Abstract
The gene encoding ribonucleotide reductase 3 (RNR3) is strongly induced in response to DNA damage. Its expression is strictly dependent upon the TAFII subunits of TFIID, which are required for the recruitment of SWI/SNF and nucleosome remodeling. However, full activation of RNR3 also requires GCN5, the catalytic subunit of the SAGA histone acetyltransferase complex. Thus, RNR3 is dependent upon both TFIID and SAGA, two complexes that deliver TATA-binding protein (TBP) to promoters. Furthermore, unlike the majority of TFIID-dominated genes, RNR3 contains a consensus TATA-box, a feature of SAGA-regulated core promoters. Although a large fraction of the genome can be characterized as either TFIID- or SAGA-dominant, it is expected that many genes utilize both. The mechanism of activation and the relative contributions of SAGA and TFIID at genes regulated by both complexes have not been examined. Here we delineated the role of SAGA in the regulation of RNR3 and contrast it to that of TFIID. We find that SAGA components fulfill distinct functions in the regulation of RNR3. The core promoter of RNR3 is SAGA-dependent, and we provide evidence that SAGA, not TAFIIs within TFIID, are largely responsible for TBP recruitment. This taken together with our previous work provides evidence that SAGA recruits TBP, whereas TFIID mediates chromatin remodeling. Thus, we described an unexpected shift in the division of labor between these two complexes and provide the first characterization of a gene that requires both SAGA and TFIID.
The activation of genes requires the assembly of large macromolecular complexes at the promoter which function to alter chromatin structure and drive the assembly of the general transcription machinery (1–4). This process is generally believed to occur in stages, the first being chromatin remodeling and exposure of the promoter followed by the assembly of the preinitiation complex (PIC).2 A rate-limiting step in this process is the delivery of the TATA-binding protein (TBP) to the core promoter. TBP is recruited to the promoter of genes as part of one of a number of multisubunit complexes (4, 5), although it is not known if monomeric TBP can bind to promoters in vivo.
TFIID and SAGA (Spt-Ada-Gcn5 acetyltransferase complex) are the two major pathways that deliver TBP to promoters in yeast. Promoters can be characterized as TAFII-independent or TAFII-dependent (6). TAFII-independent promoters have a low TAFII occupancy (7, 8), and TBP is delivered by SAGA via the Spt8-Spt3 subunits (9–14). In fact, genome-wide expression data and computational studies predict a “bipolar genome.” SAGA regulates highly inducible stress responsive genes, whereas TFIID regulates constitutive housekeeping genes (15). Furthermore, SAGA-regulated genes tend to have consensus TATA boxes, and TFIID-regulated genes do not (16, 17). Even the precise positioning of the core promoter nucleosome at SAGA-versus TFIID-dependent genes differs (18). These results have led to a model where genes are regulated by TFIID or SAGA. However, many genes in yeast (and likely many more in metazoans) rely on both SAGA and TFIID, and GCN5 and TAF1 are considered redundant throughout the genome (19). Why two TBP recruiting complexes are utilized at a single gene is not clear, and the analysis of the contributions of these two redundant complexes at a single locus has not been undertaken.
SAGA, like many transcription factor complexes, performs multiple functions in gene expression. A large body of genetic, structural, and functional characterization of SAGA subunits has identified “modules” within the complex, the SPT, TAFII, and ADA/GCN5 (for review, see Refs. 20–22). The activities of SAGA can be classified into two groups. The first is histone modification, which includes acetylation and regulation of ubiquitylation, and the second is the delivery of TBP to promoters. Because the expression of some genes is more sensitive to the deletion of subunits within one module versus another, it is believed that the importance of each activity differs among genes.
We have used RNR3 as a model gene to study the mechanism of TAFII-dependent gene regulation (23–28). The activation of RNR3 requires a transient activation function in Crt1 and the abundant transcription factor Rap1 (26, 27). Crt1 is the sequence-specific DNA-binding protein that recruits Ssn6-Tup1 to the promoter to repress transcription by positioning nucleosomes, recruiting histone deacetylases and interfering with mediator (28). DNA damage signals cause the release of repression and coverts Crt1 into a transient activator. Crt1 then recruits TFIID and SWI/SNF and ultimately disassociates from the promoter (26). TFIID maintains the remodeled state and a high level of transcription by stabilizing the binding of Rap1 directly or indirectly through the retention of SWI/SNF (27). Mutation or depletion of multiple TFIID subunits impairs the action of this gene and other ribonucleotide reductase genes (24, 29, 30). RNR3 has a consensus TATA-box, and its core promoter is insensitive to TAFII mutations; the TAFII dependence of this gene is mediated by the upstream repression/activating sequences (URS/UAS) (24). Despite being TFIID-dependent, it has the features of a SAGA-dependent gene; thus, it is likely to be regulated by both complexes. However, the contributions of TFIID versus SAGA at the gene are unclear.
Here, we carried out an analysis of the role of SAGA in regulating RNR3. Strains containing mutations in different functional modules of SAGA were analyzed for RNR3 expression. We found that RNR3 is more sensitive to inactivation of the TBP recruitment module than the histone modification module, suggesting that SAGA delivers TBP to the promoter. Consistent with this model, the core promoter of RNR3 is very sensitive to the inactivation of the TBP module of SAGA. We propose a model where the primary function of SAGA is to deliver TBP to the RNR3 core promoter and that of TFIID is to mediate the chromatin remodeling step through the UAS/URS of the gene. Thus, we have uncovered unexpected roles of these two coactivator complexes at RNR3, which likely applies to other genes in yeast and metazoans.
EXPERIMENTAL PROCEDURES
Strains and Media—Saccharomyces cerevisiae strains used in this study are described in Table 1. All strains are derived from PH499 (MATa ade2-101ochre his3-200 leu2-1 ura3-52 trp1-63 lys2-801amber). Cells were cultivated at 30 °C in YPD (yeast extract 1%, peptone 2%, and dextrose 2%) medium supplemented with 0.05 mg/ml adenine. Methyl methanesulfonate (MMS) was added to a final concentration of 0.03% for 2.5 h where indicated (+MMS). Gene deletions were carried out by homologous recombination using PCR-generated cassettes (31).
TABLE 1.
