Abstract
Normal human prostate (NHP) epithelial cells undergo senescence in vitro and in vivo, but the underlying molecular mechanisms remain obscure. Here we show that the senescence of primary NHP cells, which are immunophenotyped as intermediate basal-like cells expressing progenitor cell markers CD44, α2β1, p63, hTERT, and CK5/CK18, involves loss of telomerase expression, up-regulation of p16, and activation of p53. Using genetically defined manipulations of these three signaling pathways, we show that p16 is the primary determinant of the NHP cell proliferative capacity and that hTERT is required for unlimited proliferative life span. Hence, suppression of p16 significantly extends NHP cell life span, but both p16 inhibition and hTERT are required to immortalize NHP cells. Importantly, immortalized NHP cells retain expression of most progenitor markers, demonstrate gene expression profiles characteristic of proliferating progenitor cells, and possess multilineage differentiation potential generating functional prostatic glands. Our studies shed important light on the molecular mechanisms regulating the proliferative life span of NHP progenitor cells.
The prostatic glands contain neuroendocrine (NE)7 cells and two epithelial cells: 1) luminal cells expressing cytokeratin 8 (CK8) and CK18, androgen receptor (AR), prostate-specific antigen (PSA), prostatic acid phosphatase, CD26, CD57, and 15-lipoxygenase 2 (15-LOX2) and 2) basal cells expressing CK5/CK14, CD44, CD104 (integrin β4), Bcl-2, p63, telomerase, and glutathione S-transferase-π (1-3). It has been proposed that a common stem/progenitor cell may generate both basal and luminal cells (4). Alternatively, basal cells may function as progenitors to luminal cells (5, 6).
The adult rodent prostate possesses regenerative stem cells (SCs) (6-8). Whether adult human prostate contains definitive SCs is less certain, although there exists strong evidence that the basal cell layer harbors regenerative cells (1, 6, 9-12), and several candidate populations of human prostate stem/progenitor cells, preferentially localized in the basal layer, have been reported. These include the cells that preferentially express CD44 (13), α2β1(i.e. α2β1hi) (14), or CD133 (15, 16) and the side population (17), whose phenotype is mediated by multidrug resistance family proteins, such as MDR-1 and ABCG2 (18). Interestingly, the ABCG2+ cells in the benign prostate constitute <1% of total basal cell population and share essentially the same transcriptome as the side population cells (19). Both side population and ABCG2+ cells express SC-associated genes (19). A population of CK5 and CK18 double positive (CK5+/CK18+) cells called intermediate basal cells, which constitute ∼1% of the basal cells, have also been proposed as prostate stem/progenitor cells (4, 20). Since definitive prostate SCs have not been reported, prostate epithelial cells with certain SC properties (e.g. regenerating prostatic glands in vivo) are often termed as stem/progenitor cells or, simply, progenitor cells.
Mammalian cells, including adult SCs, have a finite proliferative life span in that after a certain number of cell divisions, they enter a permanent cell cycle arrest termed senescence (21), which may constitute a critical tumor-preventing mechanism (22). Prostate epithelial cells undergo cell senescence in vitro (9, 23, 24) and in vivo (25, 26), but molecular mechanisms remain only partially understood. In animal models, prostate epithelial cell senescence appears to constitute a mechanism that inhibits progression of benign to malignant lesions (27), although our recent work demonstrates that senescent human prostate epithelial cells can also enhance tumorigenicity via cell fusion (28).
In this study, we first aim to characterize multiple primary strains of normal human prostate (NHP) epithelial cells with respect to their phenotypic and proliferative properties, since systematic studies have not been reported. We then explore the molecular mechanisms of NHP cell senescence by focusing on three essential molecules, p16INK4a (p16), p53, and telomerase. Although p16 has previously been shown to be up-regulated during prostate epithelial cell senescence (23, 24), the critical question of whether it is actually required for senescence has not been addressed. We provide definitive evidence that p16 is the primary factor that limits the proliferative capacity of NHP progenitor cells. Likewise, although hTERT has been utilized to immortalize prostate tumor cells (29-31), it is unclear whether telomerase is required for indefinite NHP cell proliferation. We show herein that both telomerase and p16 inhibition are required for NHP cell immortalization. Finally, we demonstrate that immortalized NHP cells retain gene expression profiles characteristic of proliferating progenitor cells and can differentiate into functional prostatic glands in vivo.
MATERIALS AND METHODS
Cells and Reagents—Twelve primary NHP cell strains (Table 1) were derived either from normal organ donors (NHP1, NHP3, NHP4, and NHP6 to -10; obtained from Cambrex (Rockland, ME)) or from uninvolved benign prostate tissues distant from PCa foci (NHP2, -5, -11, and -12). NHP2 and NHP5 cells were previously used in our studies (9, 28, 32). NHP11 and NHP12 primary cells were obtained following our standard digestion, enrichment, and purification protocols (18, 33) (see below). NHP cells were routinely cultured on collagen-coated dishes in serum/androgen-free, partially defined PrEBM medium containing insulin, EGF, bovine pituitary extract (BPE) (Invitrogen), and antibiotics. This medium was based on that of Peehl and Stamey (34), modified by Chopra et al. (35), and referred to herein as PrEBM(EGF + Ins). PCa and 293T cells were cultured in RPMI1640 (for PCa cells) or Dulbecco's modified Eagle's medium (for 293T cells) supplemented with 7% heat-inactivated FBS. Primary antibodies used in this study are listed in Table S1. All regular culture media were purchased from Invitrogen, and all other chemicals and reagents were from Sigma unless indicated otherwise. Growth factors, including EGF, basic fibroblast growth factor, insulin, LIF, and Wnt3a, were all obtained from Peprotech (Rocky Hill, NJ). The isotype control antibody and fluorescein isothiocyanate-, phycoerythrin-, streptavidin-, or AlexaFluor-conjugated secondary antibodies were from Chemicon (Temecula, CA).
TABLE 1.
Primary NHP strains used in this study
| Strains | Donor (patient) age | Origina |
|---|---|---|
| years | ||
| NHP1 | 17 | Normal donor (H) |
| NHP2/HPCa-N | 59 | Distant from PCa (AA) |
| NHP3 | 42 | Normal donor (AA) |
| NHP4 | 17 | Normal donor (C) |
| NHP5/HPCa0-N | 56 | Distant from PCa (AA) |
| NHP6 | 26 | Normal donor (C) |
| NHP7 | 14 | Normal donor (C) |
| NHP8 | 41 | Normal donor (C) |
| NHP9 | 18 | Normal donor (AA) |
| NHP10 | 16 | Normal donor (C) |
| NHP11/HPCa1-N | 51 | Distant from PCa (H) |
| NHP12/HPCa2-N | 73 | Distant from PCa (C) |
Cells were obtained, prepared, and maintained as detailed under “Materials and Methods.” HPCa-N, cells derived from the uninvolved, noncancerous prostate tissues (N) distant from human prostate cancer (HPCa); H, Hispanic; AA, African-American; C, Caucasian.
Preparation of NHP11 and NHP12 Cells from Uninvolved Benign Prostate Tissues—Tissues were obtained with the patients' consent, and experiments on human samples were approved by an institutional review board. The basic cell derivation protocol was modified from our established experimental procedures (18, 33, 36). Briefly, benign prostate tissues were minced into ∼1-mm3 pieces and subjected to an overnight digestion in a mixture containing type I collagenase and DNase (both at 50 units/ml). Upon digestion, epithelial organoids were enriched by a brief centrifugation, followed by trypsin digestion to release epithelial cells. Samples were then subjected to a discontinuous Percoll gradient purification step to remove the majority of mononuclear blood cells and dead cells. Finally, cell preparation was subjected to a negative selection process using MACS (Miltenyi, Auburn, CA) and the antibody mixture (anti-CD3, -CD14, -CD16, -CD19, -CD20, -CD31, -CD45, -CD56, and -CD140b) to remove the Lin+ cells, including hematopoietic, endothelial, and other stromal cells (smooth muscle, myoepithelial, fibroblast, etc.). The whole procedure took 6-8 h to accomplish and generally resulted in >98% pure human epithelial cells as judged by Ber-EP4 immunostaining.
