Abstract
Myotrophin, a 12-kDa ankyrin repeat protein, stimulates protein synthesis and cardiomyocyte growth to initiate cardiac hypertrophy by activating the NF-κB signaling cascade. We found that, after internalization into myocytes, myotrophin cotranslocates into the nucleus with p65 to stimulate myocyte growth. We used structure-based mutations on the hairpin loops of myotrophin to determine the effect of the loops on myotrophin and p65 localization, induction of protein synthesis, and cardiac hypertrophy. Loop mutants, most prominently glutamic acid 33→alanine (E33A), stimulated protein synthesis much less than wild type. Myotrophin-E33A internalized into myocytes but did not translocate into the nucleus and failed to promote nuclear translocation of p65. In addition, two cardiac hypertrophy marker genes, atrial natriuretic factor and β-myosin heavy chain, were not up-regulated in E33A-treated cells. Myotrophin-induced myocyte growth and initiation of hypertrophy thus require nuclear co-translocation of myotrophin and p65, in a manner that depends crucially on the myotrophin hairpin loops.
Myotrophin, a 12-kDa protein, is the smallest known ankyrin (ANK)2 repeat protein (1) and contains four ANK repeats (2, 3). Myotrophin, also known as V-1 protein, was identified in spontaneously hypertensive rat hearts, cardiomyopathic human hearts (4), and neonatal rat cerebella (5). Myotrophin expression is ubiquitous in mammalian tissues (6). It is known to participate in the catecholamine synthesis signaling pathway (7), in regulation of actin polymerization (8), and in regulation of capping and uncapping by actin capping protein at the barbed ends of actin filaments (9). Myotrophin plays an important role in stimulation of cardiac myocyte growth and cardiac hypertrophy (4, 10). Sil et al. (11) reported an increase in myotrophin levels in human cardiomyopathic heart compared with normal heart. Overexpression of myotrophin in transgenic mice initiates cardiac hypertrophy that progresses toward heart failure (12, 13). Significantly, an increase in myotrophin level in blood plasma is observed after acute myocardial infarction (14) and in patients with heart failure (15). These data have established myotrophin as an initiator of cardiac hypertrophy and eventual heart failure.
Myotrophin has been shown to interact with NF-κB proteins (6, 16), and myotrophin-induced myocyte growth appears to be mediated through activation of the NF-κB pathway (10), although the details of these interactions are poorly characterized. Knuefermann et al. (16) showed that myotrophin physically associates with p65 and prevents the reassociation of the p65-p50 heterodimer, resulting in activation of the NF-κB pathway. Separate studies have established the involvement of NF-κB signaling in cardiac hypertrophy and dilated cardiomyopathy (17-19) and the importance of NF-κB activation in the pathogenesis of cardiac remodeling and heart failure (20-22). Recently, Gupta et al. (13) showed that progression of cardiac hypertrophy to heart failure is associated with elevated levels of NF-κB activity in transgenic mice overexpressing myotrophin. However, the molecular mechanism of NF-κB activation by myotrophin has not been determined.
To dissect the structural basis of myotrophin function, we previously determined the solution structure of myotrophin (2, 23), which consists of four ANK repeats. ANK repeat motifs are often involved in protein-protein and protein-nucleic acid interactions (24, 25). We reported that two full repeats (ANK1 and ANK2) of myotrophin contain hairpin-like loops protruding out from the core structural framework and are comparable with the crystal structure of another ANK-repeat protein, 53BP2, that complexes with p53 (26). We hypothesized that the hairpin loops act as protein recognition sites in myotrophin-mediated signaling (2). Residues in these loops are highly variable among ANK repeat proteins. Specifically, the loops of myotrophin differ from those of 53BP2 and likely allow myotrophin and 53BP2 to recognize different proteins. Recently, it has also been reported that myotrophin binds directly to the actin-capping protein through these two loops and inhibits the binding of actin-capping protein to the barbed ends of actin filaments, thus influencing actin assembly and cell motility (9). We hypothesize that residues of these two hairpin loops are involved in the role of myotrophin in the stimulation of myocyte growth.
To characterize the mechanism of NF-κB activation by myotrophin, we have investigated the cellular localization of myotrophin during myotrophin-driven NF-κB activation to determine its status at the time of the hypertrophic phenomenon. We have also identified key determinants in the hairpin loops of myotrophin that regulate the stimulatory activity of myotrophin. These observations provide a better understanding of the intrinsic mechanism of myotrophin-induced myocyte growth.
EXPERIMENTAL PROCEDURES
Materials—Timed pregnant American Wistar Kyoto rats were obtained from Harlan, Inc. The rats were fed TEKLAD regular rat chow, given water ad libitum, and housed in Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) certified facilities. The experimental procedures for animals were performed following institutional guidelines. Media and other reagents for tissue culture were purchased either from Invitrogen or from Sigma-Aldrich. Collagenase type II was purchased from Worthington Biochemicals. Factor Xa, Escherichia coli TB1 host, and purified maltose-binding protein (MBP) were obtained from New England Biolabs, Inc. Reagents for cloning purposes were purchased from Roche Applied Science. PDTC and protease inhibitor mixture were purchased from CalBiochem. Mouse monoclonal anti-p65 antibody was from Santa Cruz Biotechnology, Inc., and Alexa® 568 conjugated goat anti-mouse IgG was from Molecular Probes, Inc. Vectashield with DAPI was purchased from Vector Laboratories. All other chemicals were of molecular biology grade and were purchased from Sigma-Aldrich.
Expression and Purification of Recombinant Myotrophin Protein—The Rattus norvegicus (Norway rat) myotrophin open reading frame (NCBI GenBank™ accession number U21661) was taken from a previously published construct (6) and cloned into the pMAL-c2X bacterial expression vector (New England Biolabs, Inc.). The MBP-myotrophin fusion was overexpressed in isopropylthio-β-d-galactoside-induced (0.3 mm for 3 h at 30 °C) E. coli TB1. MBP-myotrophin was purified using amylose resin in 20 mm Tris-HCl (pH 7.4), 200 mm NaCl. MBP was cleaved from myotrophin by Factor Xa (incubation for 16 h) and purified to homogeneity through a Sephadex G-50 column (Amersham Biosciences) in 20 mm Tris-HCl (pH 7.4), 100 mm NaCl. The entire purification process was performed at 4 °C, and purity of recombinant protein was analyzed by SDS-PAGE.
Generation of Recombinant Myotrophin Mutants—Mutants were created by site-directed mutagenesis using a QuikChange II XL site-directed mutagenesis kit (Stratagene). The pMAL-c2X-myotrophin/pET3a-51-myotrophin (6) construct was used as the template for generating the mutants. Recombinant mutant proteins were overexpressed and purified as described above (except for NMR studies, which are described below).
NMR Spectroscopy—Untagged, full-length Myotrophin and Myotrophin-E33A cloned into the pET3a vector (6) were expressed in BL-21(DE3) pLysS cells in minimal medium containing 0.4% glucose and 0.1% 15NH4Cl, as described previously (2, 23). Each protein was purified from 1 liter of culture by anion exchange chromatography, using a Resource Q column (GE Healthcare) and a gradient of 0.1-1 m NaCl. The proteins were further purified by preparative gel filtration chromatography using a Superdex-75 16/60 column (GE Healthcare). Purified protein was concentrated to ∼1.1 mm using Vivaspin centrifugal concentrators (3-kDa molecular weight cut-off) and dialyzed overnight into degassed buffer containing 50 mm potassium phosphate (pH 6.2) and 5 mm β-mercaptoethanol. 15N/1H heteronuclear single quantum coherence NMR spectra were collected at 25 °C on 250-μl samples supplemented with 10% D2O. The spectra were acquired using a Bruker Avance 600 MHz spectrometer equipped with a cryoprobe. The spectra were processed using NMRPipe (27) and visualized with Sparky (28).