Strains used in this study
| PH499 | MATa ade2-101 ochre his3-200 leu2-1 ura3-52 trp1-63 lys2-801 amber |
| YJR374 | PH499 but taf1-2::TRP1 |
| YJR997 | PH499 but spt3Δ::KanMx |
| YJR1002 | PH499 but gcn5Δ::KanMx |
| YJR1007 | PH499 but spt7Δ::KanMx |
| YJR 1100 | PH499 but spt3Δ::KanMx, Δgcn5::HIS3 |
| YJR986 | PH499 but (34 bp dA:dT at -90/-60) RNR3:LEU2 |
| YJR991 | YJR374 but (34 bp dA:dT at -90/-60) RNR3:LEU2 |
| YJR1000 | YJR997 but (34 bp dA:dT at -90/-60) RNR3:LEU2 |
| YJR1005 | YJR1002 but (34 bp dA:dT at -90/-60) RNR3:LEU2 |
| YJR1010 | YJR1007 but (34 bp dA:dT at -90/-60) RNR3:LEU2 |
Reporter Plasmids and RNR3 Derivates—The strategy and constructs containing insertions of dA::dT tracts at –90 and –60 within the core promoter of RNR3 were described in a previous publication (32). RNR3-lacZ-derived reporter plasmids were described in a previous manuscript (24). The promoter activity was measured by Northern blotting using a probe to lacZ.
Northern Blotting and Chromatin Mapping—Cells from 10 ml of yeast culture (A600 = 0.5 ∼ 1) were harvested for total RNA extraction. Fifteen micrograms of total RNA was separated on 1.2% formaldehyde-containing agarose gels and transferred to Hybond-XL membrane (GE Biosciences) by capillary blotting. Probes comprising the entire open reading frames of RNR3 or Scr1 were prepared by the polymerase chain reaction and gel-purified. The signal of scR1 (small cytoplasmic RNA) in each sample was used to correct for recovery and loading of RNA. Additional details have been published elsewhere (33). Micrococcal nuclease mapping of nucleosomes by indirect end-labeling was carried out using a published protocol (24, 34).
Chromatin Immunoprecipitation—The chromatin immunoprecipitation assay was performed essentially as described in previous publications (25, 35). 100 ml of yeast culture (A600 = 0.5–1.0) was treated or not with 0.03% MMS for 2.5 h before cross-linking with formaldehyde (1% v/v) for 15 min at room temperature. Cross-linking was quenched with 125 mm glycine. Whole cell extracts were prepared by glass bead disruption and sheared into fragments averaging 400 bp in size using a Bioruptor (Diagenode, Philadelphia PA). Whole cell extracts containing equal amounts of DNA were immunoprecipitated (IP) with antibodies against the various factors. Extracts were supplemented with sarkosyl to a final concentration of 0.4% (w/v) when TAF1 and Swi2 cross-linking was examined. RNAPII antibody was obtained from commercial sources (8WG16, Covance, Berkeley, CA). The following antibodies were raised in rabbits and are described in previous publications (25, 26): Crt1 (amino acids 1–240), TBP (full length), TAF1 (amino acids 1–225), Tup1 (full length), and Swi2 (amino acids 1–851). The protein-immune complexes were recovered using 30 μl of protein A-Sepharose CL-4B beads (GE Biosciences). After washing and reversing the cross-links, the IP DNA and input DNA were analyzed by semiquantitative PCR with primers directed toward RNR3 (32–36). The percentage IP was calculated from these values, and data are expressed relative to the cross-linking observed in untreated wild type cells. At least three independent experiments were analyzed, and data are presented as the means and S.D. of these repetitions. Chromatin immunoprecipitation data were analyzed by Student's t test. For detecting nucleosome eviction over the RNR3 promoter, cells were processed as described above, except that the chromatin was extensively sheared into fragments averaging about 200 bp in length. Chromatin was precipitated using an antibody to the core domain of H3 (Abcam, Cambridge, MA). Antibody to acetylated H3 (9, 14) was obtained from Millipore (Billerica, MA).
RESULTS
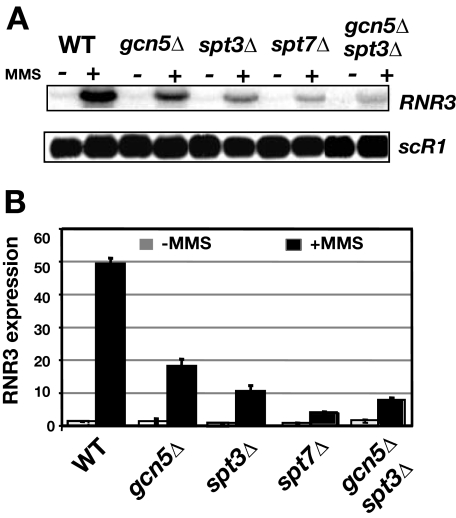
Identification of the Role of SAGA in the Activation of RNR3—SAGA is the histone acetyltransferase complex responsible for the DNA damage-induced increase in histone H3 acetylation at the RNR3 promoter (25). Previously we examined mutants of the Gcn5-Ada module, GCN5 and ADA2. However, as described above, SAGA is modular in nature, and its subunits carry out distinct functions (10–12, 37, 38) such as the covalent modification of histones and TBP recruitment. Because RNR3 has a consensus TATA-box, a feature of SAGA-regulated genes (16), we explored the role of SAGA in recruiting TBP to the promoter. We constructed strains containing deletions of GCN5, SPT3, and SPT7 and examined the induction of RNR3 after treatment with the DNA damage agent MMS. Similar to our previous results, deleting GCN5 partially reduced RNR3 activation to a level ∼40% that of the level observed in wild type cells (Fig. 1). Similar results were observed in ada2Δ and ada3Δ mutants (Ref. 25 and data not shown). Thus, RNR3 expression is partially dependent upon the histone acetylation function of SAGA. Deletion of SPT3, which is required for TBP recruitment to other SAGA-regulated genes, had a much stronger effect on activation than deleting GCN5. Deleting SPT8, another subunit of TBP recruitment module of SAGA, had a similar effect on RNR3 expression as deleting SPT3 (data not shown). Finally, deleting SPT7, which is required for SAGA integrity, had the strongest effect on activation (Fig. 1). This phenotype suggests disrupting both the histone modifying functions (GCN5) and TBP recruitment functions (SPT3) of the complex result in a stronger defect in RNR3 activation. To test this, a double gcn5Δ/spt3Δ mutant was constructed and analyzed. This double mutant had a stronger activation defect than the single mutants but still slightly less than the spt7Δ mutant. Furthermore, a comparison of the strengths of the phenotypes of the various mutants suggests that the dominant function of SAGA at RNR3 is TBP recruitment through the Spt3 module and not histone modification via the Gcn5/ADA module.
