Abstract
The life span of model organisms can be modulated by environmental conditions that influence cellular metabolism, oxidation, or DNA integrity. The yeast nicotinamidase gene pnc1 was identified as a key transcriptional target and mediator of calorie restriction and stress-induced life span extension. PNC1 is thought to exert its effect on yeast life span by modulating cellular nicotinamide and NAD levels, resulting in increased activity of Sir2 family class III histone deacetylases. In Caenorhabditis elegans, knockdown of a pnc1 homolog was shown recently to shorten the worm life span, whereas its overexpression increased survival under conditions of oxidative stress. The function and regulation of nicotinamidases in higher organisms has not been determined. Here, we report the identification and biochemical characterization of the Drosophila nicotinamidase, D-NAAM, and demonstrate that its overexpression significantly increases median and maximal fly life span. The life span extension was reversed in Sir2 mutant flies, suggesting Sir2 dependence. Testing for physiological effectors of D-NAAM in Drosophila S2 cells, we identified oxidative stress as a primary regulator, both at the transcription level and protein activity. In contrast to the yeast model, stress factors such as high osmolarity and heat shock, calorie restriction, or inhibitors of TOR and phosphatidylinositol 3-kinase pathways do not appear to regulate D-NAAM in S2 cells. Interestingly, the expression of D-NAAM in human neuronal cells conferred protection from oxidative stress-induced cell death in a sirtuin-dependent manner. Together, our findings establish a life span extending the ability of nicotinamidase in flies and offer a role for nicotinamide-modulating genes in oxidative stress regulated pathways influencing longevity and neuronal cell survival.
Genetic manipulations and environmental conditions have been shown to modulate life span in various experimental model systems (1, 2). The environmental conditions include restricted calorie and nutrient availability that affect cellular metabolism as well as stress conditions such as heat and osmotic shock. Accordingly, the genetic manipulations are documented in stress response proteins and in metabolic proteins such as those along the insulin signaling pathway (2, 3). In the yeast Saccharomyces cerevisiae, several of these experimental alternations have been found to affect the replicative life span. Recent studies identified the pyrazinamidase/nicotinamidase gene pnc1 as a key effector of calorie restriction and mild stress-induced life span extension. These conditions induce increased pnc1 transcription and activity (4-6). In addition, overexpression of PNC1 was shown to be sufficient for extending the replicative life span of yeast.
In yeast, PNC1 functions in the NAD salvage pathway by converting nicotinamide to nicotinic acid (see Fig. 1A) (7-9). Nicotinamide is a component of vitamin B3/niacin and serves as a precursor in NAD biosynthesis (10, 11). NAD serves as a coenzyme in reversible redox reactions associated with cellular metabolism in all living cells. Recently, NAD and nicotinamide have emerged as regulators of a class of enzymes known as “NAD consumers” that have been linked to cellular stress resistance, life span, and various diseases (12-14). These enzymes include ADP-ribosyl transferases, poly-ADP-ribosyl polymerases, cADP-ribose synthetases, and Sir2 (silent information regulator 2) protein deacetylases. Reactions performed by these enzymes can rapidly deplete cellular NAD and generate nicotinamide, which acts as a potent feedback inhibitor of the NAD consumers. NAD depletion and nicotinamide buildup is ameliorated by salvage enzymes such as PNC1 that convert nicotinamide to metabolites that can be recycled back to NAD (15). Increasing evidence suggests that nicotinamide recycling is essential for both maintenance of intracellular NAD and regulation of NAD consumers in response to various internal and external stimuli (9, 16, 17).
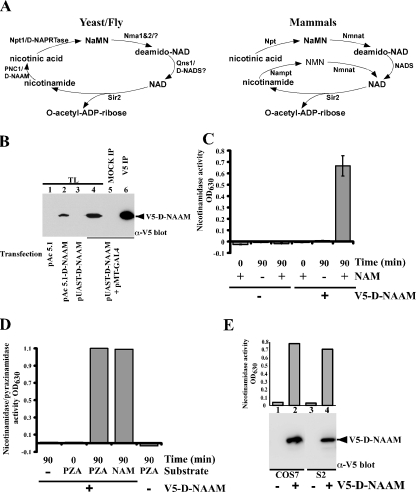
FIGURE 1.
The Drosophila homolog of yeast PNC1 encodes an active nicotinamidase. A, comparison of the NAD+ salvage pathway between mammals and yeast/flies. The nomenclature used is as in Rongvaux et al. (9) and Revollo et al. (8): PNC1, nicotinamidase; Npt1, nicotinic acid phosphoribosyltransferase; Nma1&2, nicotinic acid mononucleotide adenylyltransferase 1 and 2; Qns1, NAD synthetase; Nmnat, nicotinic acid mononucleotide adenylyltransferase; NADS, NAD synthetase; NaMN, nicotinic acid mononucleotide; NMN, nicotinamide mononucleotide. D-NAAM is our designation for Drosophila nicotinamidase. B, Drosophila S2 cells were transfected with carboxyl-terminally tagged V5-His-D-NAAM using the indicated expression vectors, and D-NAAM expression was determined in whole cell lysates (TL) or in V5 immunoprecipitates by V5 immunoblotting. C and D, V5 immunoprecipitates from S2 cells expressing a control vector or pAc 5.1-V5-His-D-NAAM were assayed for amidase activity using nicotinamide (NAM, C) or pyrazinamide (PZA, D) as a substrate. The activity is provided in A630 units corresponding to released ammonia. E, Drosophila S2 and mammalian COS-7 cells were transfected with control or pAc 5.1-V5-His-D-NAAM (S2 cells) or pExchange 5A-V5-His-D-NAAM (COS-7 cells) and nicotinamidase activity in V5 immunoprecipitates was analyzed as in C (top panel). D-NAAM recovery was determined using V5 immunoblotting (bottom panel).
The life span extending functions of pnc1 in yeast are thought to be mediated at least in part by increasing the activity of the NAD+-dependent protein deacetylase Sir2, and pnc1 is considered the key regulator of Sir2 in the yeast response to calorie restriction and stress (5, 18). PNC1 is thought to increase Sir2 function through two complementing mechanisms: 1) by increasing the NAD+/NADH ratio, providing higher concentration of the NAD+ cofactor, and 2) by decreasing the concentration of nicotinamide, which acts as a noncompetitive feedback inhibitor of Sir2. It is important to note that although increased Sir2 activity has been shown to extend the life span of several model organisms (19-21), it is not yet established whether all of the life span-extending functions of PNC1 can be attributed to increased Sir2 activity.
Yeast Sir2 is a prototype NAD+-dependent histone/protein deacetylase conserved through evolution (22-24). Mammals express at least seven Sir2 orthologs, designated sirtuins (Sirt1-7), varying in cellular expression and function (25). Mammalian sirtuins have been shown to deacetylate and regulate a large array of substrates including p53, FOXO, tubulin, and metabolic proteins such as PGC-1α, PPAR-γ, GDH, and acetyl-CoA synthetase (7, 22, 26-28). Thus, sirtuins have a significant potential to impact metabolic pathways involving glucose homeostasis, the insulin/IGF-1 signaling pathway, and stress response pathways associated with DNA damage and oxidative stress (1). Pointing to a possible role of sirtuins in mammalian aging and metabolism, two independent studies have demonstrated recently that oral administration of resveratrol (a natural compound enriched in grapes and identified as a direct activator of sirtuins) can reverse the pathophysiologic effects of high fat diet and restore life span (29, 30). However, it remains to be determined to what extent these beneficial effects of resveratrol result from its ability to activate sirtuins (31).
pnc1 homologs have been reported recently in Caenorhabditis elegans and Arabidopsis. van der Horst et al. (32) identified a C. elegans homolog of pnc1 and demonstrated that knockdown of the gene decreases the adult worm life span. Although increasing pnc1 dosage did not result in increased adult life span, it did confer increased survival under oxidative stress. The increased survival was reduced in worms treated with Sir2 RNA interference, suggesting that in worms the effects of PNC1 are also mediated at least in part through Sir2.