Fluorophore Labeling of Myotrophin—Recombinant myotrophin was labeled with Alexa Fluor 488 using the Alexa Fluor 488 microscale protein labeling kit (Invitrogen). In brief, prior to labeling, purified protein was dialyzed against phosphate buffered saline (pH 7.2). An aliquot (50 μl) of dialyzed protein (1 mg/ml) was incubated with aqueous solution of reactive dye for 15 min at room temperature, and labeled protein was purified using supplied gel resin in a spin filter. The protein concentration and degree of labeling were measured using the company supplied protocol. Labeled wild type and mutant myotrophins were stored in aliquots at -20 °C.
Preparation of Neonatal Rat Ventricular Myocyte—Neonatal myocytes were isolated and cultured on laminin-coated wells according to the procedure described by Sen et al. (4). In brief, hearts from 3-day-old normal Wistar Kyoto rat pups were aseptically taken in DMEM/F-12 medium (Invitrogen). The ventricles were separated, minced, and incubated. Isolated myocytes were plated on laminin-coated wells (20 mg/35-mm well) at a density of 106 cells/35-mm well. For localization experiments, the cells were plated on a laminin-coated glass coverslip. On culture day 3 (or 4), the myocytes were incubated in DMEM/F-12 medium alone and were used for the experiment. Throughout the experimental procedure, bromodeoxyuridine was used at a concentration of 0.1 mm/liter to inhibit the growth of cardiac fibroblasts. The myocyte population (∼80-90% myocytes) was confirmed by α-actinin staining.
Cell Treatment—To study the effect of myotrophin (wild type or mutant) on myocyte growth, neonatal myocytes were treated with 40 nmol of recombinant myotrophin for 0-16 h. To inhibit the activation of NF-κB, the myocytes were pretreated with 100 μm PDTC (10). Following PDTC pretreatment, the cells were treated with recombinant myotrophin (wild type or mutant) for the desired period of time. For localization of myotrophin, Alexa 488-labeled myotrophin was used for cell treatment.
Measurement of Protein Synthesis—Total protein synthesis in myotrophin-treated myocytes was determined according to Sen et al. (4). In brief, on culture day 3, neonatal rat ventricular myocytes were preincubated in serum-free DMEM/F-12 medium. 40 nmol of myotrophin was added, and the incubation was continued from 0-16 h. 3[H]Leucine (10 μCi) diluted in DMEM was added to each well, and the samples were incubated at 37 °C. At the end of the incubation, the medium was removed by aspiration, and total cellular protein was extracted to measure the incorporation of 3[H]leucine as described previously (4, 10). At least three independent sets of experiments were performed.
Localization by Confocal Microscopy—After treatment of myocytes with Alexa 488-labeled myotrophin (wild type or mutant), cell fixation and permeabilization were performed according to the protocol of Rouet-Benzineb et al. (29). The primary antibody was anti-p65 mouse monoclonal antibody, and the secondary antibody was Alexa® 568 goat anti-mouse IgG (HL) conjugate. The Cells were mounted in Vectashield mounting medium containing DAPI. Fluorescence of Alexa 488-labeled myotrophin and NF-κB-p65 staining was observed using a Leica TCS-SP-AOBS spectral laser scanning confocal microscope (Leica Microsystem) equipped with UV1364, two argon ion NA 1.4 (488 nm) lasers, and a krypton/argon ion laser (568 nm). A PlanApo 63× oil immersion objective lens was used. To compare the staining patterns, images of Alexa 488 (green), Alexa 568 (red), and DAPI (blue) channels were merged to determine overlap in the staining patterns of labeled myotrophin, antibody to NF-κB p65, and DAPI, respectively.
In Vitro Myotrophin and NF-κB-p65 Interaction Studies—Pure recombinant NF-κB-p65 was obtained from Panomics (Fremont, CA) and was subjected to interaction with recombinant MBP-myotrophin (wild type/E33A) in equimolar (15 pmol) ratio by incubating with amylose resin (New England Biolabs Inc.) in binding buffer (20 mm Tris-HCl, 200 mm NaCl, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride) overnight at 4 °C. The following day, the mixtures were centrifuged at 5000 × g for 5 min, and the precipitates were washed three times with binding buffer. Laemmli sample buffer (Bio-Rad) was then added to the amylase-protein mixtures, resolved in SDS-PAGE, and immunoblotted with anti-NF-κB-p65 monoclonal antibody (Santa Cruz Biotechnology) and with anti-v-1/myotrophin monoclonal antibody (Transduction laboratories). Expression levels were also quantified with ImageJ software, and the obtained values were expressed in arbitrary units.
Electrophoretic Mobility Shift Assay (EMSA)—An EMSA was performed using a double-stranded NF-κB oligonucleotide as a probe, as described previously (10). The sequence of NF-κB probe used in this study was 5′-AGT TGA GGG GAC TTT CCC AGG C-3′. Binding reaction was initiated using 10 μg of nuclear extract made from untreated, wild type-treated, or E33A-mutant myotrophin-treated neonatal myocytes. Competition assays were performed using 100-fold molar excess of cold NF-κB oligonucleotide. Supershift analysis was performed using anti-p65 antibody. EMSA profile obtained from three independent experiments was quantified with ImageJ software.
Transient Transfection and Luciferase Assay—Primary neonatal rat cardiomyocytes were transiently transfected with the 2X NF-κB-Luc construct (10) by using Lipofectamin reagent (Invitrogen) according to the manufacturer's instructions. After transfection, the cells were washed in DMEM/F-12 medium for four times and maintained in serum-free medium with or without myotrophin (wild type and E33A), as indicated in the figure legend following the protocol of Gupta et al. (10). Luciferase activity and normalization of the protein content of each extract were determined by using the Luciferase assay system (Promega) according to the manufacturer's protocol.
Determination of Cell Size—Images of rat ventricular myocytes treated for 72 h either with wild type or with E33A mutant myotrophin protein were acquired using a Leica DMR, upright wide field microscope, a 20× objective, and a Retiga EXi CCD digital camera (Q-Imaging). Phase contrast images were taken from different fields in each slide, and cell area measurements were determined using ImagePro Plus 6.1 software.
Reverse Transcription-PCR Analysis of Atrial Natriuretic Factor (ANF) and β-Myosin Heavy Chain (β-MHC) Gene Expression—Myocytes treated either with wild type or with mutant myotrophin were scraped from culture plates at 4 °C, and total RNA was isolated with a Qiagen RNeasy kit. Cloned avian myeloblastosis virus first strand cDNA synthesis kit (Invitrogen) was used to synthesize the first strand with oligo(dT) primer. For each sample, a total of 5 μg of RNA was used. Synthesis of the second strand was performed in a Bio-Rad iCycler by using gene-specific primers for ANF and β-MHC and the Roche Applied Science PCR core kit. A profile of gene expression was determined by analysis on a 1% agarose gel after equi-volume sample loading. Expression of glyceraldehyde-3-phosphate dehydrogenase was used as an internal control. Expression levels were quantified with ImageJ software, and the obtained values were expressed in arbitrary units.