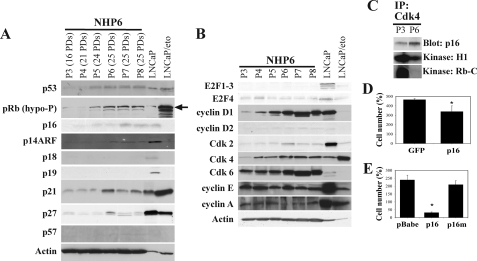
Immunoprecipitation and Coupled Immune Complex Kinase Assays—For immunoprecipitation, young (P3) and senescent (P6) NHP6 cells were harvested by trypsin and washed once in ice-cold phosphate-buffered saline. Then cells were resuspended in the homogenizing/lysis buffer (20 mm HEPES-KOH, pH 7.5, 10 mm KCl, 1.5 mm MgCl2, 0.2% CHAPS, 1 mm sodium EDTA, 1 mm sodium EGTA, and 1 mm dithiothreitol) containing 250 mm sucrose and protease inhibitors and homogenized in a glass Pyrex homogenizer (type A pestle). At the end, cells were centrifuged (12,000 × g for 20 min at 4 °C) to obtain clarified supernatant. Primary antibody (i.e. rabbit polyclonal Cdk4; 5 μg) or the control (Rb IgG) was preconjugated (in two aliquots) to protein G-Sepharose, followed by blocking with bovine serum albumin. Then the cell supernatant was added to the antibody-bound beads and incubated at 4 °C under rotating conditions overnight. Following extensive washing (4 times) in the homogenizing buffer, the immune complex-Sepharose beads were split into three portions. One portion was directly used in Western blotting for Cdk-associated p16. The other two portions were used in coupled immune complex kinase assays using either Rb C-terminal domain (sc-4112; Santa Cruz Biotechnology, Inc., Santa Cruz, CA) or histone H1 as substrate. Briefly, the immune complex on the beads was resuspended in 25 μl of kinase buffer (50 mm HEPES, pH 7.8, 10 mm MgCl2, 5 mm MnCl2, 0.1 mg/ml NaF, and 0.1 mm sodium orthovanadate) containing 50 μm ATP, 5 mm dithiothreitol, 1 μg of substrate, and 10 μCi of [γ-32P]ATP. Reactions were incubated for 30 min at 30 °C and stopped by adding an equal amount of 2× sample loading buffer. Reaction products were then fractionated on 15% SDS-PAGE followed by autoradiography or phosphorimaging analysis. The Cdk-associated kinase activity was determined by densitometric scanning of the substrate band.
Peptide Nucleic Acid Fluorescence in Situ Hybridization (FISH) Analysis—Hybridization of metaphase spreads (28) was performed with TRITC-OO-(TTAGGG)4 or peptide nucleic acid probes (Applied Biosystems) at a concentration of 0.5 μg/ml. A minimum of 25 metaphases was scored blinded for each sample, and pairwise comparisons for statistical significance were made by Student's t test. Images were captured with Metamorph Premiere (Molecular Devices) and processed with Metamorph and Adobe Photoshop CS.
Tissue Recombination (TR) Assays—Basic procedures (12) can be found in an illustrated tutorial on the World Wide Web. Briefly, the rat urogenital sinus mesenchyme (rUGM) was isolated from E17 or E18 rat embryos. NHP cells (200,000 cells) were then mixed with one rUGM in collagen and incubated overnight prior to transplantation. The following day, tissue recombinants were surgically transplanted under the renal capsule of male NOD/SCID mice supplemented with testosterone pellet. Grafts were collected and analyzed 3 or 6 months later. Paraffin-embedded sections (4 μm) or cryosections (8 μm) were used in immunostaining for various marker proteins.
cDNA Microarray Analysis—Total RNA was extracted from NHP cells (see Fig. 12A) using an RNeasy RNA purification kit (Qiagen, Valencia, CA), including on-column DNase digestion to completely remove contaminating genomic DNA. RNA concentrations were determined using NanoDrop, and the quality was assessed using a BioAnalyzer. Microarray experiments were carried out using the 44 K 60-mer “Human Whole Genome Oligo Microarray Kit” from Agilent (Agilent Technologies, Santa Clara, CA) with 500 ng of total RNA as starting material according to the manufacturer's protocol. Samples 3, 4, 5, 7a, 9, 11, and 13 were all hybridized to sample 1 (see Fig. 12A) in triplicate; therefore, a total of 21 G4112F-014850 Agilent microarrays (each array containing 45,015 probe sets) were used. Hybridization was performed at 65 °C for 17 h, and hybridized arrays were scanned with Agilent's dual laser-based scanner. Feature Extraction software GE2-v5_91 was used to link a feature to a design file and determine the relative fluorescence intensity between the two samples.
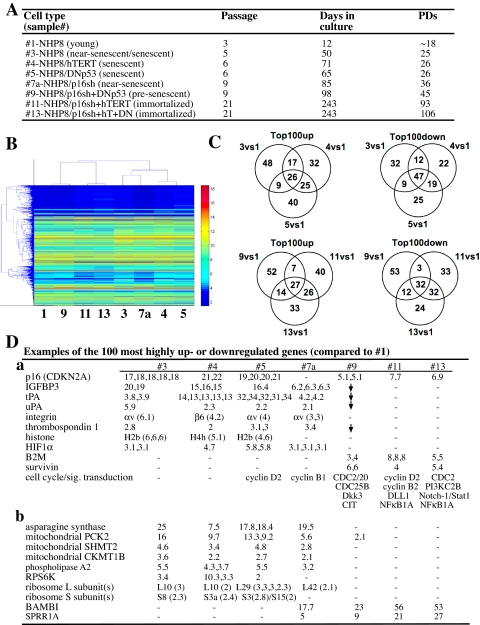
FIGURE 12.
Gene expression profiling of young, senescent, and immortalized NHP cells. A, NHP cells used in microarray analysis. “Near senescent” means approaching full senescence (e.g. 7a-NHP8/p16sh cells at 36 PDs; these cells had a total life span of ∼39 PDs). “Presenescent” means further away from senescence (e.g. 9-NHP8/p16sh + DNp53 cells at 45 PDs; these cells had a total life span of 54 PDs). B, hierarchical clustering of the samples based on the intersection of the significant genes (i.e. 1585 genes) using 1-correlation as distance and average linkage and the absolute intensity values on each channel. The genes (y axis) are sorted in ascending order of mean intensity. C, Venn diagram presentations of the 100 most highly up-regulated and down-regulated genes. See “Materials and Methods” for details. D, examples of 100 most highly up-regulated (a) or down-regulated (b) genes. The numbers indicate -fold changes, and some genes (e.g. p16) show changes with multiple probe sets. -, no increase in the 100 most highly up/down-regulated gene lists. ↓, decreased expression levels. CIT, Rho-interacting, serine/threonine kinase 21; CKMT1B; creatine kinase, mitochondrial 1B; Dkk3, Dickkopf homolog 3; delta-like 1; PCK2, phosphoenolpyruvate carboxykinase 2; RPS6K, ribosomal protein S6 kinase; SHMT2, serine hydroxymethyltransferase 2.