In Arabidopsis, at least two nicotinamidase genes have been identified (AtNIC1 and NIC2), and null alleles have been associated with increased sensitivity to salt, abscisic acid, and DNA-damaging agents (33, 34).
Although PNC1 appears the primary enzyme that metabolizes nicotinamide in yeast, Arabidopsis, and low metazoans, higher organisms and mammals evolved a somewhat different NAD salvage pathway (see Fig. 1A and Refs. 7-9). The functional counterpart of yeast PNC1 is proposed to be nicotinamide phosphoribosyltransferase (Nampt/PEBF/visfatin), which converts nicotinamide into nicotinamide mononucleotide instead of nicotinic acid, with nicotinamide mononucleotide being subsequently converted to NAD. Nampt function has been correlated with insulin regulation, cell survival, and life span and has also been shown to affect the activity of mammalian sirtuins, including the mammalian ortholog of yeast Sir2, SIRT1 (7, 8, 35). Nampt appears to function both intracellularly and as a secreted enzyme; however, our current knowledge of its exact regulation and function is limited (12, 36-38). Nevertheless, the nicotinamide-Sir2 link seems conserved through evolution, because mammalian sirtuins are also inhibited by nicotinamide. Thus, nicotinamidase orthologs, through affecting the activity of sirtuins and other NAD consumers, could prove key players in physiological pathways controlling cellular metabolism, cell death and survival, and ultimately aging (7).
In the present study, we describe the identification and biochemical characterization of the Drosophila nicotinamidase, D-NAAM, and demonstrate life span-extending properties of the gene. The nicotinamidase activity of D-NAAM was cell type autonomous and displayed comparable activity when purified from mammalian or Drosophila cell lines. In contrast to yeast, D-NAAM expression and activity in Drosophila cells was not affected by calorie and nutrient availability or by inhibitors of the PI3K3 or TOR pathways. Rather, D-NAAM was found to be responsive primarily to oxidative stress and anisomycin. Low oxidative stress increased D-NAAM mRNA expression by up to 6-fold and also increased protein expression and cellular nicotinamidase activity. Importantly, transgenic flies overexpressing D-NAAM exhibited an increase in both the mean and maximal life span of up to 30%. The increased life span was reversed in Sir2 mutant flies, suggesting a role of sir2 in D-NAAM-induced life span extension. Interestingly, D-NAAM was found to be functional in mammalian cells, and its expression in human neuronal cells conferred resistance to oxidative stress-induced cell death in a manner dependent on sirtuin activity. Our results establish a life span extending the ability of nicotinamidase in flies and suggest a role for nicotinamide-modulating genes in oxidative stress pathways influencing longevity and neuronal cell survival.
EXPERIMENTAL PROCEDURES
DNA Constructs and Antibodies—D-NAAM was PCR-amplified from a Drosophila gene collection clone LD05707 and subcloned into pAc 5.1, pUAST, pExchange 5A, or pEGFP vectors as Acc65I-BstEII, Acc65I-Acc65I, or SacII-XhoI fragment, respectively. pEGFP-Sirt1 was a kind gift from Dr. Izumi Horikawa (25). V5 epitope antibodies were from Invitrogen. D-NAAM antibodies were generated by PickCell Laboratories (Amsterdam, The Netherlands) against the last 20 carboxyl-terminal D-NAAM residues. Crude serums from two immunized rabbits were tested for D-NAAM reactivity and were affinity-purified using the immunizing peptide and a SulfoLink kit (Pierce). Preimmune serum from the above rabbits served as a control antibody as indicated in the figure legends.
Tissue Culture, Transfection, and Cell Death Assay—Embryonic Drosophila S2 cells, mammalian COS-7 cells, and human neuroblastoma SH-SY5Y cells were maintained in their appropriate growth media. S2 cells were transfected using CellFectin (Invitrogen), COS-7 cells using Lipofectamine (Invitrogen), and SH-SY5Y cells using lipofection and Lipofectamine. For neuronal cell death assay, 60-70% confluent cells expressing the indicated vectors were treated with 100 or 300 μm 6-(2-hydroxy-1-methyl-2-nitrosohydrazino)-N-methyl-1-hexanamine (NOC-9) or sodium nitroprusside, and cell death was analyzed using trypan blue dye exclusion 24 h post-treatment, and cell apoptosis was analyzed using a deoxynucleotidyltransferase-mediated dUTP nick end labeling assay as previously described (39, 40). The mean survival was determined by counting eight randomly selected nonoverlapping fields with each containing ∼10-30 cells (viable and nonviable). Each experiment was replicated 4-6 times with comparable results. In experiments using EGFP-tagged proteins, images were acquired with “blinded” assessment with a Leitz DMIRB microscope (Leica) and a Fuji/Nikon Super CCD (6.1 megapixels), and cell death was analyzed in all cells or was analyzed separately for EGFP-positive and -negative cells. In experiments using sirtinol, the cells were treated with varying concentrations of sirtinol 1 h prior to exposure to NOC-9.
Nicotinamidase/Pyrazinamidase Assay—Cellular and whole fly protein extracts were prepared in extraction buffer containing 50 mm Tris-Cl, pH 7.5, 100 mm NaCl, 1% Triton X-100, 1 mm dithiothreitol, 1 mm EDTA, 1 mm EGTA, 2 mm Na3VO4, 50 mm β-glycerophosphate, and a protease inhibitor mixture (GE Healthcare, Piscataway, NJ). D-NAAM for the assay was immunopurified from 1 mg of protein extracts using D-NAAM or V5 antibodies conjugated to protein A beads. As a control, preimmune serum was used for the D-NAAM antibody, and non-transgenic fly extracts or nontransfected cell extracts were used for the V5 immunopurifications. The beads were washed twice with extraction buffer, twice with extraction buffer containing 500 mm LiCl, and twice with nicotinamidase reaction buffer containing 5 mm Tris-Cl, pH 7.5, 150 mm NaCl, and 1 mm MgCl2. For the assay, the beads were incubated in reaction buffer alone or in reaction buffer containing 4 mm nicotinamide or pyrazinamide for 0 min or for 90 min as indicated in the figure legends at 30 °C in a rotating mixer. Following the incubation, the mixture was spun down, and the supernatant was collected and analyzed for ammonia content using ammonia detection kit (Wako Chemicals, Richmond, VA) according to the manufacturer's instructions. The amount of the nicotinamidase used in the assay and the reaction conditions were set in a way that the final ammonia readings were in a linear range, i.e. final A630 readings were in the range of 0.1-1. In all experiments, the A630 readings in control immunoprecipitates, 0-min incubations, and nicotinamide-omitted samples were consistently below 0.01.
D-NAAM Expression Analysis—For analyzing effects of culture conditions on D-NAAM expression, S2 cells were maintained in serum-free complete growth medium (Drosophila SFM medium; Invitrogen) or were cultured in medium lacking proteins (DS2 medium; CellGrow) for growth factor depravation. For nutrient depravation, the SFM and DS2 media were diluted 1:4 with phosphate-buffered saline as indicated in the figure legends. For assaying stress effects, the cells were grown in complete medium and treated with stress factors as indicated in the figure legends. D-NAAM expression was analyzed using real time PCR for mRNA and Western blotting for protein expression. Real time PCR was performed according to established methods using 107 S2 cells. ToTALLY RNA kit (Ambion, Austin, TX) and RNeasy (Qiagen) were used for RNA extraction and purification, and Applied Biosystems kits (Roche Applied Science) were used for primer design/synthesis, reverse transcription, and the real time PCR. For standardization, D-NAAM real time PCR results were normalized against Drosophila ribosomal protein L32 using a standard probe from Applied Biosystems. Each experiment was performed in duplicate, and the real time PCR was done in triplicate. The fold change in RNA expression was calculated using the ΔΔCt method. The D-NAAM expression in S2 cells growing in complete medium was taken as the standard point for calculating fold change.