Statistical Analysis—The results were expressed as values ± S.E., and Student's t test was performed. The differences were considered significant at p < 0.05.
RESULTS
Internalization and Localization of Myotrophin—To localize myotrophin within cells and to follow its trafficking in myocytes after adding myotrophin to culture medium, we labeled myotrophin using Alexa Fluor-488. To verify that labeled myotrophin retained its biological activity, neonatal rat ventricular myocytes were incubated with both unlabeled recombinant myotrophin as well as Alexa 488-labeled myotrophin. We observed increased myocyte protein synthesis in the presence of unlabeled and Alexa 488-labeled myotrophin (69 and 71% over control, respectively), thus confirming that the fluorescent label did not interfere with myotrophin function.
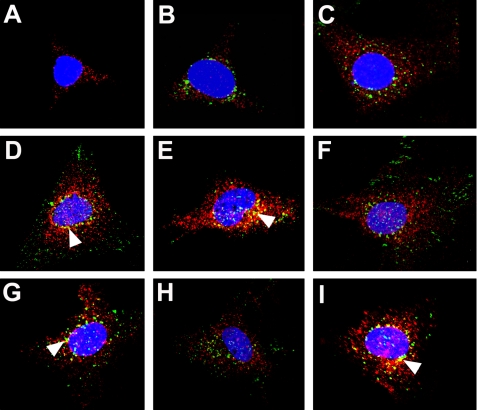
We followed the localization of Alexa 488-labeled myotrophin movement after addition to medium. Initially, no myotrophin was observed inside the cell, whereas p65 localized to the cytoplasm (Fig. 1A). After 5 min of incubation, myotrophin was internalized within myocytes and resided in the cytoplasm, with p65 (Fig. 1B). The cytoplasmic localization of myotrophin and p65 continued for 10 min (Fig. 1C). From 20 min through 16 h, however, both myotrophin and p65 were also found within the nucleus (Fig. 1, D-I). Moreover, both the proteins co-localized in cytoplasm, particularly in perinuclear regions (Fig. 1, D-I, arrowheads).
FIGURE 1.
Myotrophin co-translocates to the nucleus with p65. Neonatal rat ventricular myocytes were grown on coverslips, serum-starved, and treated with Alexa 488-labeled wild type myotrophin (green). The cells were fixed, blocked, and stained for NF-κB-p65 (red) by Alexa 568-conjugated goat anti-rabbit IgG and examined by confocal microscopy. The nuclei were stained with DAPI (blue). Co-localization of myotrophin and p65 (yellow; marked by arrowheads) is evident in certain cases. At zero time (A), p65 was observed only in the cytoplasm. Internalized Alexa 488-labeled wild type myotrophin was observed in the cytoplasm along with p65 after 5 min (B) and 10 min (C) of myotrophin treatment. Nuclear co-translocation of both Alexa labeled wild type myotrophin and p65 was observed after 20 min (D), 30 min (E), 60 min (F), 3 h (G), 4 h (H), and 16 h (I) of myotrophin treatment.
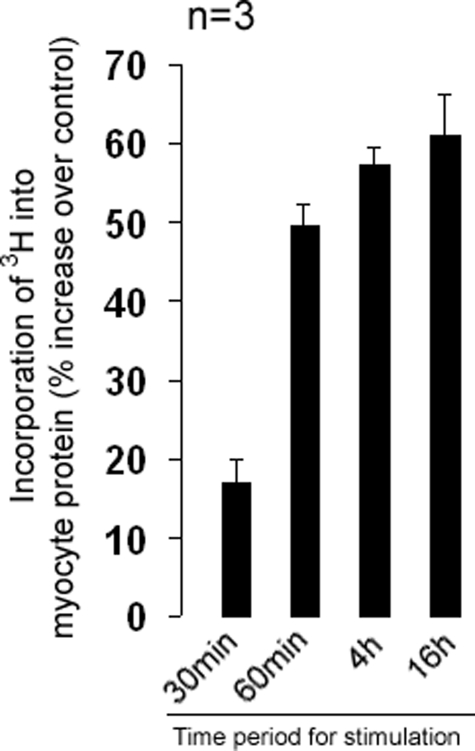
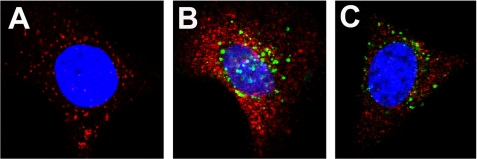
Concomitant with the nuclear localization of myotrophin and p65, we measured an increase in protein synthesis. Protein synthesis in myocytes increased 17% over control after 30 min of myotrophin treatment. After 4 and 16 h, we measured an increase to 57 and 61% over control, respectively (Fig. 2). When the nuclear translocation of p65 was blocked by pretreating cells with PDTC, an inhibitor of NF-κB, both myotrophin and p65 remained in the cytoplasm (Fig. 3), suggesting that the nuclear translocation of both proteins is coupled to NF-κB activation.
FIGURE 2.
Stimulatory effect of wild type myotrophin on myocyte protein synthesis. Neonatal rat ventricular myocytes were treated with myotrophin for different time periods starting from 5 min up to 16 h in presence of 3[H]leucine. The data show the means (± S.E.) of three different sets of results. An increase in myocyte protein synthesis was observed in cells treated with myotrophin from 30 min onwards.
FIGURE 3.
An inhibitor of NF-κB activation blocks co-translocation of myotrophin and p65. A, neonatal rat ventricular myocytes were pretreated with PDTC for 30 min prior to myotrophin treatment. p65 (red) was observed in the cytoplasm. B, when cells were treated with Alexa 488 labeled wild type myotrophin (green) without PDTC pretreatment, nuclear translocation of both myotrophin (green) and p65 (red) was observed. C, this translocation was inhibited when cells were pretreated with PDTC before treatment with myotrophin. Co-localization of myotrophin and p65 was evident from yellow fluorescence. The nucleus is stained with DAPI (blue).