FIGURE 1.
Analysis of RNR3 expression in different classes of SAGA mutants. A, Northern blot of RNR3 levels in total RNA from untreated (–) or cells treated with MMS for 2.5 h (+). scR1, an RNA polymerase III-transcribed gene, was used as a loading control. B, quantification of RNR3 signals corrected for scR1 levels. The level of RNR3 in untreated wild type (WT) cells was set to 1.0. The values shown in gray and black bars were calculated from untreated and MMS-treated samples, respectively. Results are presented as the means and S.D. of three samples.
Spt3 and Spt7 Are Required for TBP Recruitment and PIC Formation—Activation of RNR3 requires cells to detect and transduce DNA damage signals to the RNR3 promoter. DNA damage checkpoint-dependent release of Crt1 is required for expression of the RNR genes (26, 35, 39); thus, the activation defects in the mutants could be indirect and caused by the inactivation of the DNA damage checkpoint pathway. To verify that mutating SAGA does not indirectly affect RNR3 expression by disrupting checkpoint function, we examined the cross-linking of Crt1 to the promoter. MMS treatment reduced Crt1 cross-linking in wild type cells (Fig. 2A), consistent with the activation of the checkpoint and release of the repressor. Likewise, MMS treatment reduced Crt1 cross-linking in the three SAGA mutants, suggesting that SAGA is not required for checkpoint activation and that SAGA directly regulates RNR3 at the level of transcription. The small reduction in Crt1 cross-linking in undamaged cells in the mutants is within experimental error and is not likely to be significant.
FIGURE 2.
Examination of repressor release and PIC formation in SAGA mutants. Cross-linking of Crt1 (A), RNAPII (B), TBP (C), and TAF1 (D) to RNR3. Data from untreated and MMS-treated cells are shown in gray and black bars, respectively. The results are the means and S.D. of at least three independent experiments performed on different chromatin preparations. The percentage IP (IP signal/input signal) calculated from untreated wild cells (WT) was set to a value of 1.0, and other data points are expressed relative to this value. The cross-linking of PIC components were measured over the promoter, and that of Crt1 was measured over the URS/UAS. In panels C and D, the small increase in MMS-induced cross-linking of TBP and TAF1 in the spt3Δ and spt7Δ mutants was not significant, based upon Student's t test.
A feature of a prototypical TFIID-dependent gene is that TBP recruitment to its promoter is not or is weakly, dependent upon SAGA (38, 40). Disabling the TBP recruitment module (spt3Δ) had a stronger effect on RNR3 activation than disabling the histone acetylation module (Fig. 1), suggesting that SAGA participates in the recruitment of TBP to this gene. We examined the recruitment of TBP, TAF1, and RNA polymerase II (RNAPII) to the promoter of RNR3 using the chromatin immunoprecipitation assay. The data show that the MMS-induced cross-linking of RNAPII correlated well with the levels of RNR3 mRNA in each of the mutants; deletion of GCN5 partially reduced recruitment, whereas deleting SPT3 or SPT7 essentially eliminated it (compare Fig. 1 with Fig. 2B). Next, we examined TBP recruitment to the promoter. Consistent with the idea that the Spt3 module of SAGA is required for TBP recruitment to RNR3, mutation of SPT3 or SPT7 severely compromised the recruitment of TBP (Fig. 2C). Surprisingly, TBP recruitment was not affected in the gcn5Δ mutant. This indicates that TBP recruitment does not require H3 acetylation and suggests that a SAGA complex lacking Gcn5 is capable of recruiting TBP to RNR3. Similar results were observed at GAL1 and other SAGA-dependent genes, and a stable subcomplex of SAGA is detected in a gcn5Δ mutant (10, 11, 38, 41). The most striking result is that TBP recruitment is normal, but RNAPII recruitment is reduced in this mutant. This suggests that Gcn5, presumably through histone modification, is required for stable or multiple rounds of RNAPII recruitment. A similar phenotype is observed when histone acetylation is constitutively increased by deleting the two histone deacetylases RPD3 and HOS2 (36). Thus, reducing or misregulating histone acetylation adversely affects RNAPII recruitment, suggesting that dynamic histone modifications are required for high levels of RNAPII recruitment to RNR3.
We have previously shown that activation of RNR3 correlates with increased recruitment of TAFIIs (25, 26). We analyzed the recruitment of TAF1p, an integral component of TFIID, to RNR3 in the three classes of SAGA mutants. MMS treatment caused a 3-fold recruitment of TAF1p in wild type cells, and deletion of GCN5 had minimal to no effect on recruitment (Fig. 2D). The difference in cross-linking between the wild type cells and the gcn5Δ mutant under induced conditions is slight and probably not statistically significant (Student's t test, p ≤ 0.11). Examination of the TAF1p cross-linking in the spt3Δ and spt7Δ mutants revealed no significant increase in response to MMS treatment, suggesting TAFII recruitment is defective in these mutants (Fig. 2D). Thus, the recruitment of TAF1 mirrors that of TBP in the SAGA mutants and SAGA is required for TFIID recruitment.
SAGA Is Required for Nucleosome Eviction and SWI/SNF Recruitment—We have previously used micrococcal nuclease mapping to identify the requirement for GCN5 in the disruption of nucleosome positioning at RNR3 (25). Deleting GCN5 reduced the extent of nucleosome remodeling under the activated condition but did not eliminate it. Furthermore, deleting GCN5 actually increased the recruitment of SWI/SNF to the promoter. Here, we extended the analysis to the other SAGA mutants and also monitored the eviction of the core promoter nucleosome using antibodies to the core domain of H3. As reported previously (27, 32), MMS treatment caused a significant reduction in the occupancy of the nucleosome over the core promoter (Fig. 3A). We interpret this as eviction of the nucleosome caused by remodeling. The recruitment of Swi2, the catalytic subunit of the SWI/SNF complex, is increased under this condition, indicating a link between SWI/SNF recruitment and nucleosome eviction (Fig. 3B). Deleting SPT3 or SPT7 eliminated the eviction of the core promoter nucleosome and eliminated the MMS-induced recruitment of SWI/SNF (Fig. 3, A and B). Thus, Spt3 and Spt7 are required for SWI/SNF recruitment and nucleosome eviction. As noted before (25), deleting GCN5 actually increased SWI/SNF recruitment both in the treated (p ≤ 0.03) and untreated (p ≤ 0.04) cells (Fig. 3B). The increase in recruitment in untreated cells is surprising but also correlated with a reduction in H3 cross-linking under the same conditions (Fig. 3A). Next, we analyzed H3 acetylation of the core promoter nucleosome in the SAGA mutants. Deletion of GCN5 or SPT7 strongly reduced the MMS-induced increase in H3 acetylation (Fig. 3C). Furthermore, the level of acetylation was reduced in these mutants in untreated cells too. This suggests that a basal level of acetylation is present before DNA damage. In contrast, deleting SPT3 had a lesser effect on H3 acetylation, suggesting that the histone acetylation function of SAGA is partially intact in this mutant. These results mirror those described at the GAL1 locus, where deleting SPT3 had a relatively minor effect on histone acetylation (9–11).