Genetic Crosses, Longevity Assays, and Statistical Analysis—The UAS-Gal4 system was used to drive overexpression of D-NAAM. Overexpression was driven with both the ubiquitous tub-Gal4 driver and the pan-neural elav-Gal4 driver. pUAS-D-NAAM-V5 DNA was injected into w1118 embryos (Duke university), and the resulting positive transformants were identified and chromosomal insertions balanced using standard techniques. Transformants were crossed directly to tub-Gal4 and or elav-Gal4 when their insertion was on a different chromosome from the Gal4 driver. This allowed for obtaining flies heterozygous for the UAS-D-NAAM insertion on one chromosome and for the Gal4 driver on another chromosome. F1 test flies of the genotypes (D-NAAM33/+, tub-Gal4/+; D-NAAM42/+, tub-Gal4/+; D-NAAM31/+, tub-Gal4/+; and elav-Gal4/+, D-NAAM42/+) were selected by absence of balancers. Genetically matched wild type controls lacking both driver and UAS-transgene were generated by self-crossing F1 test flies. When the D-NAAM transgene and Gal4 driver were on the same chromosome, lines were first out-crossed to w1118 flies to obtain D-NAAM33/+, D-NAAM31/+, and elav-Gal4/+ flies, and these lines were crossed to collect test flies (D-NAAM33/elav-Gal4 and D-NAAM31/elav-Gal4) based on eye color and matched wild type controls by the absence of eye color. To overexpress D-NAAM in a Sir2 mutant, we used Sir24.5/CyO, Sir25.26/CyO (obtained from Stephen Helfand), D-NAAM42/TM3, and elav-Gal4/TM3 to generate two parental stocks: Sir24.5/CyO, elav-Gal4/TM3; and Sir25.26/CyO, D-NAAM42/TM3. These flies were crossed to generate flies overexpressing D-NAAM in a Sir2 mutant background (Sir24.5/Sir25.26, D-NAAM42/elav-Gal4). To control for elav-Gal4 or UAS-D-NAAM42 insertion sites, these parental stocks were crossed to the opposite Sir2 allele, i.e. to generate (Sir24.5/Sir25.26, elav-Gal4/+; and Sir24.5/Sir25.26, D-NAAM42/+, respectively). The Sir2 mutant control was obtained by directly crossing Sir24.5/CyO to Sir25.26/CyO to generate Sir24.5/Sir25.26 flies. Flies overexpressing D-NAAM42 in neurons and their genetically matched controls were generated as described above.
All of the fly stocks were maintained on standard cornmeal molasses medium. Test and control flies were verified with PCR analysis for transgenes/mutations and Western blotting for D-NAAM overexpression. For the longevity experiments, the flies were mated and raised on sucrose medium (41). Virgin male and female flies were separated into aliquots of 20/vial and transferred into fresh vials every 2 days while scoring for viability. Recording of deceased flies began after one initial food transfer to avoid anesthetization effects. Deceased flies were recorded and replaced with marker flies to maintain population density. χ2 tests compared the proportion of flies surviving between test and control flies to assign significance to the curves. Median survival was compared using Kruskal-Wallis tests. Mean and maximal survival of the top 10 and 20% were calculated for males and females separately using a one-way analysis of variance and orthogonal contrasts. All of the analyses were done using SPSS, version 13.5.
RESULTS
Identification and Biochemical Characterization of Drosophila Nicotinamidase, D-NAAM—To identify PNC1 homologs in higher organisms, we used the yeast PNC1 protein sequence to blast search the Swiss-Prot and GeneBank data bases, identifying two open reading frames in C. elegans (accession number NP_499876 and NP_001023531) and one in Drosophila melanogaster (accession number NP_732446). Alignment of the Drosophila protein, which we designate D-NAAM (Drosophila nicotinamie amidase), with yeast PNC1 and PNCA protein from Pyrococcus horikoshii shows a moderate overall homology (supplemental Fig. S1); however, the homology is significantly higher when looking at regions in PNCA regarded as critical for catalytic activity (42) (i.e. β sheets β1-6 and α helixes α1-4), suggesting functional conservation.
A DNA clone obtained from the Drosophila gene collection containing the full-length D-NAAM cDNA (LD05707) was verified for sequence accuracy and subcloned with a carboxyl-terminal V5-His6-epitope tag into the Drosophila expression vector pAc 5.1 for expression in Drosophila Schneider2 (S2) cells and the pUAST vector for P-element mediated germ-line transformation (43). D-NAAM protein expression was examined in S2 cells transfected with pAc 5.1-V5-His6-D-NAAM or pUAST-V5-His6-D-NAAM cotransfected with pMET-Gal4 that encodes the yeast Gal4 transcription factor needed for expression from the pUAST vector (Fig. 1B). The tagged protein in S2 cells migrated as a 48-kDa band on SDS-PAGE and was efficiently purified by immunoprecipitation using V5 antibodies.
To examine the enzymatic activity of D-NAAM, the tagged protein was immunopurified using V5 antibodies and subjected to a nicotinamidase activity assay developed in this study by modifying a previously described calorimetric assay (44) (Fig. 1, C and D). The assay measures nicotinamidase activity using either nicotinamide or pyrazinamide as a substrate and determining ammonia release as the reaction end product. Nicotinamidase activities measured in this assay had low background readings even after 90-min incubations in the absence of a substrate or the enzyme (V5 immunoprecipitate from mock transfected cells was used as a negative control). These experiments established that D-NAAM encodes a bona fide nicotinamidase and that D-NAAM does not distinguish between nicotinamide and pyrazinamide as a substrate, similarly to the bacterial PNCA.
To test whether D-NAAM functions autonomously or whether it requires specific cofactors present exclusively in Drosophila cells, we expressed D-NAAM in mammalian COS-7 cells and compared its nicotinamidase activity with that of D-NAAM expressed in Drosophila S2 cells (Fig. 1E). COS-7 cells expressed comparable protein levels and nicotinamidase activity as S2 cells, indicating that D-NAAM does not require Drosophila cell-specific cofactors for expression or activity.
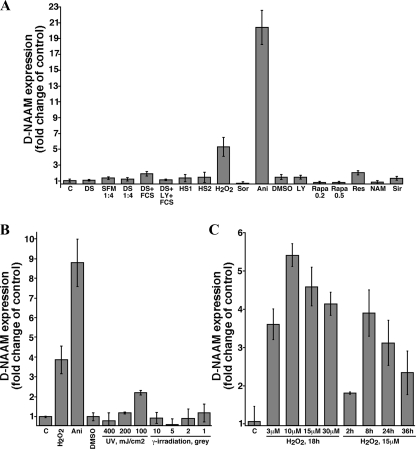
D-NAAM Expression and Activity Are Regulated by Oxidative Stress—PNC1 in yeast has been demonstrated to be regulated by calorie and nutrient availability as well as by mild stress conditions such as heat shock and osmotic stress (5). To examine the regulation of D-NAAM in Drosophila S2 cells, we measured the expression and activity of D-NAAM under varying environmental conditions (Figs. 2 and 3). Using real time PCR experiments, we identified primarily two factors that significantly affected D-NAAM mRNA expression: hydrogen peroxide and anisomycin (Fig. 2A). Interestingly, various other stress factors, such as osmotic and heat shock, high salt, or UV and γ-irradiation did not significantly affect D-NAAM expression levels (Fig. 2, A and B), although they induced comparable cell death to oxidative stress (data not shown). These results suggest a unique regulation of D-NAAM by pathways sensitive to oxidative stress and anisomycin. The effects of hydrogen peroxide on D-NAAM mRNA expression were dose- and time-dependent, reaching up to a 5-fold increase at 10 μm and following 8-16 h of treatment (Fig. 2C). Anisomycin treatment resulted in a higher increase, reaching up to 20-fold (Fig. 2, A and B). Importantly, we did not observe any affect of nutrient restriction or compounds that inhibit signaling molecules along the insulin/IGF-1 pathway such as PI3K inhibitors or TOR inhibitors on D-NAAM expression (Fig. 2A). These results indicate that D-NAAM regulation in Drosophila cells does not follow the same pattern as in yeast, at least with regards to dietary restrictions and responses to high salt and osmotic shock.
