in both eIF1A and eIF5 that increase initiation at non-AUG codons in vivo strengthen this shift in the presence of a UUG codon and weaken it in the presence of an AUG codon. In addition, mutants of eIF1A that have a leaky scanning phenotype strongly shift the equilibrium toward the state in which eIF1A makes fewer stabilizing interactions (25). When eIF1A is bound more tightly, a reduction in the conformational freedom of its C terminus is also observed, indicating that a structural change has taken place in which this region has become more confined (24, 25). These data are consistent with a model in which the more tightly bound state is the closed one and the less tightly bound state is the open, scanning-competent complex.
Primer extension studies also support a role for eIF1A in maintaining this equilibrium. A mutation in eIF1A leads to the formation of a PIC that stalls on the mRNA between the 5′-cap and the start codon (19). This error is thought to be due to a scanning defect, consistent with a role for eIF1A in maintaining an open, scanning-competent complex.
Recent structural studies may have actually captured pictures of the open and closed states. Cryoelectron microscopy (cryo-EM) reconstructions of the yeast 40 S ribosomal subunit alone and with eIF1 and/or eIF1A bound demonstrate that these factors alter the accessibility of the mRNA binding channel. When both factors are bound, the “latch” region of the channel is no longer visible, and a new connection is formed between the head and shoulder on the back side of the subunit (26). This conformation would likely promote mRNA binding and allow the ribosome to move more easily across the mRNA. A similar result was achieved in a cryo-EM reconstruction of the 40 S subunit bound to eIF1A and the multifactor complex (MFC), which is composed of eIF1, eIF3, eIF5, and TC (27). These results are consistent with earlier hydroxyl radical cleavage studies that suggested that structural changes occur in the mRNA binding region of the ribosome upon association of eIF1 (28).
The unstructured N and C termini of eIF1A have been identified as particularly important for proper maintenance of the equilibrium between the open and closed states. Interestingly, the two termini take on opposite roles, with mutations in the N-terminal tail (NTT) causing leaky scanning phenotypes and acting as suppressors of Sui– mutations and with mutations in the C-terminal tail (CTT) producing Sui– phenotypes (19, 25). Like a molecular version of Dr. Doolittle's two-headed pushmipullyu, the ends of eIF1A are moving toward opposing goals, but instead of reaching a stalemate, the factor is able to walk the fine line between moving past near-cognate codons and identifying the subtly different cognate start codon.
Response to Start Codon Recognition
Once the start codon has finally been found, how does translation initiation proceed if eIF1 is preventing conversion of GTP to GDP and subsequent commitment of the complex? Fluorescence resonance energy transfer-based studies revealed that a conformational change takes place in the PIC upon start codon recognition, causing the C termini of eIF1 and eIF1A to move away from each other (29). This conformational change is followed by dissociation of eIF1 from the complex, which in turn allows phosphate release to occur (4, 29). eIF2·GDP, most likely in complex with eIF5, can then dissociate from the PIC, which allows downstream events to proceed.
Recent in vivo and in vitro data have substantiated the importance of eIF1 release by demonstrating that mutations in eIF1 that produce Sui– phenotypes lower the affinity of the factor for the 40 S subunit, cause it to dissociate more easily, and lead to an increased rate of Pi release (30). These Sui– phenotypes can be reduced by overexpression of the mutant factors presumably because mass action causes an increase in PIC-bound eIF1 (30). Structural data also support the proposal that the PIC rearranges to the closed state after eIF1 dissociation. Cryo-EM reconstructions of the 40 S complexes revealed that eIF1 and eIF1A synergistically induce the open conformation and that neither alone is able to produce this state. In fact, in the 40 S subunit·eIF1A complex, there is actually an increase in the density of the latch of the mRNA binding channel compared with the unoccupied 40 S subunit, suggesting that this complex is even more closed than the naked subunit (26). Release of eIF1 after start codon recognition would leave eIF1A alone in the PIC, and based on these cryo-EM data, it would be unable to stabilize the open state of the complex, causing it to revert to the closed, scanning-arrested form.