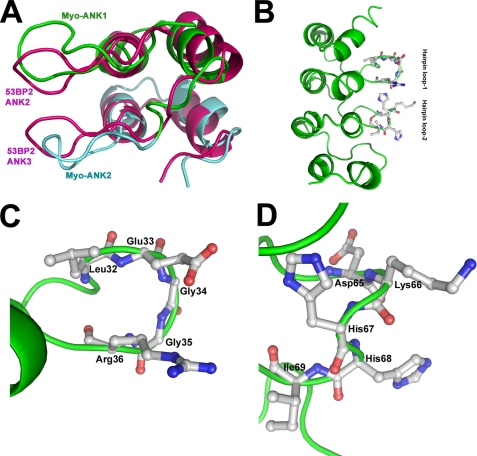
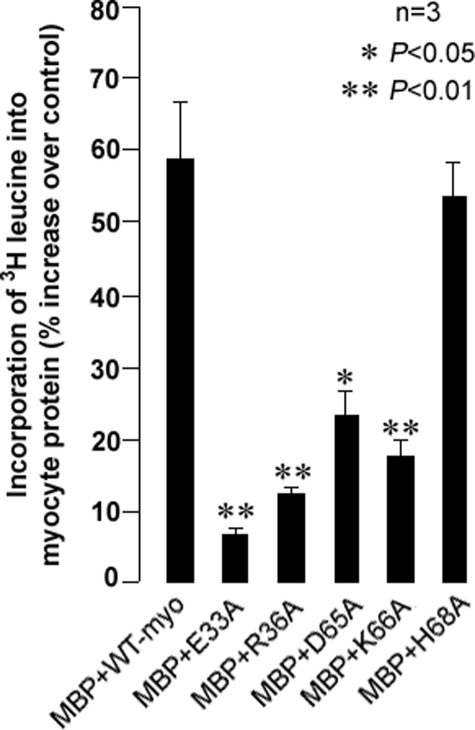
Effect of Mutant Myotrophin on Myocyte Protein Synthesis and Effect of Mutation on Structural Integrity of the Protein—The solution structure of myotrophin (2) is comparable with the crystal structure of the ANK repeats of 53BP2 (Fig. 4A). The prominent hairpin loops in ANK repeats 1 and 2 of myotrophin, however, differ from their counterparts in 53BP2, although the proteins have overall structural similarities (2). In 53BP2, the loops are closely packed together through extensive backbone and side chain interactions. In myotrophin, only a few intra-tip contacts exist (23). Considering that the hairpin loops may contribute to the functional specificity of myotrophin, we targeted them for mutagenesis to investigate their relationship to the biological activity of myotrophin. We selected several amino acids for site-specific mutation (Fig. 4, B-D). We did not mutate leucine 32 in the first hairpin loop or histidine 67 or isoleucine 69 of the second hairpin loop, because these residues may pack against the helical core of the protein; mutation of these residues may thus lead to destabilization of the protein. Similarly, glycines 34 and 35 were not mutated as these residues stabilize the conformation of the first hairpin loop (Fig. 4, C and D). A total of five sites were selected for mutation to alanine: glutamic acid 33 and arginine 36 on the first hairpin loop and aspartic acid 65, lysine 66, and histidine 68 on the second loop (Fig. 4, C and D). These residues were replaced with alanine, and the mutants were generated and purified as MBP fusion proteins.
FIGURE 4.
A, comparison of the ANK1 and ANK2 regions of myotrophin with those of ANK2 and ANK3 of 53BP2 (adopted from Ref. 2). B, ribbon representation of overall structure of myotrophin showing two hairpin loops protruding from the core structure. C and D, magnified views of hairpin loop 1 (C) and hairpin loop 2 (D) of myotrophin showing the amino acid side chains in each loop. Molecular graphics were rendered using Pymol 0.9 (www.pymol.sourceforge.net).
Neonatal rat ventricular myocytes were treated with MBP-fused mutant and wild type myotrophin proteins, and the increase in myocyte protein synthesis was quantified in terms of 3[H]leucine incorporation. Myocytes treated with purified MBP were used as controls. Treatment with MBP-fused wild type myotrophin resulted in an increase in protein synthesis compared with cells treated with MBP alone (Fig. 5). All mutants, except MBP-H68A, promoted less myocyte protein synthesis than wild type MBP-myotrophin (Fig. 5). The E33A mutant, located on the first hairpin loop, exhibited the least stimulation of protein synthesis (6% increase over control; Fig. 5). The E33A mutant was cleaved from MBP by Factor Xa and used for further experiments.
FIGURE 5.
Effect of mutants of hairpin loop on myocyte protein synthesis. Compared with the MBP-wild type myotrophin fusion, incorporation of 3[H]leucine into neonatal rat ventricular myocytes was suppressed in cells treated with MBP-myotrophin mutants at glutamic acid 33 (E33A), arginine 36 (R36A), aspartic acid 65 (D65A), and lysine 66 (K66A), but not in histidine 68 (H68A). The data show the means (± S.E.) of three independent sets of results.
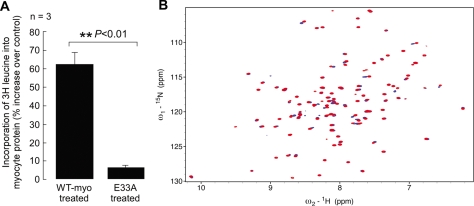
Myocytes treated with purified E33A increased protein synthesis only 6% over control, whereas wild type myotrophin stimulated synthesis 65% over control after 16 h of treatment (Fig. 6A). Myocytes treated simultaneously with wild type and E33A myotrophin, in 1:1 and 1:2 ratios, exhibited increased protein synthesis similar to cells treated with wild type myotrophin alone (data not shown), suggesting that the mutant had no dominant negative effect.
FIGURE 6.
Effect of E33A mutant myotrophin on increase in myocyte protein synthesis and structural comparison of wild type and E33A mutant protein. A, incorporation of 3[H]leucine into neonatal rat ventricular myocytes showed significantly (p < 0.05) less increase in protein synthesis in E33A-treated cells compared with wild type (WT) myotrophin-treated cells. The data show the means (± S.E.) of three independent sets of results. B, 15N/1H heteronuclear single quantum coherence spectra collected from 15N-labeled wild type and E33A myotrophin. The spectrum of wild type myotrophin is shown in blue, overlaid with the E33A spectrum in red. The spectra of the two proteins are mostly identical or very similar. The well dispersed spectrum of the E33A mutant shows that the mutation does not disrupt the overall folding of the protein or cause global changes to the protein fold but rather only limited local changes.
To verify that the E33A mutation does not destabilize or cause large scale structural perturbations to myotrophin, we collected nuclear magnetic resonance spectra from 15N-labeled wild type myotrophin and myotrophin-E33A. 15N/1H heteronuclear single quantum coherence spectra from the two proteins (Fig. 6B) are well dispersed and extremely similar, showing that the mutation does not cause myotrophin-E33A to be misfolded.
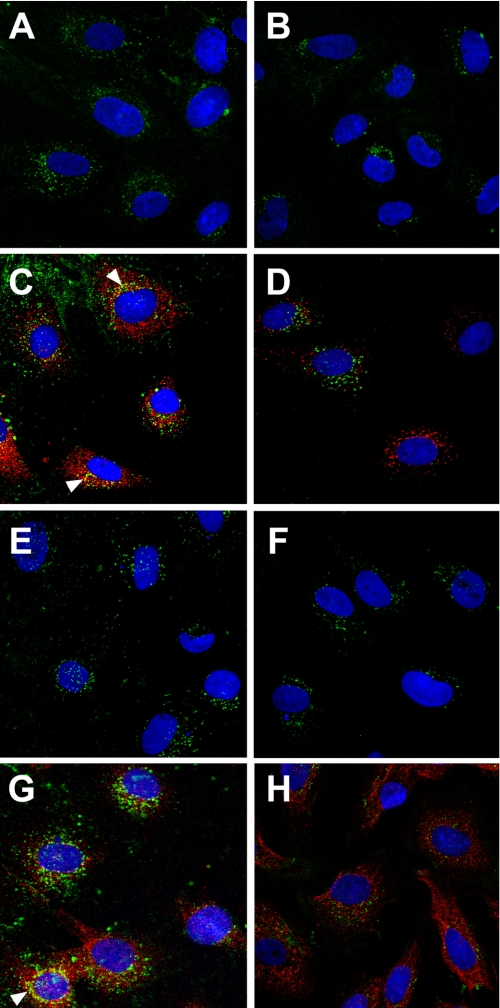
Internalization and Localization of Myotrophin E33A Mutant Protein—When myocytes were treated with Alexa 488-labeled wild type myotrophin or Alexa 488-labeled E33A for 5 min., the proteins were internalized within the cell and were observed in the cytoplasm (Fig. 7, A and B). We also observed cytoplasmic localization of p65 at this time point (Fig. 7, C and D). Both the E33A and p65 remain within the cytoplasm after 1 h of treatment (Fig. 7, F and H), whereas wild type myotrophin and p65 localization are observed in both cytoplasm and nucleus after 1 h. (Fig. 7, E and G). Co-localization of wild type myotrophin with p65 in the cytoplasm, particularly in the perinuclear region, was observed (Fig. 7, C and G), but such co-localization was not evident in E33A-treated cells (Fig. 7, D and H). When the cells were examined globally by confocal microscopy, both E33A and p65 remained in the cytoplasm, even after 16 h of treatment with Alexa 488-labeled E33A (data not shown).