FIGURE 2.
D-NAAM mRNA expression is regulated by oxidative stress. A, Drosophila S2 cells were cultured under the indicated growth conditions, and the expression of D-NAAM mRNA was analyzed using real time PCR. The data are presented as fold change in D-NAAM expression after standardizing to ribosomal protein L32. The treatments were as follows: C, control, complete SFM media; DS, DS2 media (media lacking protein factor additives); SFM 1:4, SFM medium diluted 1:4 with phosphate-buffered saline (nutrient deficient media), 18 h; DS 1:4, DS medium diluted 1:4 with phosphate-buffered saline, 18 h; FCS, 10% fetal calf serum, 4 h; LY, 10 μm LY294002, 18 h (PI3K inhibitor); HS1, heat shock, 2 h at 37 °C and 24 h recovery; HS2, heat shock, 2 h at 37 °C; H2O2, 15 μm, 18 h; Sor, 0.5 m sorbitol, 18 h; Ani, 10 μg/ml anisomycin, 18 h; DMSO, 0.1% Me2SO, 18 h (control); Rapa, rapamycin, 0.2 or 0.5 μg/ml, 18 h; Res, 100 μm resveratrol, 18 h; NAM, 40 mm nicotinamide, 18 h; Sir, 50 μm sirtinol, 18 h. B, Drosophila S2 cells growing in complete SFM medium were treated as indicated or cells were irradiated and left to recover for 24 h. The cells were analyzed for changes in D-NAAM mRNA expression as in A. C, Drosophila S2 cells were treated with the indicated concentrations of H2O2 for the indicated times, and the D-NAAM mRNA levels were analyzed as in A.
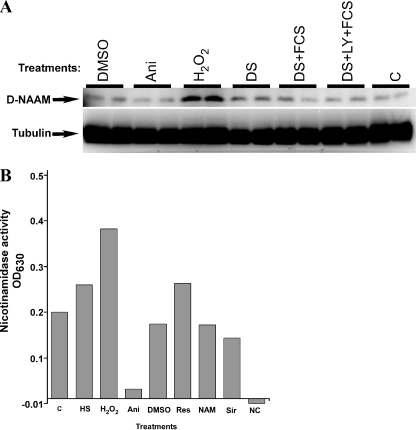
FIGURE 3.
Oxidative stress enhances D-NAAM protein expression and activity. A, Drosophila S2 cells were cultured under the indicted growth condition (as in Fig. 2A) for 18 h, and total cell lysates were analyzed for D-NAAM protein expression using a carboxyl-terminal D-NAAM peptide antibody (top panel). Tubulin immunoblotting was used to confirm protein loading (bottom panel). Cell treatments and the protein expression analysis were performed in duplicate. DMSO, dimethyl sulfoxide. B, D-NAAM was immunopurified from S2 cells cultured under the indicated conditions using the carboxyl-terminal D-NAAM antibody and assayed for nicotinamidase activity as in Fig. 1. NC, negative control, no substrate (NAM) was added in the nicotinamidase assay. The experiment was performed in duplicate and is representative of three independent experiments. The treatments were as follows: C, control; HS, heat shock, 2 h at 37 °C; H2O2, 15 μm, 18 h; Ani, 10 μg/ml anisomycin, 18 h; DMSO, 0.1% Me2SO, 18 h (control); Res, 100 μm resveratrol, 18 h; NAM, 40 mm nicotinamide, 18 h; Sir, 50 μm sirtinol, 18 h.
To confirm the results obtained with the mRNA experiments at the protein level, we developed antibodies against a carboxyl-terminal D-NAAM peptide and examined D-NAAM protein expression in S2 cells grown under various media and stress conditions (Fig. 3A). Similarly to what we observed with D-NAAM mRNA expression, only oxidative stress significantly affected D-NAAM protein expression, inducing up to a 3-fold increase. As anticipated from its function as a protein synthesis inhibitor, the ability of anisomycin to induce D-NAAM mRNA expression was not reproduced at the protein level. These experiments also pointed out that D-NAAM is not a highly stable protein, showing a half-life of ∼8 h (data not shown). Finally, the activity of D-NAAM in S2 cells exposed to various growth conditions was determined by immunopurifying endogenous D-NAAM and assaying for nicotinamidase activity (Fig. 3B). The increased activity observed with oxidative stress correlated with increased D-NAAM expression, suggesting that the differences in activity reflected increased D-NAAM protein expression rather than changes in specific activity. Confirming this notion, we did not observe any effects of oxidative stress on D-NAAM activity when it was expressed from an expression vector or when the activity was standardized for D-NAAM protein recovery (data not shown). Combined, these results indicate that oxidative stress regulates D-NAAM at the transcriptional level, increasing its mRNA and protein expression and thus its cellular nicotinamidase activity.
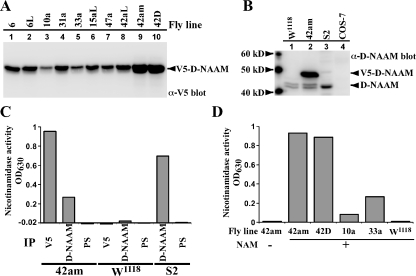
Transgenic Flies Overexpressing D-NAAM Exhibit Increased Nicotinamidase Activity—As noted above, PNC1 overexpression extends yeast replicative life span. We hypothesized that D-NAAM may have similar effect in Drosophila. To test this hypothesis, 10 independent germ-line transformants carrying a pUAST-D-NAAM vector were generated and crossed with tub-Gal4 driver flies to achieve ubiquitous expression of D-NAAM via the UAS-Gal4 system. These flies expressed varying amounts of D-NAAM protein (Fig. 4A and supplemental Fig. S2), consistent with different insertion sites. The transformants were grouped as low (lanes 3 and 5), medium (lanes 1, 2, 4, and 6-8), or high (lanes 9 and 10) expressers. The high expressing D-NAAM transgenic lines exhibited up to a 5-fold increase in D-NAAM protein expression when compared with the endogenous protein level (Fig. 4B). The D-NAAM antibody stained a doublet band in fly extracts, suggesting alternative splicing or a post-translational modification of D-NAAM. Importantly, the level of D-NAAM expression in the different transformants directly correlated with increased nicotinamidase activity in these flies (Fig. 4, C and D). The difference in activity seen when using V5 and D-NAAM antibodies reflected the difference in their immunoprecipitation efficiencies (data not shown). As expected, nicotinamidase activity was not detected in control preimmune serum immunoprecipitates from transgenic flies or from V5 immunoprecipitates from control, w1118 flies. Similar to PNC1 overexpression in yeast, D-NAAM overexpressing flies showed, in an initial analysis, increased NAD+/NADH levels, resulting mainly from decreased NADH levels (supplemental Table S1). These experiments confirm that overexpression of D-NAAM increases the cellular nicotinamidase activity in adult flies and that this expression potentially affects NAD metabolism.
FIGURE 4.
Increased nicotinamidase activity in transgenic Drosophila lines overexpressing D-NAAM. A, expression of V5-D-NAAM protein in pUAST-D-NAAM transgenic lines in tubulin-Gal4 driver background was analyzed using V5 immunoblotting. B, D-NAAM protein expression in control w1118 flies, a high expressing D-NAAM transgenic line 42am (tubulin-Gal4 driver), S2 cells, and control COS-7 cells were analyzed using a carboxyl-terminal D-NAAM antibody. C and D, nicotinamidase activity in protein extracts of the indicated fly lines or S2 cells was analyzed by immunoprecipitating (IP) D-NAAM using V5 or a carboxyl-terminal D-NAAM antibody and assaying amidase activity as in Fig. 2B. Preimmune serum for the D-NAAM antibody (PS) was used as a control antibody. The difference seen in activity between V5 and D-NAAM immunoprecipitates reflects the difference in immunoprecipitation efficiencies (data not shown). Note that the low activity seen in the w1118 sample is a result of assay linearity limitations (the total amount of protein to be used for the immunoprecipitation was determined in a way to allow remaining in the linear range of the assay for the high expressing D-NAAM lines). NAM, nicotinamide.