We used the Lowess-normalized data provided with the data files (columns gDyeNormSignal and rDyeNormSignal). Using the annotation provided by Agilent (available on the company website). We focused on the 18,841 distinct gene symbols, with each symbol corresponding to one or more probe sets. In the case of multiple probe sets per gene, they were treated as additional replicates for the analysis. For each one of the 18,841 genes, we identified the corresponding probe sets (one or more) and applied a paired t test between the group of interest and the control (sample 1). The paired t test yields t-statistic values as well as p values to assess statistical significance of differential expression. For each comparison between the group of interest and control, a β-uniform analysis was performed on the correspondingp values (37) to control for the false discovery rate that is typically used to account for multiple testing in high throughput data. In this case, we used a rather small value of the false discovery rate, equal to 0.0001. All comparisons yielded a varying and large number of significant genes. Table 2 shows the number of significant genes found by β-uniform analysis.
TABLE 2.
| Comparison | Number of significant genes (false discovery rate = 0.0001) |
|---|---|
| Type 3 versus control (type 1) | 5073 (p value cut-off = 0.0028) |
| Type 4 versus control (type 1) | 4063 (p value cut-off = 0.0023) |
| Type 5 versus control (type 1) | 3176 (p value cut-off = 0.0018) |
| Type 7a versus control (type 1) | 4691 (p value cut-off = 0.0025) |
| Type 9 versus control (type 1) | 3618 (p value cut-off = 0.0019) |
| Type 11 versus control (type 1) | 3281 (p value cut-off = 0.0019) |
| Type 13 versus control (type 1) | 3068 (p value cut-off = 0.0018) |
RESULTS
Primary NHP Epithelial Cells are CK5 and CK18 Double Positive Intermediate Basal Cells That Also Express Many Other Progenitor Cell Markers—We studied 12 primary NHP epithelial cell strains (Table 1), derived either from normal organ donors without prior history of prostatic diseases (including NHP1, -3, -4, and -6-10) or from uninvolved, benign prostate tissues distant from PCa foci (NHP2, -5, -11, and -12). We routinely cultured the purified prostate epithelial cells on collagen-coated dishes in epithelium-specific medium termed PrEBM(EGF + Ins) (9) (see “Materials and Methods”). Under these culture conditions, we generally obtained enough pure NHP epithelial cells for experiments at passage 2 (P2), at which time the cells had undergone 16-19 cumulative population doublings (PDs). NHP2, NHP6, and NHP7 cells, at P2, were all CK5+/CK18+ (i.e. double positive) epithelial cells that also expressed the reported prostate stem/progenitor cell markers p63, hTERT, α2β1, and CD44, and none expressed luminal differentiation markers 15-LOX2, AR, PSA, prostatic acid phosphatase, and CD57 or NE cell markers neuron-specific enolase (NSE), chromogranin A, or synaptophysin (1, 9, 38). Immunophenotypic characterizations of all other NHP strains, including NHP1, NHP3 to -5, and NHP8 to -12 cells (Table 1), at P1 or P2, using antibodies for various markers (Table S1), similarly showed that essentially all of these NHP cells were CK5+/CK18+p63+hTERT+α2β1+CD44+ and negative for luminal and NE cell markers, as illustrated for NHP8 (Fig. 1, P2), NHP9 (Fig. S1), and NHP10 (Fig. S2) cells. For example, NHP8 cells at P2 were essentially all double positive for CK5 and CK18 and were also positive for CD44, α2β1, p63, and hTERT (Fig. 1). These observations suggest that in PrEBM(EGF + Ins) culture conditions, the primary NHP epithelial cells are CK5+/CK18+ intermediate basal-like cells (4, 20) that also express many other reported progenitor cell markers (see Introduction).
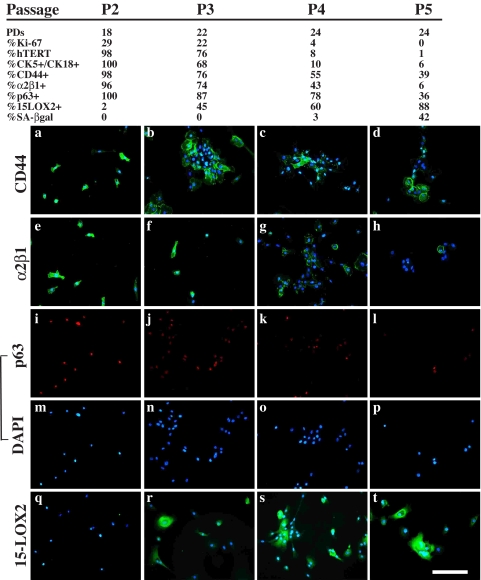
FIGURE 1.
Cultured NHP8 cells lose progenitor cell markers as they lose proliferative capacity. NHP8 cells at different passages were immunostained for the molecules indicated on top and counterstained by 4′,6-diamidino-2-phenylindole (DAPI). The passage number, cumulative PDs, and the percentages of proliferating (i.e. Ki-67+), hTERT+, CK5+/CK18+, p63+, CD44+, α2β1+, 15-LOX2+, and SA-β-galactosidase+ cells are indicated. Shown below are some representative microphotographs. Shown are cells at P2 (panels a, e, i, m, and q), P3 (panels b, f, j, n, and r), P4 (panels c, g, k, o, and s), and P5 (panels d, h, l, p, and t).
NHP Epithelial Cells Lose Proliferative Capacity in Culture and Undergo Senescence; Loss of Progenitor Markers Accompanies Senescence—We next assessed the proliferative capacity of serially passaged NHP cells and determined changes in various progenitor cell markers. As illustrated in Fig. 2A, NHP6 cells, derived from a 26-year-old normal organ donor (Table 1), underwent ∼17 cumulative PDs at P2 and, by P5, had undergone a total of ∼26 PDs, which did not further increase at P6-P7. NHP6 cells in culture showed a gradual decrease in cumulative BrdUrd labeling such that by P6, cells no longer incorporated BrdUrd and cell proliferation at the population level essentially stopped as a 5-day continuous BrdUrd pulse led to <10% labeling (Fig. 2, B and C). Starting from P5, many NHP6 cells displayed large and flat morphology and stained positive for SA-β-galactosidase (Fig. 2C), and at P6 and P7, senescent NHP6 cells increased to ∼50 and 90%, respectively. Similar passage-related senescence was observed in other NHP cells examined (Fig. 1) (9) (also see below). In general, NHP cells fully senesced at ∼20-26 PDs and, chronologically, within ∼2 months after initial plating. These observations indicate that 1) primary NHP cells in culture undergo senescence asynchronously, and the percentage of senescent cells correlates with cumulative PDs and 2) there is a noticeable gap between the cessation of PDs and the full manifestation of senescent phenotype, consistent with cell-cycle arrest preceding senescence. On these observations and considerations, we have used several terms in the forgoing discussions, which include young (P2-P3, low PDs), senescent or fully senescent (at the end of total PDs), near senescent (approaching the total PDs), and presenescent (intermediate between young and near senescent) (9, 28).
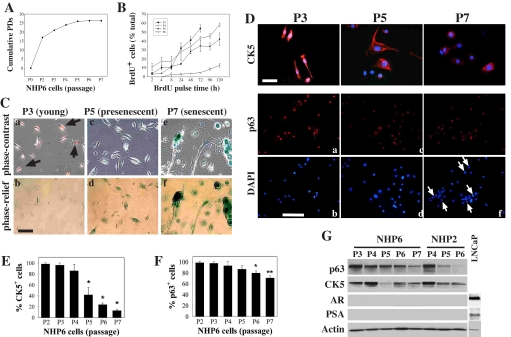
FIGURE 2.