From the description above, it would seem as if eIF1 were the key player in start site selection and eIF1A an overeager factor that must be restrained until the proper moment, but this would be too harsh a judgment of eIF1A. In fact, eIF1A seems to be intimately involved in giving eIF1 permission to leave its post. NTT mutations in eIF1A that suppress the Sui– phenotypes of mutations in other factors slow both the conformational change that occurs upon start codon recognition and the subsequent release of eIF1 (30). The sum of the data presented here and above suggests that an increase in stabilizing interactions between eIF1A and the PIC is a key step in start codon recognition that is likely required for efficient eIF1 release (29, 30).
Factor Interactions
Just as for the termini of eIF1A, the two ends of eIF1 play important roles in the events surrounding start codon recognition. The ability of eIF1 to promote MFC binding to the ribosome is mediated through its two ends. The presence of a FLAG tag at either end reduces binding of the factor to eIF3 and the 40 S subunit and eliminates its ability to bind to eIF2 and eIF5 (31), and mutations in both the N and C termini have been found to reduce the efficiency of PIC formation in vivo and in vitro (30). Because of the unstructured nature of both the CTT and NTT of eIF1A and the NTT of eIF1, these regions have a large degree of conformational flexibility, potentially allowing them to interact with a variety of other factors during translation initiation. It is likely that the unstructured nature of the termini of eIF1 and eIF1A increases their reach both physically across the ribosome and functionally throughout the initiation pathway. The terminal regions are not conserved in the bacterial ortholog of eIF1A, IF1, suggesting that they play roles specific to eukaryotic translation, consistent with their abilities to interact with factors without bacterial counterparts (eIF2, eIF3, and eIF5).
The CTT of eIF1A promotes MFC recruitment to the 40 S subunit, whereas the NTT, which also interacts with components of the MFC, is not necessary for recruitment of these factors to the PIC (19). This discrepancy suggests that the NTT interactions might be necessary at a later step of translation initiation such as scanning or start codon identification (32). Based on these observations, it was suggested that eIF1A has a modular structure, in which the central OB-fold serves as a 40 S binding domain, whereas the termini are able to interact with a variety of factors. Considering that the termini of eIF1 and eIF1A are involved in both start site selection and protein-protein interactions, these interactions may be key for transfer of the “go” signal from the start codon to the factors responsible for GTP hydrolysis. Prior to start codon recognition, eIF1 and eIF1A interact on the 40 S subunit (14). After start codon recognition, eIF1 is released, and eIF1A interacts, either directly or indirectly, with eIF5 (29). One appealing but unproved possibility is that when eIF1 is ejected from the PIC, breaking its interaction with eIF1A, eIF1A is freed to interact with eIF5, and this new interaction is involved in triggering Pi release from eIF2. An arginine in the N terminus of eIF5 has been proposed to act as an arginine finger to activate GTP hydrolysis by eIF2 (33, 34), and thus, it seems possible that eIF5 might hinder Pi release by virtue of its binding in the GTPase active site and that the interaction with eIF1A might cause it to alter position or conformation enough to allow Pi to dissociate.
It has been suggested that eIF1 may also serve as a link between the start codon and eIF5 (35). NMR studies have shown an interaction between eIF5 and eIF1 at the latter's NTT and a region composed of basic and hydrophobic residues. The direct interaction between eIF1 and eIF5 is not observed under all conditions (35, 36), but the two may be in close proximity on the PIC. Both bind to eIF3c in the MFC, which could promote a direct or indirect interaction. Although most of the body of eIF3 is located on the solvent-exposed side of the 40 S subunit (37, 38), whereas the factors are bound to the interface side, an unidentified portion of eIF3 has been shown to reach around toward the intersubunit face where it might be close enough to interact with eIF1 or eIF5 (37). Mutations in eIF3 with Sui– and suppressor of Sui– phenotypes suggest that this factor also plays a role in start site selection (18).