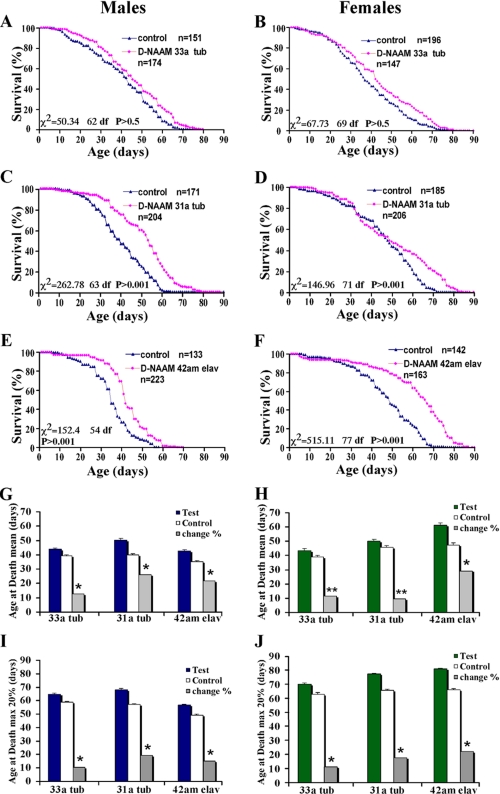
D-NAAM Overexpression Extends Drosophila Life Span—To determine the effect of nicotinamidase overexpression on fly life span, low, middle, and high D-NAAM-expressing flies using the tub-Gal4 or an elav-Gal4 driver (allows expression of the transgene exclusively in post-mitotic neurons) were analyzed for life span and compared with genetically matching w1118 control flies not expressing D-NAAM (see “Experimental Procedures,” Fig. 5, supplemental Table S2, and supplemental Fig. S2). Up to 200 flies were used in each group of male and female flies, which were collected as virgins and analyzed separately to avoid mating effects. Low expressing lines such as D-NAAM33 (see supplemental Fig. S2 for D-NAAM expression levels in male and female transgenic flies analyzed for life span) had a small but significant increase in life span compared with their controls (Fig. 5, A and B, and supplemental Table S2). Importantly, the higher expressing D-NAAM lines such as D-NAAM31 or D-NAAM42 displayed a more robust increase in life span as compared with their respective matching controls (Fig. 5, C-F, and supplemental Table S2). The life span extension was observed both in males and females and reached up to a 30% increase in the mean life span and in the maximal life span calculated for the top 20% survivors in each group (Fig. 5. G-J). Statistical analyses verified that these increases were significant, with most of the experiments reaching p values below 0.005. Comparable results were obtained with other D-NAAM transformants (supplemental Table S2). The fact that multiple UAS-D-NAAM insertions exhibit extended life span using three different driver lines strongly argues that the overexpression of D-NAAM, and not insertion site effects, is responsible for the observed life span extension. In support, no increase in life span was measured in adult flies carrying only the UAS-D-NAAM transgene, arguing against effects of heterosis (data not shown). Importantly, the increased life span observed using elav-Gal4 driver lines that overexpress D-NAAM strictly in neuronal tissues suggests that increased D-NAAM activity in neuronal tissues is sufficient to affect the life span of adult flies. This result is in agreement with previous studies showing that Sir2 overexpression in neuronal tissues is sufficient to increase fly life span (19) and with studies linking NAD+ production to neuroprotection. We cannot yet conclude whether this effect is mediated exclusively by affecting the neuronal tissue or involves secondary effects mediated by the neuronal cells, e.g. secretion of specific hormones.
FIGURE 5.
D-NAAM overexpression extends Drosophila life span. A-F, survival curves for male (A, C, and E) and female (B, D, and F) adult flies expressing low (D-NAAM33; A and B) or middle (D-NAAM31; C and D) D-NAAM protein levels ubiquitously via the tubulin-Gal4 driver or of a high expressing line expressing D-NAAM strictly in neurons via the elav-Gal4 driver (D-NAAM42; E and F; see Fig. 4A for D-NAAM expression levels in the specific lines). A genetically matching control line was used for each test line. Indicated are the n values for each experiment and the statistical parameter values. See supplemental Table S2 for the complete longevity analysis. G-J, mean (G and H) and the maximal life span of the top 20% survivors of test and control flies (I and J). The gray bars represent the percentage of change between the matching control and the test line for each pair. The single asterisk denotes a p value <0.005, and two asterisks denote a p value <0.05.
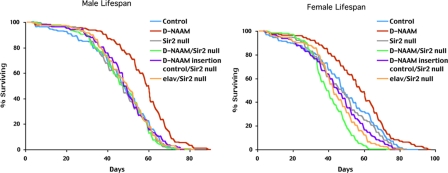
D-NAAM-induced Longevity Is Reversed in Sir2 Null Mutants—Because pnc1-induced longevity in yeast is dependent on Sir2 (5), we sought to determine whether D-NAAM-induced longevity has a similar requirement in Drosophila. Sir2 null flies were generated by breeding flies carrying two independent dSir2 excisions, Sir24.5 and Sir25.26. The transheterozygote adults are viable and reported to exhibit a life span comparable with control animals (19, 45). D-NAAM was overexpressed in this background by generating lines that carried Sir24.5 on the second chromosome and the elav-Gal4 driver on the third chromosome and crossing to a second line carrying the Sir25.26 allele on the second chromosome and the high expressing D-NAAM42 transgene on the third chromosome. Progeny of this cross overexpress D-NAAM in neurons while being deficient for Sir2. Parallel control crosses were used to test for potential background effects of elav-Gal4 and D-NAAM chromosomes or their transgene insertion sites (see “Experimental Procedures”). In male flies, overexpression of D-NAAM resulted in a significant (p < 0.001) increase of 23% (56.9 days versus 46.35 days) in mean life span versus genetically matched control flies (Fig. 6, left panel). This was comparable with the increase in life span observed using the elav-Gal4 driver on the second chromosome (Fig. 5E). In agreement with the earlier studies, the mean life span of Sir24.5/Sir25.26 flies did not differ significantly from matched wild type control flies (46.67 days versus 46.35 days). Importantly, D-NAAM overexpression in the Sir2 mutant background failed to extend life span, with the mean life span being similar to control animals (46.83 versus 46.35 days). Life span of control flies harboring only the UAS-D-NAAM transgene or the elav-Gal4 driver in a Sir2 mutant background were also similar to controls. Thus, in adult males, the extension of life span mediated by D-NAAM overexpression is completely abolished if the Sir2 gene is mutated. Similar results were observed in female flies (Fig. 6, right panel). Overexpression of D-NAAM resulted in a significant (p < 0.002) increase of 17% (56.83 versus 48.48 days) in the mean life span versus matched controls. As with male flies, there was no significant change in female mean life span in Sir24.5/Sir25.26 transheterozygotes versus control flies (47.22 versus 48.48 days). D-NAAM overexpression in Sir2 mutants did not extend life span, and in fact, these flies exhibited a significant (p < 0.001) decrease in mean life span when compared with either matched wild type controls or Sir2 mutant flies (40.35 versus 48.48 and 47.22 days, respectively). The reduced life span appears to reflect an additive negative effect of the two transgenic third chromosomes on fly life span. Female flies carrying either the UAS-D-NAAM insertion or the elav-Gal4 driver insertion in a Sir2 mutant background displayed a small reduction in mean life span (43.86 and 44.76 days versus 47.22), which when combined appear to have an additive effect on female life span. Why this is observed in females only is unclear, but as virgin females continue to produce eggs, it may reflect a potential trade off between life span and fecundity, a phenomenon observed with several other gene mutations (46). Nevertheless, as in males, pan-neural overexpression of D-NAAM increases adult female life span as long as the Sir2 gene is functional.