NHP6 progenitor cells in culture lose progenitor markers and proliferative capacity and become senescent. A, cumulative PDs of NHP6 cells cultured in serum/androgen-free PrEBM(EGF + Ins). B, cumulative BrdUrd labeling. The percentage of BrdUrd+ cells was determined by counting 500-1000 cells for each passage (values represent the mean ± S.E. from two experiments). C, representative photomicrographs showing NHP6 cells at P3, P5, and P7 stained for BrdUrd (10 μm × 4 h; red) and SA-β-galactosidase (blue). The images in the lower panels were taken using the phase relief contrast filter to better reveal the SA-β-galactosidase staining. Bar, 2 μm. D, NHP6 cells stained for CK5 (top panels; bar, 2 μm) or p63 (lower panels; bar, 10 μm). The arrows in f indicate the cells that have lost p63 labeling. E and F, quantification of filamentous CK5-positive (E) or p63-positive (F) cells. 600-1200 cells were counted for each passage, and the data represent mean ± S.E. from 2-3 independent counts. E,*, p < 0.01. F,*, p < 0.05; **, p < 0.01. G, Western blotting analysis of marker molecules in NHP6 and NHP2 cells. LNCaP cells were used as positive control for AR and PSA.
NHP6 cells, which were all positive for CK5/CK18, CD44, p63, α2β1, and hTERT at P2 (1), became partially (95%) positive for filamentous CK5 and nuclear p63 at P3 (Fig. 2, D-F). By P5, ∼40% of the NHP6 cells expressed filamentous CK5, and <90% of the cells stained positive for nuclear p63 (Fig. 2, D-F). By P7, when most NHP6 cells were fully senescent, only ∼10% of the NHP6 cells expressed filamentous CK5, and ∼70% of the cells showed weak p63 (Fig. 2, D-F). Western blotting verified decreases in p63 and CK5, which were also observed in senescing NHP2 cells (Fig. 2G). Importantly, as NHP6 cells lost the marker expression and proliferative capacity, they did not up-regulate differentiation markers AR and PSA (Fig. 2G).
Systematic analysis of NHP8 cells, derived from a 41-year-old normal donor (Table 1), revealed similar loss of progenitor markers (Fig. 1). We correlated the marker expression with the proliferative (PDs and Ki-67 staining) and senescence (15-LOX2 and SA-β-galactosidase staining) parameters (Fig. 1). At P2, when the NHP8 cells had undergone ∼18 PDs, 29% of the cells were actively proliferating, and the majority of the cells were CK5+/CK18+CD44+α2β1+p63+, with ∼2% of the cells positive for 15-LOX2 (an NHP cell senescence marker) (9), and no cells were positive for SA-β-galactosidase. By P4, when there was no longer a PD increase, only 4% of the NHP8 cells were proliferating, and all progenitor marker-expressing cells were significantly decreased with the 15-LOX2-expressing, senescing NHP cells increased to 60% (Fig. 1). By P5, when the NHP8 cell cultures became terminally senescent, there was no longer cell proliferation; ∼90% of the cells stained positive for 15-LOX2, and ∼40% of the cells became SA-β-galactosidase-positive. By this time, the majority of the NHP8 cells had lost the expression of CK5 and/or CK18 and α2β1, and ∼60% of the cells lost CD44 and p63 expression (Fig. 1).
NHP2 and NHP7 cells, like NHP6 and NHP8 cells, also lost most progenitor markers, including CK5/CK18, p63, CD44, and α2β1, as they reached the end of their proliferative life span (Fig. 2G) (9). Importantly, hTERT expression, similar to CK5/CK18, CD44, α2β1, and p63, also showed passage-related loss in NHP6 (28), NHP7 (Fig. 3), and NHP8 (Fig. 1) cells.
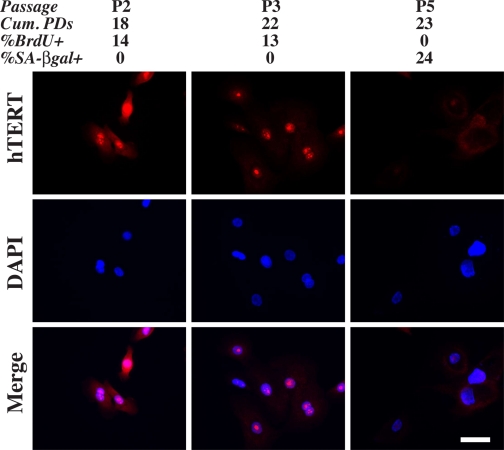
FIGURE 3.
Loss of hTERT expression accompanies NHP7 cell senescence. NHP7 cells at different passages were stained for hTERT. The percentages of hTERT+ cells at P2, P3, and P5, were 99, 70, and 2%, respectively (a total of 1000-1300 cells counted). The passage number, cumulative PDs, and percentages of BrdUrd- and SA-β-galactosidase-positive cells are indicated on top. In these experiments, the NHP7 cells at different passages were BrdUrdpulsed for 4 h. Bar, 2 μm. DAPI, 4′,6-diamidino-2-phenylindole.
These observations suggest that 1) NHP cells lose both intracellular and surface progenitor markers as they lose their proliferative capacities and enter senescence and 2) expression of the progenitor markers may be functionally linked to the proliferative status of NHP cells.
Culturing Primary NHP Cells in the Presence of Basic Fibroblast Growth Factor, Wnt3a, and LIF or on Fibroblast Feeders with or without hTERT Transduction Fails to Extend Their Proliferative Life Span—The preceding experiments suggest that the PrEBM(EGF + Ins) mixture is unable to maintain primary NHP epithelial cells as proliferating cells that express progenitor markers. To explore whether we can find suitable culture conditions that could extend the proliferative life span of NHP cells, we added basic fibroblast growth factor, Wnt3a, or LIF (leukemia-inhibitory factor), which can promote self-renewal of neural, hematopoietic, and mouse embryonic SCs, respectively, either individually or in combination, to PrEBM(EGF + Ins). As shown in Fig. S3A, these growth factors did not increase the PDs of primary NHP9 or NHP11 cells. Culturing these cells on a fibroblast feeder layer also failed to extend their life span (Fig. S3B). Human keratinocytes and mammary epithelial cells have been shown to go beyond their normal replicative life span when transduced with the catalytic subunit of telomerase (hTERT) and cultured on fibroblast feeder layers (39). However, when NHP10 or NHP11 cells were infected with an hTERT-encoding retrovirus and cultured on the fibroblast feeder layer, they similarly stopped proliferation and underwent senescence after ∼2 months (Fig. S3, C and D). These observations suggest that the above culture conditions also do not allow the long term maintenance of NHP (progenitor) cell properties.
p16 Up-regulation and p53 Activation Accompany NHP Cell Senescence; Enforced Expression of p16 Is Sufficient to Limit NHP Cell Proliferation—Subsequently, we turned our attention to the intracellular mechanisms that regulate the proliferative life span of NHP cells. We first examined the changes in cell cycle-related proteins in serially passaged NHP6 cells and found that multiple negative cell cycle regulators were up-regulated as NHP6 cells underwent senescence (Fig. 4A). Specifically, p16, an INK family cyclin-dependent kinase inhibitor (CKI) that inhibits the cyclin D1/D2-Cdk4/6 complexes, was up-regulated (Fig. 4A; also see Fig. 5C). The p16 protein was undetectable in multiple strains of NHP cells at P2-P3 (≤20 PDs) and thereafter began to accumulate (Fig. 4A and Fig. S4). p14ARF, an alternate product of the ink4a locus that activates p53 through inhibiting MDM2, was also undetectable in NHP6 cells at P3-P5 but slightly increased at ≥P6 (Fig. 4A). Unlike p16, the other three INK family members, p15, p18, and p19, were expressed at undetectable levels in both young and senescent NHP cells (Fig. 4A) (data not shown). The three Cip/Kip family members (i.e. p21, p27, and p57), which have broad inhibitory effects on all Cdks, showed variable changes. Both p21 and p27 were up-regulated at P6 and then declined slightly (Fig. 4A). p57 was expressed at very low levels in NHP6 cells and did not undergo significant changes (Fig. 4A). Both p53 and active (i.e. hypophosphorylated) pRb were up-regulated at P5, which was slightly later than p16 induction (Fig. 4A and Fig. S4). Interestingly, although p53 was mainly localized in the nucleus, the up-regulated p16 was distributed in both the nucleus and cytoplasm (Fig. S4), as previously observed in some tumor cells (40).