FIGURE 7.
Cellular localization of E33A mutant myotrophin protein. Neonatal rat ventricular myocytes were treated with Alexa 488 (green) labeled wild type myotrophin (left panel) or Alexa 488 (green) labeled E33A-mutant myotrophin (right panel). NF-κB-p65 co-localized (arrowheads) with Alexa 568-conjugated antibody (red) as described in the legend to Fig. 1 and in the text. After 5 min of stimulation, both the wild type (A) and E33A (B) myotrophin were observed in the cytoplasm along with p65 (C and D). After 1 h of treatment, wild type myotrophin co-translocated into the nucleus with p65 (E and G), whereas both E33A and p65 failed to co-translocate (F and H). The nuclei were stained with DAPI (blue).
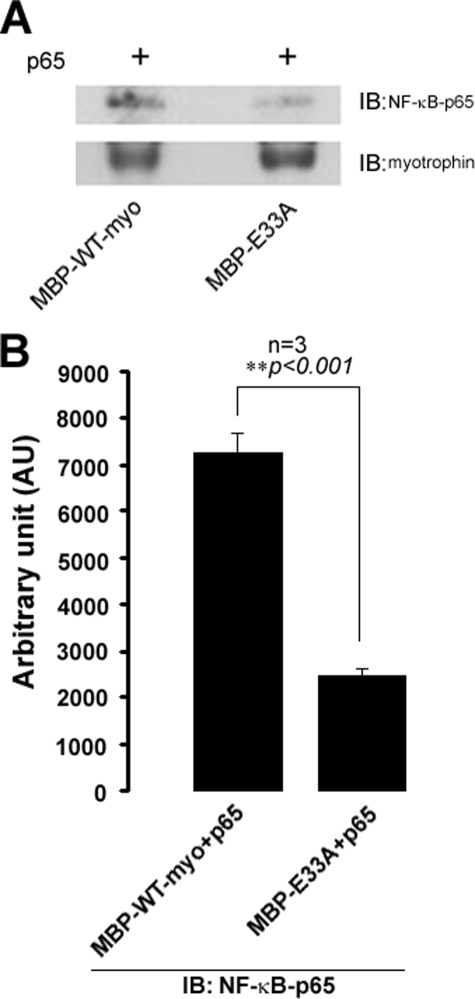
Interaction of Mytrophin and NF-κB-p65—In vitro interaction studies revealed that wild type myotrophin can physically interact with NF-κB-p65, whereas such kind of interaction was significantly reduced in the case of E33A (Fig. 8). An immunoblot profile with anti-V-1/Myotrophin monoclonal antibody showing the sample loading amount in both the cases. A significant reduction in interaction of E33A mutant with p65, compared with wild type (Fig. 8B) myotrophin, indicated that Glu33 plays an important role for co-localization and nuclear co-translocation of myotrophin and p65 to initiate the hypertrophy effect of myotrophin.
FIGURE 8.
In vitro interaction of myotrophin and NF-κB-p65. A, MBP-wild type/E33A mutant myotrophin incubated with amylose resin in equimolar (15 pmol) ratio for overnight at 4 °C, and amylose-bound protein was resolved in 10% SDS-PAGE and immunoblotted (IB) for NF-κB-p65 (upper panel). Immunoblotting for myotrophin as loading control (lower panel). B, band intensity for immunoblotting. NF-κB-p65 from three independent experiments was quantified using ImageJ software and expressed in arbitrary units.
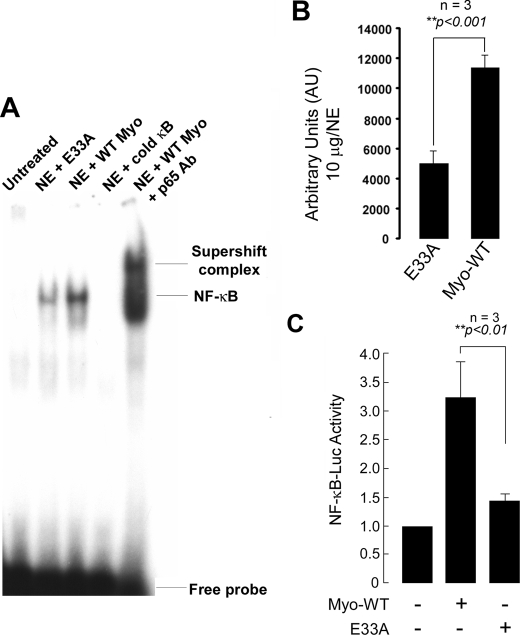
NF-κB DNA Binding Activity in Wild type and E33A Mutant Myotrophin-treated Neonatal Rat Myocytes—To investigate myotrophin-driven NF-κB activation, an EMSA using a radioactively labeled oligonucleotide containing the NF-κB-binding sequence was performed. Nuclear extracts of myocytes treated with either wild type or E33A myotrophin for a period of 1 h were used for the binding assay. A shift in mobility of the oligonucleotide probe was observed using wild type myotrophin extracts, showing that NF-κB activity was specifically stimulated. In contrast, extracts from E33A-treated exhibited substantially less mobility-shifted oligonucleotide, roughly corresponding to the decreased protein synthesis stimulated by this mutant (Fig. 9, A and B). The specific binding of NF-κB was confirmed by the addition of a 100-fold molar excess of unlabeled oligonucleotide, which abolished the binding. Antibody supershift confirmed that the shifted complex included p65. In the case of nuclear extract from untreated myocytes, we observed no NF-κB binding to the probe (Fig. 9A).
FIGURE 9.
NF-κB activation in wild type and E33A myotrophin-treated neonatal rat ventricular myocytes. A, nuclear protein was extracted from control (no stimulation), wild type (WT), and E33A myotrophin-treated neonatal rat ventricular myocytes. Binding reactions were performed with NF-κB oligonucleotide labeled with [32P]dATP. The complex formation was competed with100-fold molar excess of unlabeled NF-κB oligonucleotide. The complex formation was confirmed by supershift analysis using p65 antibody. NE, nuclear extract. B, quantification of electrophoretic mobility shift assay using ImageJ software. The values obtained from three independent experiments are expressed as arbitrary units for 10 μg/nuclear extract (p < 0.001). C, primary neonatal rat myocytes were transfected with 2× NF-κB-Luc reporter plasmid (1 μg/well) and treated with 40 nm myotrophin (wild type E33A) for 24 h or left untreated. The cells were also transfected with empty vector. After harvesting the cells, Luciferase activity was determined and normalized to the protein content of each extract. Luciferase activity expressed by cells transfected with empty vector was given an arbitrary value of 1. The results are presented as the means ± S.E. and represent three individual experiments.