FIGURE 3.
Examination of the status of chromatin and chromatin remodeling factors at RNR3. Shown is cross-linking of the core domain of H3 (A), Swi2 (B), acetylated H3 (C), and Tup1 (D) to RNR3. Data from untreated and MMS-treated cells are shown in gray and black bars, respectively. The results are the means and S.D. of at least three independent experiments performed on different chromatin preparations. The percentage IP (IP signal/input signal) calculated from untreated wild (WT) cells was set to a value of 1.0, and other data points are expressed relative to this value. The cross-linking of the proteins shown in panels A–C were measured over the promoter, and that of Tup1 (D) was measured over the URS/UAS. In panel B only the wild type and gcn5Δ cells displayed a significant MMS-induced increase in Swi2 cross-linking (p ≤ 0.01).
Finally, we examined the cross-linking of Tup1 to RNR3. The activation of RNR3 correlates with the release of Tup1, and release of the corepressor is required for activation (28, 35). MMS treatment caused the reduced cross-linking of Tup1 to the promoter (Fig. 3D) in wild type cells, and deletion of SPT3 or SPT7 did not significantly affect Tup1 cross-linking in the repressed state or its release upon MMS treatment. This further reinforces the idea that the DNA damage response and the release of the repressor are not affected by deletion of SAGA subunits. Some differences are observed in the gcn5Δ mutant. There is a slightly reduced amount of Tup1 at the promoter in the repressed state. The reduced Tup1 in the repressed state may be related to the reduced nucleosome density in this mutant (Fig. 3A) and enhanced SWI/SNF recruitment (Fig. 3B), but the cause and effect relationship is unclear at this time.
Nucleosomes adopt precise translational positions at RNR3 in the repressed state (23). Positioning requires the cooperative actions of the ISW2 complex and Ssn6/Tup1 (35). The disruption of chromatin structure can occur by the de-positioning of nucleosomes or by nucleosome eviction. Deleting ISW2 disrupts the precise translational positions of nucleosomes but does not lead to nucleosome removal that occurs during activation of the gene (35 and 42).3 Thus, we analyzed the chromatin structure over RNR3 in the spt3Δ and spt7Δ mutants in detail by micrococcal nuclease mapping to determine whether nucleosome positioning is lost in the absence of nucleosome eviction. In unstimulated wild type cells, RNR3 is packaged into a precise array of nucleosomes (Fig. 4). In particular, the TATA box region is protected from digestion by micrococcal nuclease within nuc-1, which appears as a doublet bands in naked DNA and in the remodeled state (black dot). Furthermore, distinct hypersensitive sites were observed, indicating that nucleosomes adopt precise translational positions (arrows). MMS treatment of wild type cells caused a pattern essentially identical to digested naked DNA. In particular, the appearance of the doublet bands over the TATA region indicates it is accessible to nuclease, and the intensity of the internucleosomal hypersensitive bands is reduced. Interestingly, nucleosome positioning is lost in the spt3Δ and spt7Δ mutants when the cells were treated with MMS. Perhaps some very subtle differences in the magnitude in nucleosome disruption can be detected, but nucleosome positioning is lost nonetheless. The H3 cross-linking data indicates that the density of nucleosomes is not reduced in mutants (Fig. 3A), which argues that positioning of the core promoter nucleosome is lost even though nucleosomes are not evicted. The de-positioning of nucleosomes is caused by the release of Ssn6/Tup1 (Fig. 3D), and the lack of eviction is due to the reduced SWI/SNF recruitment (Fig. 3B). Therefore, our data provide another line of evidence that nucleosome positioning can be disrupted without histone eviction. A similar observation was made at the CHA1 locus where mutation of mediator caused de-positioning of nucleosomes without eviction under the induced condition (43).
FIGURE 4.
Micrococcal nuclease mapping of nucleosome positions at RNR3. Nuclei were isolated from wild type (WT) and Δspt3 and Δspt7 mutants (either treated or not with MMS), digested with micrococcal nuclease, and subjected to indirect end labeling analysis to monitor the chromatin structure around the RNR3 promoter. Markers (M1 and M2) prepared from digested genomic DNA were loaded in adjacent lanes. The lanes marked ND contain naked DNA digested with micrococcal nuclease. The black circle marks the doublet generated by the digestion of the TATA box region by micrococcal nuclease. Arrows indicate the positions of the bands marking the locations of hypersensitive sites that are reduced or modified when the gene is active.