FIGURE 6.
D-NAAM-induced longevity requires Sir2. Survival curves of male (left panel) and female (right panel) adult flies expressing D-NAAM42 using the elav-Gal4 driver in control and Sir2 null background. Genotypes: wild type control: w1118; D-NAAM overexpression: w1118;UAS-D-NAAM42/elav-Gal4; Sir2 null: w1118; Sir24.5/Sir25.26; D-NAAM overexpression in Sir2 null: w1118;Sir24.5/Sir25.26;UAS-D-NAAM42/elav-Gal4; D-NAAM insertion control in Sir2 null: w1118; Sir24.5/Sir25.26;UAS-D-NAAM42/+; elav-Gal4 insertion control in Sir2 null: w1118;Sir24.5/Sir25.26;elav-Gal4/+.
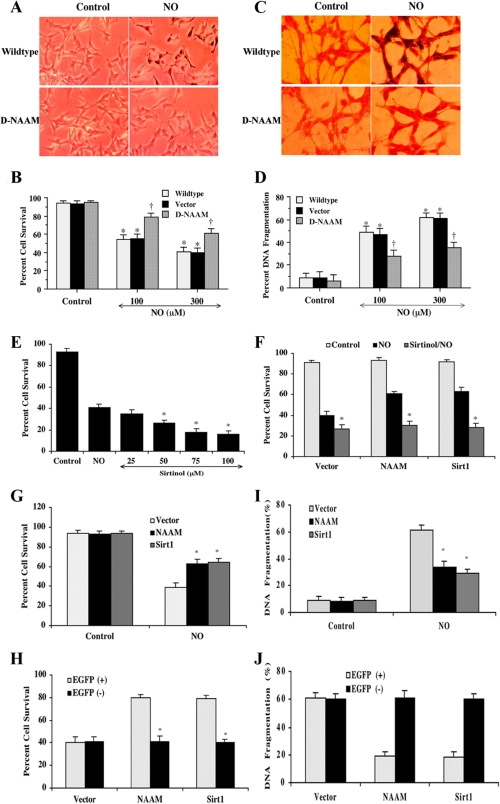
D-NAAM Expression Protects Human Neuronal Cells from Oxidative Stress-induced Cell Death—The ability of neuronally restricted D-NAAM overexpression to affect fly life span and the finding that oxidative stress was the main regulator of D-NAAM led us to hypothesize that D-NAAM might hold a role in oxidative stress resistance in neuronal cells. This hypothesis was also prompted by previous studies showing that activation of the mammalian homolog of Sir2, SIRT1, has a neuronal protective function (47-49) and that SIRT1 expression may be linked to nitric oxide production (50). As demonstrated in Fig. 1E, D-NAAM expresses well in mammalian COS-7 cells and displays comparable nicotinamidase activity levels as when expressed in Drosophila cells, suggesting that D-NAAM could be functional also in mammalian cells. Expression of D-NAAM in human SH-SY5Y neuroblastoma cells markedly inhibited cell death and apoptosis induced by treating the cells with the reactive oxygen-generating agent NOC-9 (Fig. 7) or a nonrelated nitric oxide donor, sodium nitroprusside (data not shown). The inhibition of cell death and apoptosis were not observed in control cells expressing an empty vector. These experiments indicated that increased cellular nicotinamidase activity, similarly to SIRT1 activation, has a neuronal protective function. To examine whether the protective effects of D-NAAM overexpression were related to sirtuin activation, cells were treated with the sirtuin inhibitor sirtinol 1 h prior to exposure to NOC-9 (Fig. 7, E and F). These experiments showed that sirtinol treatment abolished the protective effects of D-NAAM on neuronal cells in a similar manner to its ability to abolish the protective effects of SIRT1 overexpression, suggesting sirtuin involvement in the protective effects of D-NAAM. Finally, to test whether D-NAAM affected only the transfected cells or also had an effect on adjoining, nontransfected cells, we constructed a GFP-D-NAAM expression vector and compared cell death in GFP-positive and GFP-negative cells from the same culture dish (Fig. 7, G-J). GFP-negative cells analyzed from samples transfected with GFP-D-NAAM had similar cell death rates as cells from samples transfected with a GFP control vector, whereas the GFP-D-NAAM- and GFP-Sirt1-positive cells showed marked cell death resistance. Thus, the results showed that at least in vitro, D-NAAM overexpression exclusively affects the cells overexpressing the gene but not adjoining cells.
FIGURE 7.
D-NAAM expression protects human neuronal cells from oxidative stress-induced cell death. A-D, SH-SY5Y cells transfected with control vector or D-NAAM or nontransfected cells were exposed for 24 h to 100 or 300 μm NOC-9 and analyzed for cell death using trypan blue exclusion (A and B) or assayed for apoptotic cell death using deoxynucleotidyltransferase-mediated dUTP nick end labeling assay (C and D). For quantification of cell death and DNA fragmentation, on average, 200 cells were counted in triplicate samples (B and D). † denotes p value <0.01, and asterisks denote statistically nonsignificant difference between nontransfected and vector-transfected cells. E and F, control SH-SY5Y cells (E) or cells expressing EGFP control vector, EGFP-D-NAAM or EGFP-Sirt1 (F) were treated with the indicated concentrations of sirtinol (E) or 50 μm sirtinol (F) 1 h prior to exposure to 300 μm NOC-9 and cell survival was determined 24 h later as in B. G-J, SH-SY5Y cells were transfected with EGFP control vector, EGFP-D-NAAM, or EGFP-Sirt1; cell survival in response to 300 μm NOC-9 was analyzed as in B (G), and apoptotic cell death was analyzed as in D (I). Alternatively, EGFP positive and negative cells were analyzed separately (H and J). The asterisks denote p value <0.01.
DISCUSSION
The results presented here identify the Drosophila nicotinamidase, D-NAAM, as an oxidative stress-regulated gene that when overexpressed, significantly extends adult fly life span and confers resistance to oxidative stress in human neuronal cells. Both the extended longevity and neuroprotection were dependent on Sir2/sirtuin function. The conserved family of NAD+-dependent protein deacetylases with homology to the yeast Sir2 have been shown to affect a variety of cellular functions ranging from response to stress and calorie restriction to extending life span in Drosophila and nematodes and promoting mammalian neuronal cell survival (14, 27, 51-54). In addition, small molecule activators of sirtuins have been demonstrated recently to affect mammalian aging and health under conditions of a high fat diet (29, 30). The function of Sir2 deacetylases, including vertebrate sirtuins, is negatively regulated by nicotinamide (5, 6, 55), a by-product feedback inhibitor in deacetylation reactions involving NAD+-dependent deacetylases and a metabolite in the nucleotide salvage pathway. In this context, nicotinamidases can be viewed as master regulators of proteins that are regulated by nicotinamide such as sirtuins, and therefore, manipulation of their function could affect a large variety of cellular functions (7, 12, 15, 38). Our demonstration of a Drosophila nicotinamidase and its role in longevity and neuronal protection support this view.
The analysis of D-NAAM regulation in Drosophila S2 cells shows that both the gene and its function are regulated primarily by oxidative stress, but not by several other stress factors, growth conditions limiting calorie and nutrient availability or by pharmacological agents that block signaling along the insulin/IGF-1 receptor pathway. These findings are significantly different from the observations made in yeast showing PNC1 regulation by calorie restriction and several stress factors (5). In addition, although a recent study showed PNC1 regulation downstream of TOR through the yeast transcription factors MSN2/4 (56), we did not observe changes in D-NAAM expression in response to the TOR inhibitor rapamycin. Thus, it is possible that the yeast, as a single cell organism, combines stress and nutrient sensing pathways in same response elements, whereas in higher organisms there is more separation between these pathways. The insulin/IGF-1 pathway, through activation of the PI3K-AKT-FOXO pathway, has emerged as a key regulator of life span in response to calorie availability in various organisms including Drosophila, nematodes and to some extent, mammals (57). Because this pathway is not conserved in yeast, it is possible that in higher organisms there has been a specialization of the nicotinamidase pathway to respond to oxidative stress, whereas the PI3K-AKT-FOXO pathway became the primary responder to calorie restriction. Interestingly, these two pathways seem to cross-talk in higher organisms, and the FOXO transcription factors are deacetylation targets of sirtuins (58-63).