FIGURE 4.
NHP cell senescence involves both p16/pRb and p53 pathways. A, negative cell cycle regulators. Whole cell lysates (100 μg/lane) were used in Western blotting. The antibody for pRb recognizes only the hypophosphorylated (active) form. LNCaP cells (having wild type p53) were used as control (30 μg/lane), which showed up-regulated p53 and p21 as well as active pRb in response to etoposide (eto;10 μm for 48 h). Note that etoposide treatment decreased p16, p19, and p27 in LNCaP cells. B, Western blotting analysis of positive cell cycle regulators. Note that in LNCaP cells, etoposide treatment decreased all positive cell cycle regulators except Cdk4, which actually increased. C, P3 or P6 NHP6 cells (107 cells) were used in immunoprecipitation (IP) using anti-Cdk4 antibody. The immunoprecipitates were run on Western blot, and the blot was probed for p16 (top) or used in kinase assays using either histone H1 (middle) or Rb peptide (bottom) as the substrate. Note that immunoprecipitation using the control RgIgG antibody did not reveal a specific band (not shown). D, NHP8 cells (P3; 20 PDs) were plated at 250,000 cells/T25 and infected with either pLXSN-p16 (p16) or the control vector pLEGFP (GFP). Cell number was determined 26 days after infection and selection with G418 (200 μg/ml). Bars, mean ± S.D. E, NHP8 cells (P3; 20 PDs) were plated at 250,000 cells/T25 and infected with the empty vector (pBabe), pBabe-p16 (p16), or pBabe-p16R418 (p16m). Cell number was determined 9 days after infection and selection with puromycin (1 μg/ml). Bars, mean ± S.D. *, p < 0.01 for D and E.
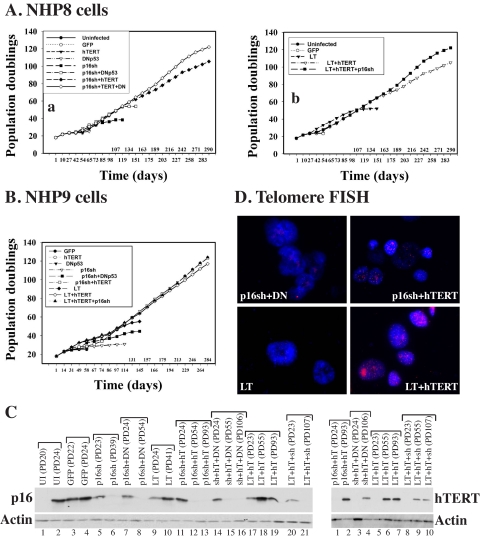
FIGURE 5.
Genetically defined life span extension and immortalization of NHP progenitor cells. NHP8 (A) or NHP9 (B) cells were infected at PD18 with the indicated vectors, either individually or in different combinations. Cumulative PDs were determined in continuous cultures. C, Western blotting (25 μg/lane) showing the down-regulation of p16 by p16sh (left) or overexpression of hTERT (right) in the hTERT (hT)-infected long term NHP cells. Note that in lanes with low PDs (i.e. recently infected cultures) p16 suppression was not complete or hTERT overexpression was not obvious, because cultures were still mixtures containing both infected and uninfected cells. UI, uninfected; p16sh or sh, cells infected with p16 shRNA; LT, large T; DN, dominant negative p53. D, telomere FISH analysis showing loss of telomere in near senescent NHP8 cells infected with p16sh plus DNp53 (DN) or large T and strong telomere FISH signals in NHP8 cells infected with either p16sh plus hTERT or large T plus hTERT (both at ∼85 PDs). Note that young NHP8 cells showed telomere FISH signals similar to those in NHP8/p16sh + hTERT cells (data not shown due to space constraints), consistent with the immunostaining results (Fig. 1, P2).
The up-regulation of CKIs and activation of pRb suggests that NHP cells undergoing senescence are probably arrested in the G1 phase. In support, several G1-positive cell cycle regulators, including cyclins D1 and D2 and Cdk4 and -6, were also up-regulated (Fig. 4B), suggesting that the up-regulated CKIs may be binding to the cyclin D1/D2-Cdk4/6 complexes and rendering them inactive. Indeed, co-immunoprecipitation revealed more p16 bound to Cdk4 in senescent NHP cells (Fig. 4C). As a result, coupled immune complex kinase assays revealed that the Cdk4-associated kinase activities, measured by two different substrates (i.e. histone H1 or the C-terminal peptide of Rb), were significantly lower in P6 than in P3 NHP6 cells (Fig. 4C). In contrast to cyclin D1/D2 and Cdk4/6, several positive cell cycle regulators mainly active in the G1-S transition or S phase, including cyclin E1, cyclin A, and Cdk2, did not significantly change or only slightly increased (for Cdk2) in senescing NHP6 cells (Fig. 4B). Among the four E2F molecules examined, E2F1 to -3 were expressed at undetectable levels, whereas E2F4 was expressed at low levels and slightly decreased in late-passage NHP6 cells (Fig. 4B).
The above results, together with loss of telomerase expression (e.g. Fig. 3), suggest that three signaling pathways (i.e. p16/pRb, p53 and telomerase/telomere) may be causally involved in NHP cell senescence. To begin dissecting their involvement, we first carried out gain-of-function experiments by overexpressing p16, using two different retroviral vectors encoding wild-type p16 (41, 42), in young NHP cells. Enforced expression of wild-type p16, in both cases, inhibited cell proliferation, resulting in reduced cell numbers (Fig. 4, D and E). Many of the infected cells were positive for SA-β-galactosidase (not shown). In contrast, a mutant p16 (p16m) that lacks the Cdk-binding activity did not show any effect (Fig. 4E).
p16 Is the Primary Determinant of NHP Cell Proliferative Life Span, and Telomerase Is Required for NHP Cell Immortalization—To systematically determine the involvement of p16, telomerase, and p53 in regulating NHP cell life span, we infected the P3 NHP8, NHP9, or NHP11 cells with retroviral vectors encoding GFP, p16 short hairpin RNA (p16sh) (43), a dominant negative p53 (DNp53) (41), hTERT, or SV40 large T (LT) (44), either individually or in different combinations. The uninfected (UI) and GFP-infected NHP8 (Fig. 5A, a) and NHP9 (Fig. 5B) cells underwent senescence after ∼24 PDs. hTERT expression alone did not affect the NHP8 (Figs. 5A (a) and 6), NHP9 (Fig. 5B), or NHP11 (not shown) cell life span, suggesting that loss of telomerase expression was not the initiating event in NHP cell senescence. Similar to hTERT, DNp53 alone did not affect or only slightly extended (by 1-2 PDs) NHP8 (Figs. 5A (a) and 6), NHP9 (Fig. 5B), or NHP11 (not shown) cell life span, suggesting that p53 induction (Fig. 4A) did not cause NHP cell senescence.