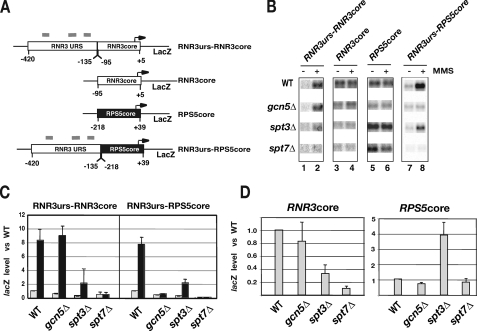
The Core Promoter of RNR3 Is Sensitive to the Inactivation of the TBP Module of SAGA—Transcription from the core promoter of RNR3 is unaffected in temperature-sensitive mutants of TAFIIs (24); therefore, it is classified as a TAFII-independent core promoter. RNR3 has a consensus TATA box, a feature of many SAGA-dependent core promoters. We speculated that the core promoter of RNR3 is SAGA-dependent. If SAGA delivers TBP to RNR3, a prediction is that the core promoter of RNR3 would be sensitive to SAGA mutations that inactivate the TBP module. To test this we analyzed the activity of reporter genes containing fusions of the RNR3 and RPS5 promoters (Fig. 5A). RPS5 is TAFII-dependent and is relatively insensitive to SAGA mutations (38, 40). First, we analyzed the expression of the RNR3 promoter containing the RNR3 upstream sequences fused to its core promoter (RNR3urs-RNR3core). Similar to the native gene, the plasmid version was very sensitive to spt3Δ and spt7Δ mutations (compare Fig. 5, B and C, with Fig. 1B). Unexpectedly, the plasmid copy was not sensitive to the gcn5Δ mutation. It is not uncommon that a plasmid copy of a gene displays different responses to mutations in chromatin modifiers and transcription factors (44, 45). In some cases this is caused by imprecise nucleosome positioning over the promoter (45). Furthermore, the plasmid copy contains the URS and core promoter regions only and is missing 40 base pairs compared with the natural gene (Fig. 5A). The change in URS promoter architecture may influence the sensitivity to the gcn5Δ mutation.
FIGURE 5.
Mapping the SAGA-dependent region of RNR3. A, lacZ-containing reporter vectors were constructed by fusing the upstream and core promoter regions of RNR3 and RPS5 together as indicated in the panel. The sequences within each of the genes are indicated below and are relative to the start site of transcription, which was set to +1. Gray bars indicate the approximate locations of the damage response elements. B, transcription was measured by Northern blotting for lacZ message as described in Li and Reese (24), and a representative Northern blot is shown. WT, wild type. C, quantification of lacZ signals corrected for scR1 levels. The level of lacZ in untreated wild type cells was set to 1.0. Results are presented as the means and S.D. of three experiments. The values shown in gray and black bars were calculated from untreated and MMS-treated samples, respectively. D, as in C, except that the data from untreated samples are shown.
More importantly, the core promoter of RNR3 (RNR3core) was very sensitive to the spt3Δ and spt7Δ mutations (Fig. 5, B and D). Given that the RNR3 core promoter is insensitive to TAFII mutations but sensitive to mutations in SAGA that disrupt TBP recruitment functions, the results support our model that SAGA is important for delivering TBP to the core promoter of RNR3. In contrast, deleting GCN5 had little to no effect on the activity of the core promoter. The minimal to no effect of the gcn5Δ mutant on the activity of the RNR3 core promoter is consistent with our data indicating that the TBP recruitment module of SAGA is functional in the absence of Gcn5 (Fig. 2C). To rule out that the lower levels of lacZ RNA are not caused by plasmid copy number differences in the mutants, we isolated DNA from the strains and detected the levels of plasmid by Southern blotting. No significant difference in copy number in the wild type cells and the mutants was observed (not shown).
The data thus far suggest that the TBP recruitment function of SAGA is dominant at RNR3, and this function is mediated through the Spt3 module. Although it is clear that SPT3 and SPT7 are required for the activity of the RNR3 core promoter (Fig. 5D), our analysis so far cannot distinguish if these two subunits also function through the URS as well. This contribution would be overshadowed by the dominant effect of the SAGA mutations on the core promoter. In fact, it is expected that the upstream regulatory regions of genes are required for SAGA function because the gene regulatory protein that recruits SAGA to allow it to deliver TBP to the core promoter binds there. We, therefore, constructed a promoter fusion between a SAGA-independent core promoter and the URS of RNR3. First, we verified that the core promoter of RPS5 (RPS5core) is insensitive to SAGA mutations. Deleting GCN5 or SPT7 had very little effect on the activity of the RPS5 core promoter; however, deleting SPT3 enhanced core promoter activity 3–4-fold (Fig. 5D). It was reported previously that inactivating the TBP binding module of SAGA (spt3Δ or spt8Δ) enhanced the uninduced level of transcription from TRP1, the +1 promoter of HIS3 (Tc), and HO (46, 47). A possible explanation for this result is that deleting SPT3 causes a shift toward the utilization of TFIID to deliver TBP to promoters, favoring expression of TAFII-dependent core promoters. Consistent with this hypothesis, RPS5, +1 of HIS3, HO, and TRP1 are sensitive to TAFII mutations, indicating that they normally utilize TFIID to recruit TBP (7, 48–50). The core promoter of RPS5 has significant transcription activity on its own, but fusing the upstream region of RNR3 to it (RNR3urs-RPS5core) led to repression in the absence of MMS treatment, as expected (compare lanes 5 and 7 of Fig. 5B). The level of expression could be restored in treated wild type cells and was even a bit higher (compare lanes 7 and 8 in wild type cells in Fig. 5B). The enhanced activity versus that of the core promoter alone may be driven by Rap1, which we have shown to activate RNR3 through the URS (27). Deleting GCN5 or SPT7 strongly reduced the activation of the construct, suggesting that histone acetylation is obligatory for expression of the construct. Deleting SPT3 clearly affected the expression of RNR3uas-RPS5core but not as much as the other mutants. Although interpretation of the results in the gcn5Δ is complicated because the RNR3urs-RNR3core does not respond to the mutation similar to the endogenous gene, this is not the case for the Δspt7 and Δspt3 mutations. A conservative interpretation of our data is that SAGA acts through the URS and core promoter of RNR3. However, the TBP recruitment data (Fig. 2C) and a comparison of the responses of the four constructs to the gcn5Δ mutation suggest that GCN5 preferentially acts through the URS of RNR3. Specifically, the core promoter of RNR3 is relatively insensitive to GCN5 mutations, and the URS of RNR3 confers GCN5-dependent transcription to the otherwise SAGA independent RPS5 core promoter.