A competitive interaction between eIF1 and eIF5 for eIF2 has also been suggested because of structural similarity between the N-terminal domain (NTD) of eIF5 and factors eIF1 and eIF2β (39). This finding led the authors to suggest that these three protein domains compete for binding to eIF2γ. Their model suggests that eIF1 interacts with eIF2γ, preventing eIF5-stimulated GTP hydrolysis until the MFC binds the 40 S ribosome, at which point the eIF5 NTD replaces eIF1 in interacting with eIF2γ, promoting reversible GTP hydrolysis. After start codon selection and GTP hydrolysis, eIF2β could take the place of the eIF5 NTD, helping eIF2 to dissociate from the PIC. Moving eIF5 away from the GTPase center of eIF2 could also serve to remove a physical barrier for Pi, allowing Pi release upon start codon recognition. This change of binding partners could be promoted by the interaction between eIF1A and eIF5.
eIF1 is not the only factor with a competitive spirit. Directed hydroxyl radical probing showed that eIF3j, a loosely bound subunit of eIF3, binds to the mRNA binding channel and the A-site of the ribosome. eIF3j displays negative cooperativity with eIF1A and, in the absence of TC, with mRNA in binding the 40 S subunit (40). The consequences of this competition are not yet fully understood, but because eIF3j is not an essential protein in yeast, it may be that this interaction is required only for the translation of select mRNAs or for fine-tuning of the process.
If these possible interactions are not enough to make your head spin, another factor has been thrown into the equation. eIF4G, part of the eIF4F 5′-cap binding complex, has been shown to interact with both eIF1 and eIF5 (41, 42). The observation that a mutation in eIF1 that produces a Sui– phenotype in vivo reduces this interaction with eIF4G led to the suggestion that eIF4G may influence scanning and start codon selection through eIF1 (42).
The pathway of communication between the start codon and the GTPase active site of eIF2 has not yet been fully defined, but the number of proposed interactions rivals those seen in the most complex of biological systems. It is unlikely that all of these connections are important in vivo. Defining those that are necessary for start site selection is a key step in the development of a molecular mechanism of translation initiation. eIF5 is emerging as the center of this tangled web of interactions. Elucidating its binding partners throughout the process of initiation would be of great interest.
Conclusions
In the past decade, a growing body of biochemical and biophysical data has added to the genetic observations that first identified eIF1 and eIF1A as regulators of start site selection. These data have elucidated the role of eIF1 as a negative regulator of phosphate release that is ejected from the PIC upon start codon recognition and revealed the complex balance that eIF1A strikes in promoting both scanning and start site selection, ultimately interacting more strongly with the PIC upon start codon recognition and stabilizing the closed state of the complex. Such advances could not have been made without combining the power of genetic studies with the detail of biochemical and biophysical work.
A recent genetic study on the roles played by eIF3 and eIF4G in mRNA recruitment and scanning suggests that eIF4G plays a role after mRNA binding (43), contrary to the accepted view that its primary function is as a scaffolding protein required for mRNA recruitment. It will not be long before the same combination of genetic, biochemical, and structural studies will elucidate the roles of eIF4G and eIF3 as well as those of the many other factors more poorly understood than eIF1 and eIF1A. Such studies may also increase our understanding of the “black box” of the current model, scanning. A full understanding of the molecular mechanics of start codon recognition will not be possible without a detailed dissection of this key process.
Supplementary Material
Acknowledgments
We thank Alan Hinnebusch, Sarah Kolitz, and Julie Takacs for comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants GM62128 and DK078633 from NIGMS and NIDDK. This minireview will be reprinted in the 2008 Minireview Compendium, which will be available in January, 2009.
Footnotes
The abbreviations used are: IF, initiation factor; eIF, eukaryotic IF; TC, ternary complex; PIC, pre-initiation complex; MFC, multifactor complex; cryo-EM, cryoelectron microscopy; NTT, N-terminal tail; CTT, C-terminal tail; NTD, N-terminal domain; PABP, poly(A)-binding protein.