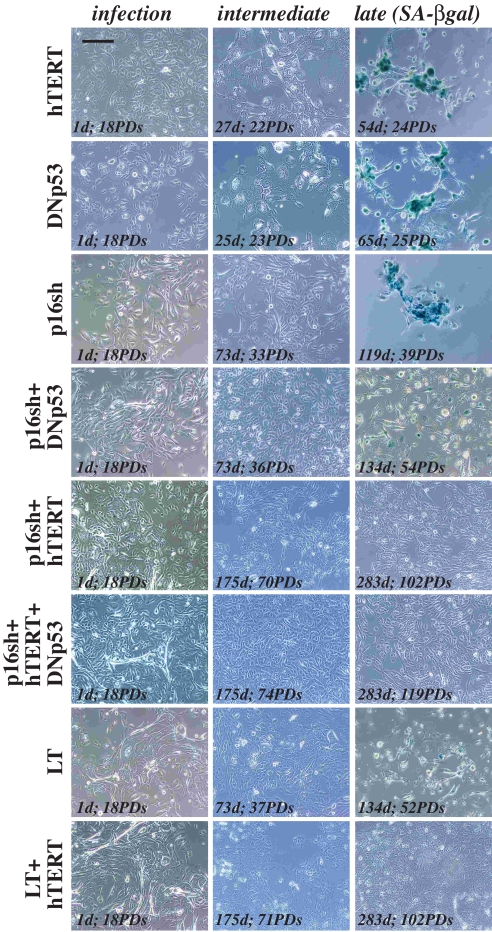
FIGURE 6.
Genetically defined immortalization of NHP progenitor cells. Representative images of NHP8 cells infected with retroviral vectors encoding the molecules indicated on the left. Three representative images are shown. Note that uninfected and GFP-infected cells (not shown) behaved like the hTERT- and DNp53-infected cells. Bar, 10 μm.
In contrast to hTERT and DNp53, p16sh alone more than doubled the chronological culture time (i.e. from 54 to 119 days) and extended the life span of NHP8 cells from 24 to ∼40 PDs (Figs. 5A (a) and 6). The uninfected and GFP-infected NHP8 (i.e. NHP8/GFP) cells approaching senescence expressed significantly increased levels of p16 (Fig. 5C, left, lanes 2-4). Early after p16sh infection, a low level of p16 protein was detected (Fig. 5C, left, lane 5), probably because at PD23 (5 PDs after infection) the cultures contained both infected and uninfected cells. By PD33, p16 protein could no longer be detected (Fig. S5A), and NHP8 cells at this time expressed wild type p53 (Fig. S5B) but not mutant p53 (Fig. S5C), as determined by negative staining by a mutant p53-specific antibody (Table S1) (28). The p16sh-infected NHP8 (i.e. NHP8/p16sh) cells at PD33 expressed high levels of p63, CD44, and α2β1 (Fig. S5, D and E) and were CK5+/CK18+ (Fig. S5G). At PD39, when NHP8/p16sh cells were senescent (Fig. 6), cells did not express p16 (Fig. 5C, left, lane 6, and Fig. S6A) but expressed high levels of nuclear wild type p53 (Fig. S6B). Senescent NHP8/p16sh cells expressed slightly reduced p63 (Fig. S6C), were no longer CK5+/CK18+ due to loss of CK18 (Fig. S6D), and mostly retained CD44 (Fig. S6E) but had completely lost α2β1 (Fig. S6F). Interestingly, many cells became positive for 15-LOX2 (Fig. S6H), a molecule that contributes to NHP cell senescence (9). p16sh also significantly extended the proliferative life span of NHP9 cells (Fig. 5B). Together, these results indicate that 1) p16 up-regulation is the primary driving force of NHP cell senescence, 2) p16sh extends NHP cell proliferative life span in the presence of up-regulated wild type p53, and 3) senescent NHP8/p16sh cells, like regular NHP8 cells (Fig. 1), lose or show reduced expression of progenitor markers.
p16sh alone was, nevertheless, insufficient to immortalize NHP cells although p16 suppression was obvious (e.g. Fig. S6A). One possibility could be due to p53 up-regulation and activation caused by slightly increased p14ARF (Fig. 4A). In support, p16sh in combination with DNp53 extended the NHP cell life span beyond that conferred by p16sh alone (i.e. from ∼40 PDs to ∼54 PDs (Figs. 5, A (a) and B, and 6)). Senescing NHP8/p16sh + DNp53 cells showed decreasing expression of all progenitor markers, including CD44, α2β1, p63, and CK5 (Fig. S7), and fully senescent cells did not express p16 (Fig. 5C, left, lane 8). Remarkably, infection of NHP8 cells with LT, which abrogates the functions of both p53 and pRb (all pocket proteins), induced life span extensions that could be superimposed on NHP8/p16sh + DNp53 cells (Figs. 5A (a and b) and 6). LT also extended the NHP9 cell life span beyond that conferred by p16sh alone (Fig. 5B). Significantly, LT extended NHP8 cell life span in the presence of increased p16 (Fig. 5C, left, lane 10), suggesting that in NHP cells, p16 functions mainly through pRb. Senescent NHP8/LT cells showed reduced p63 (Fig. S8A) and had mostly lost CD44, α2β1 and CK5/CK18 double positivity (Fig. S8B).
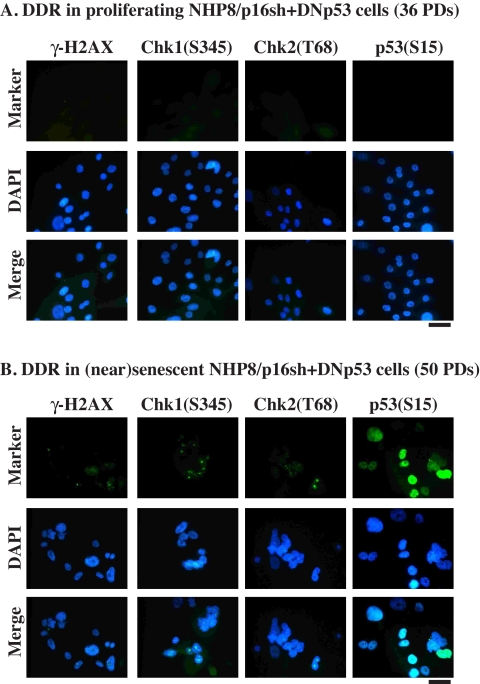
That p16sh, p16sh plus DNp53, and LT all significantly extended NHP cell life span but failed to immortalize these cells (Fig. 5, A and B) suggests that extensive cell proliferation conferred by these conditions most likely resulted in telomere erosion in the absence of telomerase expression. In support of this suggestion, telomere FISH analyses revealed that near senescent NHP8/p16sh, NHP8/p16sh + DNp53, or NHP8/LT cells showed reduced telomere signals (Fig. 5D; data not shown). Telomere shortening/erosion would activate DNA damage response (DDR). Indeed, near senescent and senescent NHP8/p16sh + DNp53 (Fig. 7) or NHP9/LT (Fig. S9) cells showed positive staining for phosphorylated, activated checkpoint response proteins H2AX (i.e. γ-H2AX), Chk1(Ser345), and Chk2(Thr68) as well as activated p53(Ser15), whereas their proliferating counterparts stained mostly negative for these markers.
FIGURE 7.
DDR in near senescent/senescent (PD50) NHP8/p16sh + DNp53 cells but not in proliferating (PD36) cells. The cells indicated were immunostained for phosphorylated H2AX, Chk1, Chk2, or p53, using the antibodies listed in Table S1. Bars, 2 μm. DAPI, 4′,6-diamidino-2-phenylindole.