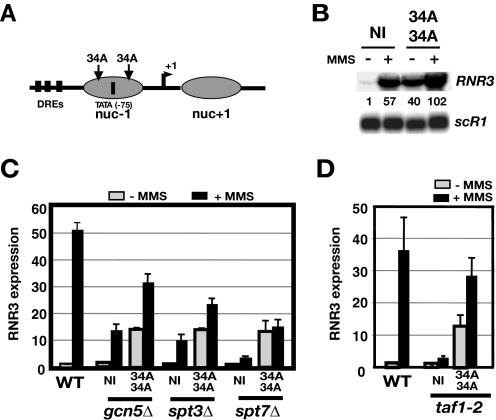
Disrupting the Core Promoter Nucleosome at RNR3 Reveals Differential Requirements for TFIID and SAGA—Disrupting the nucleosome (nuc-1) over the core promoter can alleviate the SWI/SNF and TAFII dependence of RNR3 (32). Insertion of polynucleotide tracts (PNT) upstream and downstream of the TATA box (34A/34A, Fig. 6A) suppressed taf1-2 and snf2Δ mutations, suggesting that the role of TAFIIs is to mediate chromatin remodeling through the SWI/SNF complex. One interpretation of the suppression of the taf1-2 mutant phenotype is that another activity recruited TBP to the promoter. Our data thus far indicate SAGA is responsible for this function. To test this genetically, we used the same strategy to disrupt nuc-1 and analyzed the expression of the altered promoter in SAGA mutants and a temperature-sensitive mutant of TAF1, taf1-2, as a control (Fig. 6, C and D). Our previous work showed that PNTs reduced nucleosome occupancy over the core promoter and caused a high level of constitutive expression in untreated cells (32). An example is shown in Fig. 6B. Insertion of the PNTs enhanced expression in both resting and stimulated cells. RNR3 was induced ∼37-fold in wild type cells but only about 3-fold in the taf1-2 mutant when no PNTs were inserted (Fig. 6D). However, inserting 34 bp of poly dA::dT tracts within the core promoter allowed the construct to be induced ∼28-fold in the mutant. Thus, disrupting the core promoter nucleosome restored RNR3 expression in the taf1-2 mutant to a level of approximately ∼80% that of unaltered promoter in wild type cells. In contrast, excluding nucleosome –1 by inserting PNTs within the promoter was not very effective at suppressing the activation defect of the spt7Δ mutant. In this case the level of expression was 24% that of wild type cells. Moreover, no MMS-induced increase was noted. Insertion of PNTs was not as effective at suppressing the spt3Δ mutation as the taf1-2 mutation; 43% of wild type levels versus 80% suppression in the taf1-2 mutant. Finally, disrupting the core promoter nucleosome was more effective at suppressing the gcn5Δ mutation; ∼60% of wild type levels. It should be noted, however, that all of the mutations (TFIID and SAGA) reduce the expression of the modified promoter (34A/34A). This is likely due to the fact that inserting the PNTs reduces nucleosome occupancy over the promoter but does not completely exclude a nucleosome from forming over the entire promoter and open reading frame in all of the cells in the population (32). The relatively poor suppression of the spt3Δ and spt7Δ mutants compared with the TFIID mutant suggests that SAGA carries out distinct functions from TFIID in the activation of RNR3. Based upon the data presented above, we propose it is the delivery of TBP to the core promoter.
FIGURE 6.
Disrupting nuc-1 of RNR3 poorly compensates for SAGA mutations. A, a schematic of RNR3 and the approximate location of the –1 and +1 nucleosome based upon mapping described in Li and Reese (23). Thirty-four-base pair dA::dT tracts were inserted upstream (–90) and downstream (–60) of the TATA box, which is located at –75 relative to the start site of transcription. DREs are the three damage response elements located with the UAS/URS sequence of RNR3. B, analysis of the effects of inserting polynucleotide tracts in the RNR3 promoter. The signals were normalized to uninduced wild type cells containing the unmodified promoter after correction for intensity of the loading control, scR1. NI indicates a promoter without the inserts, and 34A/34A indicates a promoter with 34 base pairs of dA::dT inserted at –90 and –60. WT, wild type. C, quantification of RNR3 signals after normalizing to scR1 levels. The values shown in gray and black bars were calculated from untreated and MMS-treated samples, respectively. The signal in untreated wild type cells containing the unmodified promoter (NI, no insert) was arbitrarily set to 1.0. Results are presented as the means and S.D. of three experiments. D, same as in C, except that the induction of RNR3 was carried out at 37 °C.
DISCUSSION
DNA-binding proteins orchestrate the expression of genes by recruiting numerous coactivators. In some cases different activators recruit a specific complex, and in others a single activator recruits several (1–2). Gal4 and Gcn4 have been studied the most extensively and provide the best examples of how an activator can recruit multiple cofactors (1, 12, 51, 52). A fundamental question in gene regulation is why activators recruit multiple coactivators that carry out redundant functions. For example, both TFIID and SAGA recruit TBP to promoters. Although a large fraction of genome can be segregated into TFIID- and SAGA-dominated, many genes have features of both classes, including RNR3 described here. We have firmly established that RNR3 is dependent upon both SAGA and TFIID, and our analysis provides some insights into how these two coactivator complexes cooperate in the regulation of genes.
The results described above suggest that SAGA performs two functions at RNR3, histone modification and TBP recruitment. Each function is dependent on distinct subunits of the SAGA complex, and in this regard, we have verified the modular nature of the complex. The severity of the individual mutations indicates that the TBP module plays a more dominant role in the regulation of RNR3 than the histone modification module. This resolves some unanswered questions about the regulation of RNR3. RNR3 is an atypical TAFII-dependent gene. Its sensitivity to TAFII mutations is mediated by the upstream activating/repressing sequences, UAS/URS (24). In fact, the core promoter of RNR3 is completely insensitive to the inactivation of TFIID-specific TAFIIs and contains a consensus TATA-box. These observations suggested that TFIID was not the core promoter selectivity factor for RNR3, and another complex delivered TBP to the promoter. We provide very strong evidence that SAGA is the factor. Inactivation of the TBP binding module specifically by deleting SPT3 or disrupting SAGA integrity by deleting SPT7 strongly impaired the activation of the RNR3. Significantly, we show that the core promoter of RNR3, when fused to lacZ, is very sensitive to mutations that disrupt the TBP recruitment function of SAGA. Thus, all of the data presented above strongly suggest that SAGA is the promoter selectivity factor that delivers TBP to RNR3. This is fully consistent with the core promoter structure of RNR3, which resembles a prototypical SAGA-dependent gene. This opens up the possibility that other TFIID-dependent genes with consensus TATA box utilize SAGA for TBP recruitment as well.