Furthermore, to determine the NF-κB activity in the cells treated with wild type and E33A myotrophin, neonatal myocytes were grown in six-well plates, transiently transfected with 2× NF-κB-Luciferase reporter gene, serum-starved, and treated with myotrophin (wild type/E33A). Wild type myotrophin stimulated NF-κB transcriptional activity by 3.2-fold (Fig. 9C) as measured by activation of the NF-κB-Luc reporter gene. NF-κB transcriptional activity was stimulated only by 1.4-fold when cells were treated with E33A myotrophin (Fig. 9C).
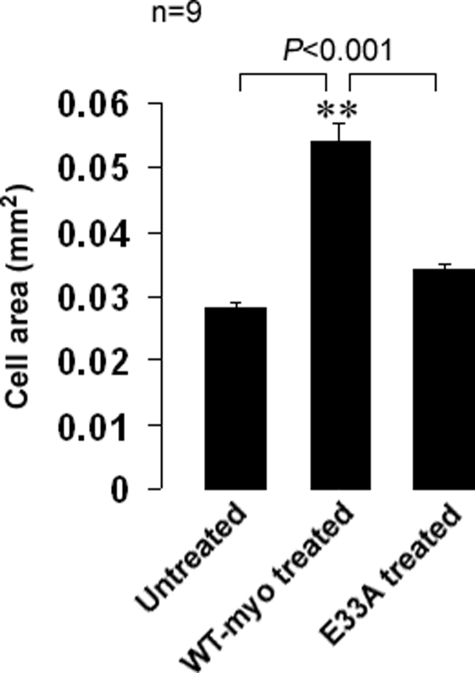
Effect of Myotrophin E33A Mutant on Changes in Cell Size—As a measure of hypertrophy, we quantitated the cell area of neonatal rat ventricular myocytes treated with either wild type or E33A myotrophin. Myocytes treated for 72 h with wild type myotrophin had an average area of 0.054 mm2 compared with 0.028 mm2 for untreated cells (>90% increase in surface area). The average cell area after treatment with E33A mutant protein was 0.034 mm2 (Fig. 10), ∼21% over untreated control.
FIGURE 10.
Effect of myotrophin treatment (wild type or E33A mutant) on cell surface area of neonatal rat ventricular cell. The cells were treated with either wild type (WT) or with E33A myotrophin protein and compared with untreated control cells. The cells were treated for 72 h, fixed and micrographs were taken randomly from different portions of the slide. The cell area was measured using ImagePro 6.1 software. The data (means ± S.E.) averaged from nine individual measurements for each group are expressed in mm2. Wild type myotrophin-treated cells showed a significant (p < 0.001) increase in cell surface area compared with untreated and E33A-treated cells.
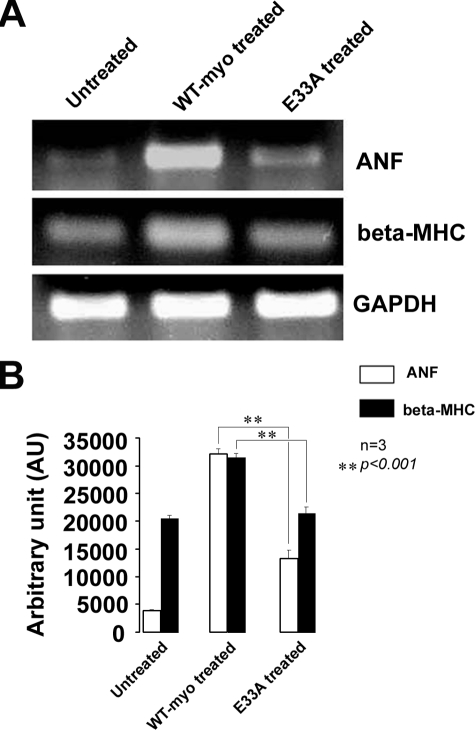
Expression of Hypertrophic Marker Genes—To confirm the attenuation of myotrophin-induced hypertrophy by the E33A mutation, we carried out reverse transcription-PCR analysis to evaluate the expression of two hypertrophic marker genes. Levels of ANF and β-MHC mRNAs are correlated with an increase in hypertrophy (10, 25, 30, 31, 10). An up-regulation (8.1-fold) of ANF transcript levels (32,067 AU) was observed in cells treated with wild type myotrophin protein compared with that of untreated cells (3,955 AU). In contrast, in E33A-treated cells the transcript level increased only 3-fold (11,987 AU) relative to control (Fig. 11). Observations were similar for the β-MHC transcript levels: a 1.6-fold increase was observed after wild type myotrophin treatment (31,653 AU), but no increase was found after E33A treatment (the values obtained were 20,356 AU and 19,569 AU in E33A-treated and untreated control, respectively) (Fig. 11).
FIGURE 11.
Expression of hypertrophic marker genes (ANF and β-MHC) in untreated control, wild type, and E33A myotrophin-treated neonatal rat ventricular myocytes. Reverse transcription-PCR banding profile (A) showed down-regulation of ANF and β-MHC in untreated control and E33A-treated cells compared with wild type (WT) myotrophin-treated cells (glyceraldehyde-3-phosphate dehydrogenase as internal control). Band intensity was quantified using ImageJ software and expressed in arbitrary units (B).
DISCUSSION
The 12-kDa ANK-repeat protein myotrophin is a potent stimulator of cardiac myocyte growth (4). Myotrophin plays an important role in the pathogenesis of cardiac hypertrophy as described by Mukherjee et al. (30). Treatment of myocytes with myotrophin results in a dose-dependent increase in cell size, which is in turn associated with reorganization of myofibrils, induction of early response genes, and enhancement of cardiac gene expression (4). Preliminary studies by Sil et al. (32) and Anderson et al. (33) predicted that myotrophin may stimulate myocytes and initiate the hypertrophic process through a receptor-mediated active process when added exogenously in cultured neonatal rat ventricular myocytes. Our data showed that myotrophin is indeed internalized after exogenous addition to cultured neonatal rat ventricular myocytes (Fig. 1, A-C). The internalized myotrophin eventually translocates into the nucleus (Fig. 1, D-I) accompanied by elevation in protein synthesis (Fig. 2). This suggests that nuclear import of myotrophin after internalization is necessary for initiation of hypertrophic mechanisms.
From the present study it is evident that the nuclear import of myotrophin, at the very least, is linked to contemporaneous NF-κB nuclear import and activation. The involvement of NF-κB activation in cardiac hypertrophy has been established in spontaneously hypertensive rats (34) and in hypertrophic growth of cardiomyocytes (35). We previously reported that myotrophin stimulates the nuclear translocation of NF-κB and activates hypertrophy marker genes (10). Furthermore, we showed that NF-κB is activated at the initiation phase of hypertrophy in myotrophin-overexpressing transgenic mice and that activation is more pronounced at the end stage of heart failure (13). In the present study we have shown that, on a cellular level, p65 initially resides in the cytoplasm with myotrophin (Fig. 1, A-C) but co-translocates into the nucleus with myotrophin thereafter (Fig. 1, D-I). The co-translocation of both proteins results in an increase in protein synthesis (Fig. 2). We observed limited co-localization of myotrophin and p65 within the cytoplasm, particularly at the nuclear periphery (Fig. 1, D--I). In accordance with the findings of Knuefermann et al. (16), this co-localization is consistent with a direct, if perhaps transient, interaction between myotrophin and p65 that was confirmed by the in vitro physical interaction of myotrophin with NF-κB-p65 (Fig. 8). Moreover, co-localization was not evident inside the nucleus, suggesting that myotrophin may interact with p65 only prior to, and as a precondition for, nuclear import. Inhibition of nuclear translocation of p65 by pretreatment with PDTC, a known inhibitor of NF-κB, prevents nuclear import of both proteins (Fig. 3). Therefore, activation of NF-κB-p65 is one of the essential step for nuclear import of myotrophin. Once imported, myotrophin and p65 individually perform nuclear functions that converge to hypertrophic effect.