Telomere shortening (due to extensive cell proliferation) in the absence of telomerase expression would conceivably limit the further proliferation of life span-extended NHP cells. Consequently, enforced expression of hTERT in NHP/p16sh, NHP/p16sh + DNp53, or NHP/LT cells resulted in cell immortalization (Fig. 5, A and B). hTERT expression, which was detected on Western blotting in the hTERT-immortalized cells (Fig. 5C, right), led to increased telomere signals (Fig. 5D) and telomerase activity (not shown). Critically, the immortalized NHP8/p16sh + hTERT and NHP8/p16sh + DNp53 + hTERT cells (Fig. 8) and NHP8/LT+hTERT cells (Fig. S10) all expressed strong p63, CD44, α2β1, and CK5, although they mostly lost CK18 expression. As expected, p16 was not detected in NHP8 cells immortalized with p16 shRNA in conjunction with other conditions (Figs. 5C (left, lanes 12, 13, 15, 16, and 21) and 8A); in contrast, both nuclear and cytoplasmic p16 were detected in the immortalized NHP8/LT + hTERT cells (Fig. S10A), again suggesting that p16 functions in NHP cells through pRb. That hTERT in conjunction with p16 shRNA, without suppressing p53, was sufficient to immortalize NHP8 cells (Fig. 5A) further supports the contention that p16 is the primary determinant of NHP cell life span. On the other hand, we consistently observed that NHP8/p16sh + hTERT + DNp53 cells proliferated faster than NHP8/p16sh + hTERT cells (Fig. 5A (a)), suggesting that, as observed for the difference between NHP/p16 cells versus NHP/p16sh + DNp53 cells, p53 activation does partially restrict NHP cell proliferative capacity. Finally, the proliferative rates of NHP9/LT + hTERT cells and NHP9/LT + hTERT + p16sh cells were very similar (Fig. 5B), indicating that p16 suppression on top of pRb inhibition (by LT) does not offer any further advantages, which once again supports that in NHP cells p16 functions through pRb.
FIGURE 8.
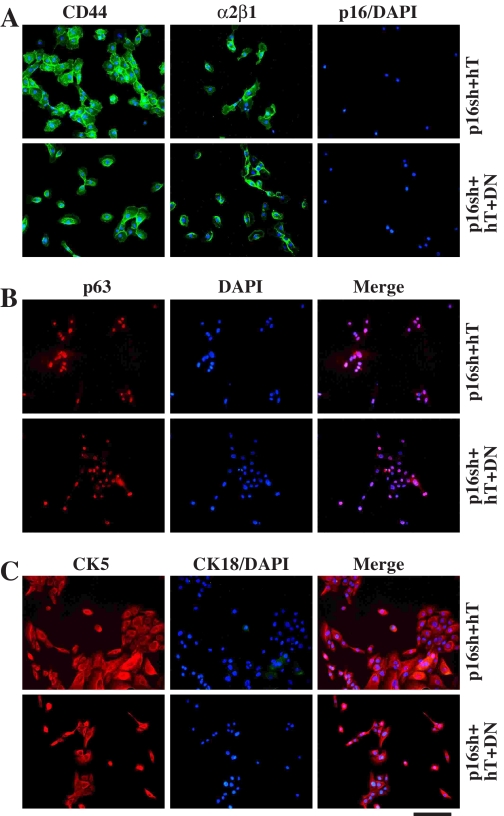
Immunophenotyping of the immortalized NHP8/p16sh + hTERT (78 PDs) and NHP8/p16sh + hTERT + DNp53 (86 PDs) cells. Representative images of the infected NHP8 cells stained for surface markers CD44 or α2β1(A), nuclear marker p63 (B), and cytoskeletal markers CK5/CK18 (C). Bar, 2 μm.
The Immortalized NHP Cells Maintain Normal DDR and Are Nontumorigenic—Immortalized NHP cells showed contact inhibition of proliferation, and most expressed basal (progenitor) cell markers CD44, α2β1, p63, and CK5 (Fig. 8). Immortalized NHP8 cells expressed wild-type p53, as determined by antibody staining and by SSCP and sequencing analyses (not shown). G-banding revealed that immortalized NHP8/p16sh + hTERT, NHP8/LT + hTERT, NHP8/p16sh + hTERT + DNp53, and NHP9/p16sh + hTERT cells all maintained grossly normal karyotypes (not shown). Furthermore, immortalized NHP8 cells demonstrated normal ATM-dependent DDR, as evidenced by the appearance of discrete foci containing phosphorylated H2AX, Chk1, and Chk2 in response to DNA double strand breakers, such as γ-irradiation and etoposide (Fig. 9 and Fig. S11). Finally, immortalized NHP cells were nontumorigenic (Table S2). Although PCa cells (PC3, Du145, and LAPC9) regenerated tumors with 10,000 or 100,000 cells implanted into the prostate of NOD/SCID mice, one million or more NHP8 cells immortalized by p16sh plus hTERT or by LT + hTERT did not give rise to any tumor (Table S2). Enforced expression of AR, previously shown to increase prostate epithelial cell tumorigenicity (45), did not confer any tumorigenic potential (Table S2).
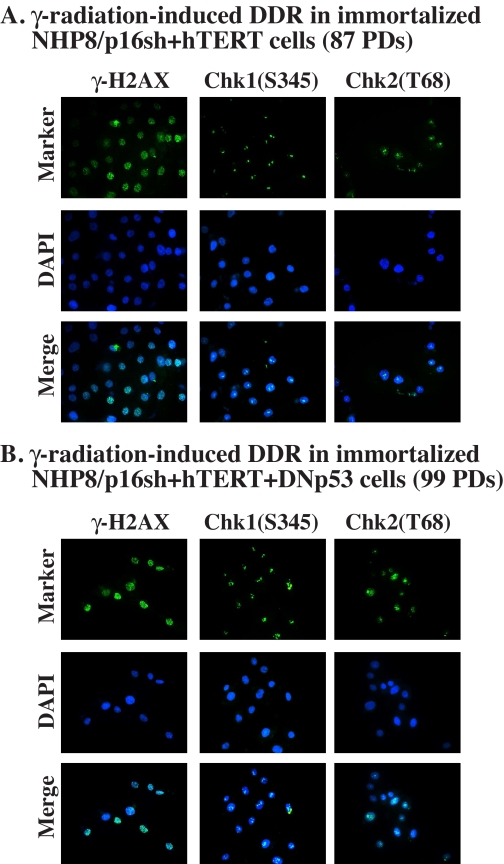
FIGURE 9.
Normal DDR in immortalized NHP8/p16sh+hTERT (A) or NHP8/p16sh + hTERT + DNp53 (B) cells. Cells at the indicated PDs were irradiated with 20-gray x-ray for 10 min and then fixed and immunostained for phosphorylated H2AX, Chk1, or Chk2 using the antibodies listed in Table S1. Note that the phosphorylated γ-H2AX was detected as discrete intranuclear granules, whereas phosphorylated Chk1 and Chk2 were generally detected as intranuclear speckles or dots. The staining intensities were significantly increased in cells irradiated for 45 min (not shown). Treatment of cells with another DNA damage agent, etoposide, induced very similar DDR (not shown). Note that cells without treatments or stained with secondary antibody alone did not reveal any specific staining. DAPI, 4′,6-diamidino-2-phenylindole.