TBP recruitment is virtually unaffected in a gcn5Δ mutant, and the activity of the RNR3 core promoter is insensitive to the gcn5Δ mutation. In addition, the URS of RNR3 can confer SAGA-dependent transcription to the otherwise SAGA-independent RPS5 core promoter (Fig. 5C). As noted above, the unexpected change in the sensitivity of the RNR3 URS when fused out of context with the RNR3 core promoter muddies the interpretation a bit. However, we believe that the data presented as a whole support the idea that Gcn5 is dispensable for core promoter selectivity and that the Gcn5-ADA module works through the URS to facilitate chromatin remodeling by the SWI/SNF complex by acetylating nucleosomes (25). This hypothesis is also supported by previous studies showing that the URS region of RNR3 establishes the repressive chromatin structure by recruiting Ssn6-Tup1 and is also required for remodeling by recruiting SWI/SNF during activation (27, 35), and because Gcn5 specifically acts on chromatin and is dispensable for TBP recruitment, it is feasible that Gcn5 acts through this element.
Although dispensable for TBP recruitment, GCN5 was required for optimal RNAPII recruitment (Fig. 2B). This suggests that acetylation affects a step subsequent to TBP binding and preferentially affects RNAPII recruitment into the PIC or is required for multiple rounds of transcription. Our study provides another example where SAGA-dependent histone acetylation is implicated in elongation. SAGA associates with the coding regions of Gcn4-regulated genes, and GCN5 is required for regulating dynamic histone methylation and nucleosome exchange during elongation of RNAPII (53). Moreover, severe hypoacetylation of coding regions by deleting both GCN5 and ELP3 reduces RNAPII recruitment with minimal reductions in TBP recruitment (54). We have previously observed that constitutively acetylating nucleosomes in the promoter by deleting combinations of histone deacetylases preferentially blocks RNAPII recruitment with a minimal effect on TBP cross-linking (36). Therefore, misregulation of normal acetylation dynamics, either reduced or constitutive acetylation, similarly reduces RNAPII recruitment. The mechanism may involve changes in histone exchange or the ability of chromatin remodeling enzymes to recognize or remodel nucleosomes during the initial steps in elongation (55).
Interestingly, SAGA and TFIID function independently of each other at RNR3, or at least their functions can be separated genetically. It is also likely that SAGA functions “upstream” of TFIID in the activation pathway. The strongest evidence to support this conclusion is the previous observation that inactivation of a temperature-sensitive mutant of TAF1 blocks PIC formation and nucleosome remodeling, but Gcn5-dependent histone H3 acetylation is unaffected (25). Thus, SAGA recruitment and nucleosome modification function can occur in the absence of TFIID. SAGA recruitment was inferred in this experiment by an increase in H3 acetylation in the TAF1 mutant, but we did not examine SAGA recruitment directly. We have not consistently cross-linked SAGA to RNR3. A meager 1.5–1.8-fold enhancement of cross-linking of Ada3, Gcn5, and Spt7 has been observed intermittently in our laboratory (not shown). We have speculated in a previous publication that the failure to achieve a high level of SAGA cross-linking is because it associates with RNR3 by a global, untargeted mechanism (25). However, knowing that SAGA is required to recruit TBP to the core promoter, an untargeted mechanism seems unlikely. The factor that recruits SAGA to RNR3 is not known. A second line of evidence suggesting that SAGA functions upstream of TFIID is the result that disrupting SAGA integrity blocks TAF1 recruitment (Fig. 2).
Regulation of genes by multiple activators and co-activators allow them to be subject to regulation by different stimuli and developmental programs. The ribonucleotide reductase genes must maintain dNTP pools in normal cells and be strongly induced during DNA damage (56). Thus, it might be expected that they contain features of both TFIID- and SAGA-controlled genes, which regulate housekeeping and stress functions, respectively. It is unlikely that this mode of regulation is unique to the RNR genes because many stress-induced genes perform essential housekeeping functions under non-stress conditions.
Acknowledgments
We thank members of the Reese laboratory and the Center for Gene Regulation at Penn State for advice and comments on this work. Drs. Fred Winston and Shelly Berger are acknowledged for providing strains used in the early stages of this work.
This work was supported, in whole or in part, by National Institutes of Health Grant GM58672 (to J. C. R.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PIC, preinitiation complex; MMS, methyl methanesulfonate; PNT, polynucleotide tracts, RNAPII, RNA polymerase II; RNR, ribonucleotide reductase; SAGA, Spt-Ada-Gcn5 acetyltransferase complex; TBP, TATA-binding protein; TAFII, TBP-associated factors; URS, upstream repression sequence; UAS, upstream activating sequence; IP, immunoprecipitated.
H. Zhang and J. C. Reese, unpublished information.
References
- 1.Cosma, M. P. (2002) Mol. Cell 10 227–236 [DOI] [PubMed] [Google Scholar]
- 2.Lemon, B., and Tjian, R. (2000) Genes Dev. 14 2551–2569 [DOI] [PubMed] [Google Scholar]
- 3.Li, B., Carey, M., and Workman, J. L. (2007) Cell 128 707–719 [DOI] [PubMed] [Google Scholar]
- 4.Thomas, M. C., and Chiang, C. M. (2006) Crit. Rev. Biochem. Mol. Biol. 41 105–178 [DOI] [PubMed] [Google Scholar]
- 5.Pugh, B. F. (2000) Gene (Amst.) 255 1–14 [DOI] [PubMed] [Google Scholar]
- 6.Green, M. R. (2000) Trends Biochem. Sci. 25 59–63 [DOI] [PubMed] [Google Scholar]
- 7.Kuras, L., Kosa, P., Mencia, M., and Struhl, K. (2000) Science 288 1244–1248 [DOI] [PubMed] [Google Scholar]
- 8.Li, X. Y., Bhaumik, S. R., and Green, M. R. (2000) Science 288 1242–1244 [DOI] [PubMed] [Google Scholar]