References
- 1.Kapp, L. D., and Lorsch, J. R. (2004) Annu. Rev. Biochem. 73 657–704 [DOI] [PubMed] [Google Scholar]
- 2.Pestova, T. V., Kolupaeva, V. G., Lomakin, I. B., Pilipenko, E. V., Shatsky, I. N., Agol, V. I., and Hellen, C. U. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 7029–7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Algire, M. A., and Lorsch, J. R. (2006) Curr. Opin. Chem. Biol. 10 480–486 [DOI] [PubMed] [Google Scholar]
- 4.Algire, M. A., Maag, D., and Lorsch, J. R. (2005) Mol. Cell 20 251–262 [DOI] [PubMed] [Google Scholar]
- 5.Kozak, M. (1986) Cell 44 283–292 [DOI] [PubMed] [Google Scholar]
- 6.Kozak, M. (1991) J. Biol. Chem. 266 19867–19870 [PubMed] [Google Scholar]
- 7.Cigan, A. M., and Donahue, T. F. (1987) Gene (Amst.) 59 1–18 [DOI] [PubMed] [Google Scholar]
- 8.Trachsel, H., Erni, B., Schreier, M. H., and Staehelin, T. (1977) J. Mol. Biol. 116 755–767 [DOI] [PubMed] [Google Scholar]
- 9.Thomas, A., Goumans, H., Voorma, H. O., and Benne, R. (1980) Eur. J. Biochem. 107 39–45 [DOI] [PubMed] [Google Scholar]
- 10.Thomas, A., Spann, W., Van Steeg, H., Voorma, H. O., and Benne, R. (1980) FEBS Lett. 116 67–71 [DOI] [PubMed] [Google Scholar]
- 11.Chaudhuri, J., Chowdhury, D., and Maitra, U. (1999) J. Biol. Chem. 274 17975–17980 [DOI] [PubMed] [Google Scholar]
- 12.Algire, M. A., Maag, D., Savio, P., Acker, M. G., Tarun, S. Z., Sachs, A. B., Asano, K., Nielsen, K. H., Olsen, D. S., Phan, L., Hinnebusch, A. G., and Lorsch, J. R. (2002) RNA 8 382–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Majumdar, R., Bandyopadhyay, A., and Maitra, U. (2003) J. Biol. Chem. 278 6580–6587 [DOI] [PubMed] [Google Scholar]
- 14.Maag, D., and Lorsch, J. R. (2003) J. Mol. Biol. 330 917–924 [DOI] [PubMed] [Google Scholar]
- 15.Kainuma, M., and Hershey, J. W. (2001) Biochimie (Paris) 83 505–514 [DOI] [PubMed] [Google Scholar]
- 16.Cui, Y., Dinman, J. D., Kinzy, T. G., and Peltz, S. W. (1998) Mol. Cell. Biol. 18 1506–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoon, H. J., and Donahue, T. F. (1992) Mol. Cell. Biol. 12 248–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valasek, L., Nielsen, K. H., Zhang, F., Fekete, C. A., and Hinnebusch, A. G. (2004) Mol. Cell. Biol. 24 9437–9455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fekete, C. A., Applefield, D. J., Blakely, S. A., Shirokikh, N., Pestova, T., Lorsch, J. R., and Hinnebusch, A. G. (2005) EMBO J. 24 3588–3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Unbehaun, A., Borukhov, S. I., Hellen, C. U., and Pestova, T. V. (2004) Genes Dev. 18 3078–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Majumdar, R., and Maitra, U. (2005) EMBO J. 24 3737–3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pestova, T. V., Borukhov, S. I., and Hellen, C. U. (1998) Nature 394 854–859 [DOI] [PubMed] [Google Scholar]
- 23.Pestova, T. V., and Kolupaeva, V. G. (2002) Genes Dev. 16 2906–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maag, D., Algire, M. A., and Lorsch, J. R. (2006) J. Mol. Biol. 356 724–737 [DOI] [PubMed] [Google Scholar]