Overexpression of the nicotinamidase gene extended life span in Drosophila similarly to its ability to increase the replicative life span in yeast (5). The increased life span appeared both in females and males and correlated with the level of D-NAAM overexpression; it was not observed in matched controls lacking the driver or the transgene. Moreover, the increased life span was dependent on Sir2 levels because life span extension was completely reversed when D-NAAM was overexpressed in a Sir2 null mutant. The observed effect of Sir2 mutation on Drosophila life span extension by D-NAAM and of sirtinol on mammalian cell death protection points to critical role of sirtuins in these functions of D-NAAM. However, the question of whether all these effects of D-NAAM are mediated by sirtuins or whether they involve other targets needs to be further evaluated.
In a recent study, van der Horst et al. (32) demonstrated that knockdown of the C. elegans homolog of pnc1 significantly shortens the life span of adult worms, establishing the role of PNC1 also in worm life span regulation. In addition, overexpression of PNC1 conferred resistance to oxidative stress in a Sirt2-dependent manner, which is consistent with our observation of neuronal protection in mammalian cells. However, the authors did not observe life span extension in worms overexpressing the PNC1 homolog. The lack of observed effect of PNC1 overexpression on life span extension could be a result of an inherent difference between worms and flies or the result of insufficient overexpression of functional protein. In the transgenic worms, PNC1 mRNA expression levels were increased up to 5-fold, but there was no indication of the protein levels. In our adult flies, the degree of life span extension correlated well with the level of protein overexpression, and maximum extension was observed when protein levels (not just mRNA levels) increased up to 5-fold. Thus, it is possible that the level of PNC1 overexpression in worms was not robust enough to observe an effect on life span.
Our and other's bioinformatics approaches did not identify any genes in mammals with significant homology to yeast, bacterial, or lower metazoan nicotinamidases, leading to the view that the NAD salvage pathway in mammals is different from the one in yeast, worms, and flies (Fig. 1A and Refs. 7-9). Nampt that converts nicotinamide into nicotinamide mononucleotide is presumed to be the functional counterpart of yeast PNC1. However, we would like to note that because of the low conservation of nicotinamidases from yeast to fly, our inability to identify an apparent nicotinamidase homolog in mammals does not explicitly exclude its existence. Regardless of the presence of a de facto nicotinamidase in mammals, our data provide strong evidence that increased nicotinamide clearance in cells provides positive effects on organism life span and cellular response to oxidative stress. This suggests that manipulation of nicotinamide metabolism through genetic approaches or pharmacological agents in vertebrates could yield similar beneficial results.
Finally, although our results clearly implicate Sir2 activity in life span extension and neuronal protection when D-NAAM is overexpressed, it remains to be determined whether nicotinamidase activation or activation of other nicotinamide modulating enzymes, such as Nampt, work exclusively through sirtuins. In this regard, a recent study suggested that the effects of nicotinamide on yeast life span are not exclusively mediated by Sir2 or other yeast sirtuins (64). This question also relates to the overall beneficial properties of direct sirtuin activators such as resveratrol in comparison with nicotinamide modulators, which may provide their benefits both through activation of sirtuins and modulation of other as of yet unidentified targets.
Supplementary Material
Acknowledgments
We thank Joseph Avruch and Ajay Rana for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-GM067134 (to G. T.) and NS053946 (to K. M.). This work was also supported by the American Diabetes Association, the American Heart Association Bugher Foundation Award, and Wayne State University institutional support (to M. V. B.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Tables S1 and S2.
Footnotes
The abbreviations used are: PI3K, phosphatidylinositol 3-kinase; NOC-9, 6-(2-hydroxy-1-methyl-2-nitrosohydrazino)-N-methyl-1-hexanamine; GFP, green fluorescent protein; EGFP, enhanced GFP; IGF, insulin-like growth factor; Nampt, nicotinamide phosphoribosyltransferase.
References
- 1.Sinclair, D. A. (2005) Mech. Ageing Dev 126 987-1002 [DOI] [PubMed] [Google Scholar]
- 2.Guarente, L., and Kenyon, C. (2000) Nature 408 255-262 [DOI] [PubMed] [Google Scholar]
- 3.Tatar, M., Bartke, A., and Antebi, A. (2003) Science 299 1346-1351 [DOI] [PubMed] [Google Scholar]
- 4.Gallo, C. M., Smith, D. L., Jr., and Smith, J. S. (2004) Mol. Cell. Biol. 24 1301-1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson, R. M., Bitterman, K. J., Wood, J. G., Medvedik, O., and Sinclair, D. A. (2003) Nature 423 181-185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bitterman, K. J., Anderson, R. M., Cohen, H. Y., Latorre-Esteves, M., and Sinclair, D. A. (2002) J. Biol. Chem. 277 45099-45107 [DOI] [PubMed] [Google Scholar]
- 7.Yang, H., Lavu, S., and Sinclair, D. A. (2006) Exp. Gerontol. 41 718-726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Revollo, J. R., Grimm, A. A., and Imai, S. (2004) J. Biol. Chem. 279 50754-50763 [DOI] [PubMed] [Google Scholar]
- 9.Rongvaux, A., Andris, F., Van Gool, F., and Leo, O. (2003) Bioessays 25 683-690 [DOI] [PubMed] [Google Scholar]
- 10.Denu, J. M. (2007) Cell 129 453-454 [DOI] [PubMed] [Google Scholar]
- 11.Denu, J. M. (2005) Trends Biochem. Sci. 30 479-483 [DOI] [PubMed] [Google Scholar]
- 12.Sauve, A. A. (2008) J. Pharmacol. Exp. Ther. 324 883-893 [DOI] [PubMed] [Google Scholar]
- 13.Csiszar, A., Pacher, P., Kaley, G., and Ungvari, Z. (2005) Curr. Vasc. Pharmacol. 3 285-291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bordone, L., and Guarente, L. (2005) Nat. Rev. Mol. Cell. Biol. 6 298-305 [DOI] [PubMed] [Google Scholar]
- 15.Belenky, P., Bogan, K. L., and Brenner, C. (2007) Trends Biochem. Sci. 32 12-19 [DOI] [PubMed] [Google Scholar]
- 16.Bogan, K. L., and Brenner, C. (2008) Annu. Rev. Nutr. 28 115-130 [DOI] [PubMed] [Google Scholar]
- 17.Berger, F., Ramirez-Hernandez, M. H., and Ziegler, M. (2004) Trends Biochem. Sci. 29 111-118 [DOI] [PubMed] [Google Scholar]
- 18.Anderson, R. M., Latorre-Esteves, M., Neves, A. R., Lavu, S., Medvedik, O., Taylor, C., Howitz, K. T., Santos, H., and Sinclair, D. A. (2003) Science 302 2124-2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rogina, B., and Helfand, S. L. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 15998-16003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tissenbaum, H. A., and Guarente, L. (2001) Nature 410 227-230 [DOI] [PubMed] [Google Scholar]