- 9.Dudley, A. M., and Winston, F. (1999) Genes Dev. 13 2940–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhaumik, S. R., and Green, M. R. (2001) Genes Dev. 15 1935–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larschan, E., and Winston, F. (2001) Genes Dev. 15 1946–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qiu, H., Hu, C., Yoon, S., Natarajan, K., Swanson, M., and Hinnebusch, A. G. (2004) Mol. Cell. Biol. 24 4104–4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warfield, L., Ranish, J. A., and Hahn, S. (2004) Genes Dev. 18 1022–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sermwittayawong, D., and Tan, S. (2006) EMBO J. 25 3791–3800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huisinga, K. L., and Pugh, B. F. (2004) Mol. Cell 13 573–585 [DOI] [PubMed] [Google Scholar]
- 16.Basehoar, A. D., Zanton, S. J., and Pugh, B. F. (2004) Cell 116 699–709 [DOI] [PubMed] [Google Scholar]
- 17.Mencia, M., Moqtaderi, Z., Geisberg, J. V., Kuras, L., and Struhl, K. (2002) Mol. Cell 9 823–833 [DOI] [PubMed] [Google Scholar]
- 18.Ioshikhes, I. P., Albert, I., Zanton, S. J., and Pugh, B. F. (2006) Nat. Genet. 38 1210–1215 [DOI] [PubMed] [Google Scholar]
- 19.Lee, T. I., Causton, H. C., Holstege, F. C., Shen, W. C., Hannett, N., Jennings, E. G., Winston, F., Green, M. R., and Young, R. A. (2000) Nature 405 701–704 [DOI] [PubMed] [Google Scholar]
- 20.Baker, S. P., and Grant, P. A. (2007) Oncogene 26 5329–5340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Timmers, H. T., and Tora, L. (2005) Trends Biochem. Sci. 30 7–10 [DOI] [PubMed] [Google Scholar]
- 22.Carrozza, M. J., Utley, R. T., Workman, J. L., and Cote, J. (2003) Trends Genet. 19 321–329 [DOI] [PubMed] [Google Scholar]
- 23.Li, B., and Reese, J. C. (2001) J. Biol. Chem. 276 33788–33797 [DOI] [PubMed] [Google Scholar]
- 24.Li, B., and Reese, J. C. (2000) EMBO J. 19 4091–4100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharma, V. M., Li, B., and Reese, J. C. (2003) Genes Dev. 17 502–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang, Z., and Reese, J. C. (2005) Mol. Cell. Biol. 25 7399–7411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomar, R. S., Zhang, S., Brunkre-Reese, D. L., Wolcott, H. N., and Reese, J. C. (2008) EMBO J. 27 1575–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang, Z., and Reese, J. C. (2004) J. Biol. Chem. 279 39240–39250 [DOI] [PubMed] [Google Scholar]
- 29.Reese, J. C., Zhang, Z., and Kurpad, H. (2000) J. Biol. Chem. 275 17391–17398 [DOI] [PubMed] [Google Scholar]
- 30.Durso, R. J., Fisher, A. K., Albright-Frey, T. J., and Reese, J. C. (2001) Mol. Cell. Biol. 21 7331–7344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brachmann, C. B., Davies, A., Cost, G. J., Caputo, E., Li J., Hieter, P., and Boeke, J. D. (1998) Yeast 14 115–132 [DOI] [PubMed] [Google Scholar]
- 32.Zhang, H., and Reese, J. C. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 8833–8838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reese, J. C., and Green, M. R. (2003) Methods Enzymol. 370 415–430 [DOI] [PubMed] [Google Scholar]
- 34.Zhang, Z., and Reese, J. C. (2006) Methods Mol. Biol. 313 245–255 [DOI] [PubMed] [Google Scholar]
- 35.Zhang, Z., and Reese, J. C. (2004) EMBO J. 23 2246–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma, V. M., Tomar, R. S., Dempsey, A. E., and Reese, J. C. (2007) Mol. Cell. Biol. 27 3199–3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barbaric, S. R., and Horz, W. (2003) Mol. Cell. Biol. 23 3468–3476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhaumik, S. R., and Green, M. R. (2002) Mol. Cell. Biol. 22 7365–7371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang, M., Zhou, Z., and Elledge, S. J. (1998) Cell 94 595–605 [DOI] [PubMed] [Google Scholar]
- 40.Li, X.Y., Bhaumik, S. R., Zhu, X., Li, L., Shen, W. C., Dixit, B. L., and Green, M. R. (2002) Curr. Biol. 12 1240–1244 [DOI] [PubMed] [Google Scholar]
- 41.Qiu, H., Hu, C., Zhang, F., Hwang, G., Swanson, M., Boonchird, C., and Hinnebusch, A. G. (2005) Mol. Cell. Biol. 25 3461–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whitehouse, I., Rando, O. J., Delrow, J., and Tsukiyama, T. (2007) Nature 450 1031–1035 [DOI] [PubMed] [Google Scholar]
- 43.He, Q., Battistella, L., and Morse, R. H. (2008) J. Biol. Chem. 283 5276–5286 [DOI] [PubMed] [Google Scholar]
- 44.Tabtiang, R. K., and Herskowitz, I. (1998) Mol. Cell. Biol. 18 4707–4718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martinez-Campa, C., Politis, P., Moreau, J. L., Kent, N., Goodall, J., Mellor, J., and Goding, C. R. (2004) Mol. Cell 15 69–81 [DOI] [PubMed] [Google Scholar]
- 46.Belotserkovskaya, R., Sterner, D. E., Deng, M., Sayre, M. H., Lieberman, P. M., and Berger, S. L. (2000) Mol. Cell. Biol. 20 634–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu, Y., Eriksson, P., Bhoite, L. T., and Stillman, D. J. (2003) Mol. Cell. Biol. 23 1910–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moqtaderi, Z., Bai, Y., Poon, D., Weil, P. A., and Struhl, K. (1996) Nature 383 188–191 [DOI] [PubMed] [Google Scholar]
- 49.Shen, W. C., and Green, M. R. (1997) Cell 90 615–624 [DOI] [PubMed] [Google Scholar]
- 50.Tsukihashi, Y., Kawaichi, M., and Kokubo, T. (2001) J. Biol. Chem. 276 25715–25726 [DOI] [PubMed] [Google Scholar]
- 51.Swanson, M. J., Qiu, H., Sumibcay, L., Krueger, A., Kim, S. J., Natarajan, K., Yoon, S., and Hinnebusch, A. G. (2003) Mol. Cell. Biol. 23 2800–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bryant, G.O., and Ptashne, M. (2003) Mol. Cell 11 1301–1309 [DOI] [PubMed] [Google Scholar]
- 53.Govind, C. K., Qiu, H., Govind, S., and Hinnebusch, A. G. (2005) Mol. Cell. Biol. 25 5626–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kristjuhan, A., Walker, J., Suka, N., Grunstein, M., Roberts, D., Cairns, B. R., and Svejstrup, J. Q. (2002) Mol. Cell 10 925–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carey, M., Li, B., and Workman, J. L. (2006) Mol. Cell 24 481–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Elledge, S. J., Zhou, Z., and Allen, J. B. (1992) Trends Biochem. Sci. 17 119–123 [DOI] [PubMed] [Google Scholar]