- 25.Fekete, C. A., Mitchell, S. F., Cherkasova, V. A., Applefield, D., Algire, M. A., Maag, D., Saini, A. K., Lorsch, J. R., and Hinnebusch, A. G. (2007) EMBO J. 26 1602–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Passmore, L. A., Schmeing, T. M., Maag, D., Applefield, D. J., Acker, M. G., Algire, M. A., Lorsch, J. R., and Ramakrishnan, V. (2007) Mol. Cell 26 41–50 [DOI] [PubMed] [Google Scholar]
- 27.Gilbert, R. J., Gordiyenko, Y., von der Haar, T., Sonnen, A. F., Hofmann, G., Nardelli, M., Stuart, D. I., and McCarthy, J. E. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 5788–5793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lomakin, I. B., Kolupaeva, V. G., Marintchev, A., Wagner, G., and Pestova, T. V. (2003) Genes Dev. 17 2786–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maag, D., Fekete, C. A., Gryczynski, Z., and Lorsch, J. R. (2005) Mol. Cell 17 265–275 [DOI] [PubMed] [Google Scholar]
- 30.Cheung, Y. N., Maag, D., Mitchell, S. F., Fekete, C. A., Algire, M. A., Takacs, J. E., Shirokikh, N., Pestova, T., Lorsch, J. R., and Hinnebusch, A. G. (2007) Genes Dev. 21 1217–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh, C. R., He, H., Ii, M., Yamamoto, Y., and Asano, K. (2004) J. Biol. Chem. 279 31910–31920 [DOI] [PubMed] [Google Scholar]
- 32.Olsen, D. S., Savner, E. M., Mathew, A., Zhang, F., Krishnamoorthy, T., Phan, L., and Hinnebusch, A. G. (2003) EMBO J. 22 193–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Das, S., Ghosh, R., and Maitra, U. (2001) J. Biol. Chem. 276 6720–6726 [DOI] [PubMed] [Google Scholar]
- 34.Paulin, F. E., Campbell, L. E., O'Brien, K., Loughlin, J., and Proud, C. G. (2001) Curr. Biol. 11 55–59 [DOI] [PubMed] [Google Scholar]
- 35.Reibarkh, M., Yamamoto, Y., Singh, C. R., del Rio, F., Fahmy, A., Lee, B., Luna, R. E., Ii, M., Wagner, G., and Asano, K. (2008) J. Biol. Chem. 283 1094–1103 [DOI] [PubMed] [Google Scholar]
- 36.Fletcher, C. M., Pestova, T. V., Hellen, C. U., and Wagner, G. (1999) EMBO J. 18 2631–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siridechadilok, B., Fraser, C. S., Hall, R. J., Doudna, J. A., and Nogales, E. (2005) Science 310 1513–1515 [DOI] [PubMed] [Google Scholar]
- 38.Srivastava, S., Verschoor, A., and Frank, J. (1992) J. Mol. Biol. 226 301–304 [DOI] [PubMed] [Google Scholar]
- 39.Conte, M. R., Kelly, G., Babon, J., Sanfelice, D., Youell, J., Smerdon, S. J., and Proud, C. G. (2006) Biochemistry 45 4550–4558 [DOI] [PubMed] [Google Scholar]
- 40.Fraser, C. S., Berry, K. E., Hershey, J. W., and Doudna, J. A. (2007) Mol. Cell 26 811–819 [DOI] [PubMed] [Google Scholar]
- 41.Asano, K., Shalev, A., Phan, L., Nielsen, K., Clayton, J., Valassek, L., Donahue, T. F., and Hinnebusch, A. G. (2001) EMBO J. 20 2326–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He, H., von der Haar, T., Singh, C. R., Ii, M., Li, B., Hinnebusch, A. G., McCarthy, J. E., and Asano, K. (2003) Mol. Cell. Biol. 23 5431–5445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jivotovskaya, A. V., Valasek, L., Hinnebusch, A. G., and Nielsen, K. H. (2006) Mol. Cell. Biol. 26 1355–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Battiste, J. L., Pestova, T. V., Hellen, C. U., and Wagner, G. (2000) Mol. Cell 5 109–119 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.