Structure-based mutagenesis of myotrophin emphasizes the functional connections between putative protein-protein interaction sites on myotrophin, nuclear import, NF-κB activation, and hypertrophic processes. The two myotrophin ankyrin repeats, ANK1 and ANK2 (residues 28-60 and 61-93, respectively), contain two hairpin loops that resemble ANK2 and ANK3 of 53BP2, which interacts with p53 (26) and induces apoptosis (36). However, the detailed structures and sequences of the hairpin loops differ in the two proteins (Fig. 4). The two loops of myotrophin were previously reported to be involved in control of actin polymerization through binding to the actin capping protein (9). We found that mutations in the loops resulted in suppression of myotrophin-driven increase in protein synthesis (Fig. 5). The E33A mutation had the maximal effect in this regard (Fig. 6A) without altering the structural integrity of the protein (Fig. 6B), confirming that the phenotype of E33A is due to site-specific effects of the mutation. The E33A mutant was internalized into the cell but failed to translocate into the nucleus or to promote nuclear translocation of p65 (Fig. 7). EMSA results and NF-κB-Luc activity confirmed the suppression of NF-κB activation by E33A (Fig. 9). E33A-treated neonatal rat ventricular myocytes did not increase in cell area, unlike myocytes treated with wild type myotrophin (Fig. 10). Furthermore, the transcript levels of two hypertrophic marker genes (10, 30, 31), ANF and β-MHC, were significantly lower in E33A mutant-treated cells than in cells treated with wild type myotrophin (Fig. 11). These data all point to the inability of E33A to induce hypertrophy. It is likely that the other loop mutants that exhibit reduced stimulation of myocyte protein synthesis will behave similarly to E33A with respect to nuclear translocation, activation of NF-κB, and myocyte growth.
Myotrophin is thought to directly interact with p65 for induction of p65-p65 dimerization and dissociation of the inactive p65-p50 heterodimer during myotrophin-driven NF-κB activation (9). The increased ratio of p65-p65 homodimer determines the level of NF-κB targeted gene expression (37, 38). Our data show that mutations in the hairpin loops at Glu33 of myotrophin do not affect cellular uptake but significantly diminish the physical interaction between myotrophin and NF-κB-p65 (Fig. 8), suggesting that the loops are not involved in interactions between myotrophin and a putative receptor or elements of the internalization machinery. However, the integrity of the loops is critical for all the post-internalization roles of myotrophin, that is, promotion of nuclear import of both myotrophin itself and NF-κB-p65 that leads to myocyte growth. Inhibiting nuclear import of p65 blocks the nuclear translocation of myotrophin. On the other hand, upon mutation of the hairpin loop of myotrophin, both myotrophin and p65 failed to translocate into nucleus. Knuefermann et al. (16) reported that myotrophin interacts and remained associated with p65 to prevent the reassociation of NF-κB proteins during sustained activation of NF-κB. Our in vitro interaction studies revealed that Glu33 is important for the physical interaction of myotrophin and p65 (Fig. 8). Therefore, the E33A mutant showed attenuation of NF-κB activation, leading to a reduced hypertrophy effect. Understanding the roles of other NF-κB family members in this interaction and subsequent activation of the cascade will require further elaborate investigation. To date, all these data suggest that the hairpin loop of myotrophin is involved in activation of NF-κB; once activated by myotrophin, NF-κB-p65 may function as a nuclear transport factor for myotrophin translocation. In this capacity, p65 could play a role comparable with that of importin β-1, which is responsible for nuclear translocation of calcineurin (40). Down-regulation of NF-κB-p65 nuclear translocation in the E33A mutant leading to attenuation of myocyte growth is similar to attenuation of angiotensin II-induced cardiomyocyte hypertrophy by down-regulation of NF-κB-p65 nuclear import driven by focal adhesion kinase-related nonkinase (41). Apart from Glu33, involvement of other residues in the hairpin loops, such as Arg36, Asp65, and Lys66, in NF-κB activation and in stimulating myocyte growth needs to be investigated further.
Additional functions of myotrophin inside the nucleus, if any, are not yet characterized and require further investigation. The expression of NF-κB target genes depends not only on the activity of NF-κB itself but also on other factors that interact with NF-κB (13). In this context, one possibility is that myotrophin modulates transcription as a cofactor, similar to the cardiac ankyrin repeat protein (39) during hypertrophy and its progression to heart failure. The presence of calcineurin in the nucleus is important for full NF-AT (nuclear factor of activated T cells) transcriptional activity. When the nuclear entry of calcineurin is blocked, development of myocardial hypertrophy can be prevented (40). Similarly, we hypothesize that an agent that binds and block the hairpin loops of myotrophin or specific residues like Glu33 would attenuate myotrophin-induced cardiomyocyte hypertrophy. Myotrophin-induced cardiac hypertrophy can be partially regressed by silencing the NF-κB cascade using a short hairpin RNA against NF-κB-p65 (13), but an inhibitor that binds to myotrophin might be more effective.
Taken together, our data showed that nuclear co-translocation of myotrophin and p65 is an essential step for myotrophin-induced, NF-κB-mediated myocyte growth. The residues of hairpin loop (such as Glu33) of myotrophin regulate the cross-talk between myotrophin and p65. Thus inhibition of this association will enable researchers to develop a therapeutic strategy to prevent myotrophin-induced myocyte growth and cardiac hypertrophy.
Acknowledgments
We acknowledge Dr. Judith Drazba of the Imaging Core Facility at Lerner Research Institute, Cleveland Clinic for confocal microscopy and David Young for laboratory assistance and statistical analysis. We are also thankful to Michele Barnard for expert secretarial help.
This work was supported, in whole or in part, by National Institutes of Health Grant HL R01 47794 (to S. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: ANK, ankyrin; ANF, atrial natriuretic factor; β-MHC, β-myosin heavy chain; EMSA, electrophoretic mobility shift assay; MBP, maltose-binding protein; PDTC, pyrrolidine dithio-carbamate; DMEM, Dulbecco's modified Eagle's medium; DAPI, 4′,6′-diamidino-2-phenylindole; AU, arbitrary unit(s).