- 21.Kaeberlein, M., McVey, M., and Guarente, L. (1999) Genes Dev. 13 2570-2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haigis, M. C., and Guarente, L. P. (2006) Genes Dev. 20 2913-2921 [DOI] [PubMed] [Google Scholar]
- 23.Longo, V. D., and Kennedy, B. K. (2006) Cell 126 257-268 [DOI] [PubMed] [Google Scholar]
- 24.Denu, J. M. (2005) Curr. Opin. Chem. Biol. 9 431-440 [DOI] [PubMed] [Google Scholar]
- 25.Michishita, E., Park, J. Y., Burneskis, J. M., Barrett, J. C., and Horikawa, I. (2005) Mol. Biol. Cell 16 4623-4635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hallows, W. C., Lee, S., and Denu, J. M. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 10230-10235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guarente, L. (2007) Cold Spring Harb. Symp. Quant. Biol. 72 483-488 [DOI] [PubMed] [Google Scholar]
- 28.Starai, V. J., Celic, I., Cole, R. N., Boeke, J. D., and Escalante-Semerena, J. C. (2002) Science 298 2390-2392 [DOI] [PubMed] [Google Scholar]
- 29.Lagouge, M., Argmann, C., Gerhart-Hines, Z., Meziane, H., Lerin, C., Daussin, F., Messadeq, N., Milne, J., Lambert, P., Elliott, P., Geny, B., Laakso, M., Puigserver, P., and Auwerx, J. (2006) Cell 127 1109-1122 [DOI] [PubMed] [Google Scholar]
- 30.Baur, J. A., Pearson, K. J., Price, N. L., Jamieson, H. A., Lerin, C., Kalra, A., Prabhu, V. V., Allard, J. S., Lopez-Lluch, G., Lewis, K., Pistell, P. J., Poosala, S., Becker, K. G., Boss, O., Gwinn, D., Wang, M., Ramaswamy, S., Fishbein, K. W., Spencer, R. G., Lakatta, E. G., Le Couteur, D., Shaw, R. J., Navas, P., Puigserver, P., Ingram, D. K., de Cabo, R., and Sinclair, D. A. (2006) Nature 444 337-342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaeberlein, M., and Rabinovitch, P. S. (2006) Nature 444 280-281 [DOI] [PubMed] [Google Scholar]
- 32.van der Horst, A., Schavemaker, J. M., Pellis-van Berkel, W., and Burgering, B. M. (2007) Mech. Ageing Dev. 128 346-349 [DOI] [PubMed] [Google Scholar]
- 33.Wang, G., and Pichersky, E. (2007) Plant J. 49 1020-1029 [DOI] [PubMed] [Google Scholar]
- 34.Hunt, L., Holdsworth, M. J., and Gray, J. E. (2007) Plant J. 51 341-351 [DOI] [PubMed] [Google Scholar]
- 35.van der Veer, E., Ho, C., O'Neil, C., Barbosa, N., Scott, R., Cregan, S. P., and Pickering, J. G. (2007) J. Biol. Chem. 282 10841-10845 [DOI] [PubMed] [Google Scholar]
- 36.Revollo, J. R., Korner, A., Mills, K. F., Satoh, A., Wang, T., Garten, A., Dasgupta, B., Sasaki, Y., Wolberger, C., Townsend, R. R., Milbrandt, J., Kiess, W., and Imai, S. (2007) Cell Metab. 6 363-375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang, H., Yang, T., Baur, J. A., Perez, E., Matsui, T., Carmona, J. J., Lamming, D. W., Souza-Pinto, N. C., Bohr, V. A., Rosenzweig, A., de Cabo, R., Sauve, A. A., and Sinclair, D. A. (2007) Cell 130 1095-1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Busso, N., Karababa, M., Nobile, M., Rolaz, A., Van Gool, F., Galli, M., Leo, O., So, A., and De Smedt, T. (2008) PLoS ONE 3 e2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chong, Z. Z., Kang, J. Q., and Maiese, K. (2004) Exp. Cell Res. 296 196-207 [DOI] [PubMed] [Google Scholar]
- 40.Kang, J. Q., Chong, Z. Z., and Maiese, K. (2003) Mol. Pharmacol. 64 557-569 [DOI] [PubMed] [Google Scholar]
- 41.Luckinbill, L. S., Arking, R., Clare, M., Cirocco, W., and Buck, S. (1984) Evolution 38 996-1003 [DOI] [PubMed] [Google Scholar]
- 42.Du, X., Wang, W., Kim, R., Yakota, H., Nguyen, H., and Kim, S. H. (2001) Biochemistry 40 14166-14172 [DOI] [PubMed] [Google Scholar]
- 43.Brand, A. H., and Perrimon, N. (1993) Development 118 401-415 [DOI] [PubMed] [Google Scholar]
- 44.Chaykin, S. (1969) Anal. Biochem. 31 375-382 [DOI] [PubMed] [Google Scholar]
- 45.Newman, B. L., Lundblad, J. R., Chen, Y., and Smolik, S. M. (2002) Genetics 162 1675-1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Partridge, L., Gems, D., and Withers, D. J. (2005) Cell 120 461-472 [DOI] [PubMed] [Google Scholar]
- 47.Wang, J., Zhai, Q., Chen, Y., Lin, E., Gu, W., McBurney, M. W., and He, Z. (2005) J. Cell Biol. 170 349-355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parker, J. A., Arango, M., Abderrahmane, S., Lambert, E., Tourette, C., Catoire, H., and Neri, C. (2005) Nat. Genet. 37 349-350 [DOI] [PubMed] [Google Scholar]
- 49.Araki, T., Sasaki, Y., and Milbrandt, J. (2004) Science 305 1010-1013 [DOI] [PubMed] [Google Scholar]
- 50.Nisoli, E., Tonello, C., Cardile, A., Cozzi, V., Bracale, R., Tedesco, L., Falcone, S., Valerio, A., Cantoni, O., Clementi, E., Moncada, S., and Carruba, M. O. (2005) Science 310 314-317 [DOI] [PubMed] [Google Scholar]
- 51.Anekonda, T. S., and Reddy, P. H. (2006) J. Neurochem. 96 305-313 [DOI] [PubMed] [Google Scholar]
- 52.Blander, G., and Guarente, L. (2004) Annu. Rev. Biochem. 73 417-435 [DOI] [PubMed] [Google Scholar]
- 53.Guarente, L. (2005) Mech. Ageing Dev. 126 923-928 [DOI] [PubMed] [Google Scholar]
- 54.Porcu, M., and Chiarugi, A. (2005) Trends Pharmacol. Sci. 26 94-103 [DOI] [PubMed] [Google Scholar]
- 55.Sandmeier, J. J., Celic, I., Boeke, J. D., and Smith, J. S. (2002) Genetics 160 877-889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Medvedik, O., Lamming, D. W., Kim, K. D., and Sinclair, D. A. (2007) PLoS. Biol. 5 e261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barthel, A., Schmoll, D., and Unterman, T. G. (2005) Trends Endocrinol. Metab. 16 183-189 [DOI] [PubMed] [Google Scholar]
- 58.Wang, Y., and Tissenbaum, H. A. (2006) Mech. Ageing Dev. 127 48-56 [DOI] [PubMed] [Google Scholar]
- 59.Giannakou, M. E., and Partridge, L. (2004) Trends Cell Biol. 14 408-412 [DOI] [PubMed] [Google Scholar]
- 60.Daitoku, H., Hatta, M., Matsuzaki, H., Aratani, S., Ohshima, T., Miyagishi, M., Nakajima, T., and Fukamizu, A. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 10042-10047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arden, K. C. (2004) Mol. Cell 14 416-418 [DOI] [PubMed] [Google Scholar]
- 62.Brunet, A., Sweeney, L. B., Sturgill, J. F., Chua, K. F., Greer, P. L., Lin, Y., Tran, H., Ross, S. E., Mostoslavsky, R., Cohen, H. Y., Hu, L. S., Cheng, H. L., Jedrychowski, M. P., Gygi, S. P., Sinclair, D. A., Alt, F. W., and Greenberg, M. E. (2004) Science 303 2011-2015 [DOI] [PubMed] [Google Scholar]
- 63.Wang, F., Nguyen, M., Qin, F. X., and Tong, Q. (2007) Aging Cell 6 505-514 [DOI] [PubMed] [Google Scholar]
- 64.Tsuchiya, M., Dang, N., Kerr, E. O., Hu, D., Steffen, K. K., Oakes, J. A., Kennedy, B. K., and Kaeberlein, M. (2006) Aging Cell 5 505-514 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.