References
- 1.Mosavi, L. K., Williams, S., and Peng Zy, Z. Y. (2002) J. Mol. Biol. 320 165-170 [DOI] [PubMed] [Google Scholar]
- 2.Yang, Y., Nanduri, S., Sen, S., and Qin, J. (1998) Structure 6 619-626 [DOI] [PubMed] [Google Scholar]
- 3.Lowe, A. R., and Itzhaki, L. S. (2007) J. Mol. Biol. 365 1245-1255 [DOI] [PubMed] [Google Scholar]
- 4.Sen, S., Kundu, G., Mekhail, N., Castel, J., Misono, K., and Healy, B. (1990) J. Biol. Chem. 265 16635-16643 [PubMed] [Google Scholar]
- 5.Taoka, M., Isobe, T., Okuyama, T., Watanabe, M., Kondo, H., Yamakawa, Y., Ozawa, F., Hishinuma, F., Kubota, M., and Minegishi, A. (1994) J. Biol. Chem. 269 9946-9951 [PubMed] [Google Scholar]
- 6.Sivasubramanian, N., Adhikary, G., Sil, P. C., and Sen, S. (1996) J. Biol. Chem. 271 2812-2816 [DOI] [PubMed] [Google Scholar]
- 7.Yamakuni, T., Hashimoto, M., Sakagami, H., Yamamoto, T., Kobayashi, M., Fujii, Y., Yamamoto, H., Rohra, D. K., Hiwatashi, Y., Honma, T., Kondo, H., Shido, O., and Ohizumi, Y. (2002) Biochem. Biophys. Res. Commun. 298 793-797 [DOI] [PubMed] [Google Scholar]
- 8.Taoka, M., Ichimura, T., Wakamiya-Tsuruta, A., Kubota, Y., Araki, T., Obinata, T., and Isobe, T. (2003) J. Biol. Chem. 278 5864-5870 [DOI] [PubMed] [Google Scholar]
- 9.Bhattacharya, N., Ghosh, S., Sept, D., and Cooper, J. A. (2006) J. Biol. Chem. 281 31021-31030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta, S., Purcell, N. H., Lin, A., and Sen, S. (2002) J. Cell Biol. 159 1019-1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sil, P., Misono, K., and Sen, S. (1993) Circ. Res. 73 98-108 [DOI] [PubMed] [Google Scholar]
- 12.Sarkar, S., Leaman, D. W., Gupta, S., Sil, P., Young, D., Morerhead, A., Mukherjee, D., Ratliff, N., Sun, Y., Rayborn, M., Hollyfield, J., and Sen, S. (2004) J. Biol. Chem. 279 20422-20434 [DOI] [PubMed] [Google Scholar]
- 13.Gupta, S., Young, D., Maitra, R. K., Gupta, A., Popovic, Z. B., Yong, S. L., Mahajan, A., Wang, Q., and Sen, S. (2008) J. Mol. Biol. 375 637-649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khan, S. Q., Kelly, D., Quinn, P., Davies, J. E., and Ng, L. L. (2007) Clin. Sci. (Lond.) 112 251-256 [DOI] [PubMed] [Google Scholar]
- 15.O'Brien, R. J., Loke, I., Davies, J. E., Squire, I. B., and Ng, L. L. (2003) J Am. Coll. Cardiol. 42 719-725 [DOI] [PubMed] [Google Scholar]
- 16.Knuefermann, P., Chen, P., Misra, A., Shi, S. P., Abdellatif, M., and Sivasubramanian, N. (2002) J. Biol. Chem. 277 23888-23897 [DOI] [PubMed] [Google Scholar]
- 17.Valen, G., Yan, Z. Q., and Hansson, G. K. (2001) J Am. Coll. Cardiol. 38 307-314 [DOI] [PubMed] [Google Scholar]
- 18.Jones, W. K., Brown, M., Ren, X., He, S., and McGuinness, M. (2003) Cardiovasc. Toxicol. 3 229-254 [DOI] [PubMed] [Google Scholar]
- 19.Purcell, N. H., and Molkentin, J. D. (2003) Circulation 108 638-640 [DOI] [PubMed] [Google Scholar]
- 20.Ritchie, M. E. (1998) Circulation 98 1707-1713 [DOI] [PubMed] [Google Scholar]
- 21.Wong, S. C., Fukuchi, M., Melnyk, P., Rodger, I., and Giaid, A. (1998) Circulation 98 100-103 [DOI] [PubMed] [Google Scholar]
- 22.Kalra, D., Baumgarten, G., Dibbs, Z., Seta, Y., Sivasubramanian, N., and Mann, D. L. (2000) Circulation 102 1302-1307 [DOI] [PubMed] [Google Scholar]
- 23.Yang, Y., Rao, N. S., Walker, E., Sen, S., and Qin, J. (1997) Protein Sci. 6 1347-1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson, C. C., Brown, T. A., and McKnight, S. L. (1991) Science 253 762-768 [DOI] [PubMed] [Google Scholar]
- 25.Michaely, P., and Bennett, V. (1992) Trends Cell Biol. 2 127-129 [DOI] [PubMed] [Google Scholar]
- 26.Iwabuchi, K., Bartel, P. L., Li, B., Marraccino, R., and Fields, S. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 6098-6102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J., and Bax, A. (1995) J. Biomol. NMR 6 277-293 [DOI] [PubMed] [Google Scholar]
- 28.Goddard T.D., Kneller. D. G. (2006) SPARKY 3, University of California, San Francisco, CA
- 29.Rouet-Benzineb, P., Gontero, B., Dreyfus, P., and Lafuma, C. (2000) J. Mol. Cell. Cardiol. 32 1767-1778 [DOI] [PubMed] [Google Scholar]
- 30.Mukherjee, D. P., McTiernan, C. F., and Sen, S. (1993) Hypertension 21 142-148 [DOI] [PubMed] [Google Scholar]
- 31.Sil, P., Gupta, S., Young, D., and Sen, S. (2004) Mol. Cell Biochem. 262 79-89 [DOI] [PubMed] [Google Scholar]
- 32.Sil, P., Kandaswamy, V., and Sen, S. (1998) Circ. Res. 82 1173-1188 [DOI] [PubMed] [Google Scholar]
- 33.Anderson, K. M., Berrebi-Bertrans, I., Kirkpatrick, R. B., McQueney, M. S., Underwood, D. C., Rouanet, S., and Chabot-Fletcher, M. (1999) J. Mol. Cell. Cardiol. 31 705-719 [DOI] [PubMed] [Google Scholar]
- 34.Gupta, S., Young, D., and Sen, S. (2005) Am. J. Physiol. 289 H20-H29 [DOI] [PubMed] [Google Scholar]
- 35.Purcell, N. H., Tang, G., Yu, C., Mercurio, F., DiDonato, J. A., and Lin, A. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 6668-6673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kobayashi, S., Kajino, S., Takahashi, N., Kanazawa, S., Imai, K., Hibi, Y., Ohara, H., Itoh, M., and Okamoto, T. (2005) Genes Cells 10 253-260 [DOI] [PubMed] [Google Scholar]
- 37.Brigelius-Flohe, R., Bilgin, B., Eickemeier, S., Hipskind, R., Singh, M., Szamel, M., and Resch, K. (1995) Biofactors 5 169-174 [PubMed] [Google Scholar]
- 38.Udalova, I. A., Richardson, A., Denys, A., Smith, C., Ackerman, H., Foxwell, B., and Kwiatkowski, D. (2000) Mol. Cell. Biol. 20 9113-9119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maeda, T., Sepulveda, J., Chen, H. H., and Stewart, A. F. (2002) Gene (Amst.) 297 1-9 [DOI] [PubMed] [Google Scholar]
- 40.Hallhuber, M., Burkard, N., Wu, R., Buch, M. H., Engelhardt, S., Hein, L., Neyses, L., Schuh, K., and Ritter, O. (2006) Circ. Res. 99 626-635 [DOI] [PubMed] [Google Scholar]
- 41.Qin, J., and Liu, Z. X. (2006) Acta Pharmacol. Sin. 27 1159-1164 [DOI] [PubMed] [Google Scholar]