The Immortalized NHP Cells Differentiate into Prostatic Glands in Vivo—To determine whether the immortalized NHP cells possess any stem/progenitor cell properties in vivo, we carried out TR assays (12) by recombining different numbers of immortalized NHP8 cells with rUGM. Similar TR assays using rUGM alone were used as controls. In total, we carried out ∼200 TRs in 60 mice. TRs were harvested either 3 months (Table 3 and Figs. 10 and 11, A and B) or 6 months (Table 3, Fig. 11, C and D, and Fig. S12) after transplantation. Comprehensive histological and IHC analyses revealed the following important points. First, TR assays using rUGM alone (n = 16) did not regenerate any glandular outgrowths (not shown). Second, regular (i.e. unimmortalized) NHP8 cells formed typical outgrowths at 6 months but not 3 months. In contrast, all immortalized NHP8 cells regenerated well differentiated glands 3 months after transplantation (Table 3, Figs. 10 and 11, and Fig. S12). Third, upon hematoxylin/eosin staining, the prostatic glands formed by regular and immortalized NHP cells were similar to those previously described for organoid-derived glands (12) and were morphologically heterogeneous with many glands filled with eosinophilic secretions. Fourth, the NHP cell-derived prostatic glands possessed well demarcated luminal and basal layers. AR and CK8 were expressed exclusively in the luminal cells, whereas CK5 was expressed exclusively in the basal layer. Unexpectedly, p63 appeared to localize in both basal and luminal cells (e.g. see Fig. 10, A (g) and B (i) and Fig. S12B (f)). Fifth, remarkably, in prostatic glands derived from regular NHP8 and most immortalized cells, NE-like cells positive for synaptophysin were observed. Most glands were negative or showed only sporadic synaptophysin-positive cells, whereas some glands showed numerous solitary or clusters of NE-like cells (Figs. 10B (k and l) and 11D (i) and Fig. S12, B (g-i) and C (h)). The presence of NE-like cells was also confirmed by staining some cryosections for chromogranin A (not shown). Sixth, enforced expression of AR in the immortalized NHP cells did not increase the levels and sizes of the outgrowths (Table 3, Fig. 11, B and C, and Fig. S12C). Seventh, prostatic glands in the 3-month outgrowths all lacked PSA expression. However, a few glands at 6 months showed weak (e.g. see Fig. S12D (d), asterisk) or even strong (Fig. 11D (f), arrow) PSA staining, consistent with a time-dependent maturation process observed with human embryonic stem cell-derived prostatic glands (46). Finally, regenerated glands were of human origin, as evidenced by positive staining for human-specific mitochondria (hMito; Fig. 10, A (h) and B (c) and Fig. S12), Ki-67 (Fig. 10A (i)), or nuclei (hNuc; Fig. 11A (f)). Positive staining for PSA (Fig. 11D (f) and Fig. S12D (d)), which is human-specific, in some glands provides further evidence for the human origin of the NHP cell-derived glands. These TR experiments suggest that immortalized NHP cells are able to differentiate into functional prostatic glands that contain both basal and luminal epithelial cells as well as NE-like cells.
TABLE 3.
TR assays in immortalized NHP cells
| Cell type (PDs)a | Nude mouseb | Regeneration and microscopic observationsc |
|---|---|---|
| Type 1, NHP8 (18 PDs) | 427 (3 mo.) | No regeneration (0 of 4) |
| 426, 428, 429 (6 mo.) | Regeneration (2 of 4 + 3 of 4 + 2 of 4 = 7 of 12); well differentiated glands | |
| Type 2, NHP8/p16sh + hTERT (75 PDs) | 431 (3 mo.) | Regeneration (3 of 4); well differentiated glands |
| 430 (6 mo.) | Regeneration (3 of 4); well differentiated glands | |
| Type 3, NHP8/p16sh + hTERT + DNp53 (90 PDs) | 434 (3 mo.) | Regeneration (2 of 4); well differentiated glands |
| 432, 433, 435 (6 mo.) | Regeneration (4 of 4 + 2 of 4 + 1 of 4 = 7 of 12); well differentiated glands | |
| Type 4, NHP8/p16sh + hTERT + DNp53 + AR (90 PDs) | 3893 (3 mo.) | Regeneration (1 of 4); well differentiated glands |
| 3894, 3587, 3545 (6 mo.) | Regeneration (2 of 4 + 3 of 4 + 0 of 4 = 5 of 12); well differentiated glands | |
| Type 5, NHP8/LT + hTERT (75 PDs) | 3441 (3 mo.) | Regeneration (1 of 4); well differentiated glands |
| 3440 (6 mo.) | Regeneration (4 of 4); well differentiated glands | |
| Type 6, NHP8/LT + hTERT + AR (75 PDs) | 3442 (3 mo.) | Regeneration (2 of 4); well differentiated glands |
| 3443 (5 mo.) | Regeneration (3 of 4); well differentiated glands |
200,000 of the indicated NHP8 cells (six types in total) were recombined with one rUGM and transplanted under the renal capsule of male nude mice supplemented with testosterone pellets. The PDs for each cell type are indicated in parentheses.
Mouse number is indicated. Each mouse kidney received two recombinants (i.e. 4 TRs/mouse). Mice were terminated at either 3 months (3 mo.) or 6 months (6 mo.) after transplantation.
FIGURE 10.
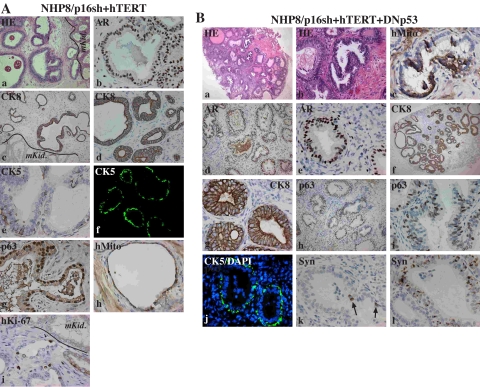
Immortalized NHP progenitor cells differentiate into prostatic glands in vivo. Shown is hematoxylin/eosin (HE) and immunohistochemical (IHC) staining of TRs of NHP8/p16sh + hTERT (A; number 431; Table 3) or NHP8/p16sh + hTERT + DNp53 (B; number 434) cells 3 months (mo.) after transplantation. Most were paraffin-embedded sections except for A (f) and B (j) (cryosections). The arrows in B (k) indicate scattered synaptophysin-positive cells. CK5 and CK8, cytokeratin 5 and 8, respectively; hMito, human mitochondria; hKi-67, human-specific Ki-67; Syn, synaptophysin; mKid, mouse kidney. DAPI, 4′,6-diamidino-2-phenylindole.
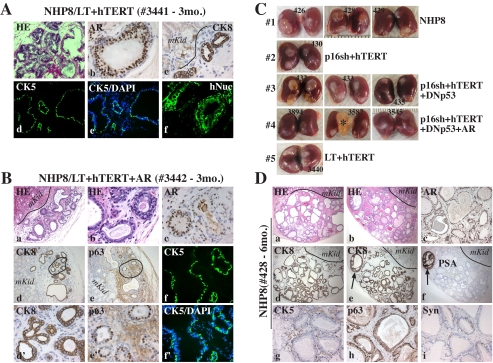
FIGURE 11.
Immortalized NHP progenitor cells differentiate into prostatic glands in vivo. Hematoxylin/eosin and immunohistochemical (IHC) staining of tissue recombinants derived from the indicated cell types either 3 months (A and B) or 6 months (C and D) after transplantation. Animal numbers are indicted (Table 3). CK8, cytokeratin 8; CK5, cytokeratin 5; hMito, human mitochondria; hKi-67, human-specific Ki-67; Syn, synaptophysin; mKid, mouse kidney. Note that AR and CK8-positive cells are localized in the luminal layer (A (b and c) and B (c and d′)), whereas CK5 specifically labels basal cells (A (d and e), B (f and f′), and D (g)). p63-positive cells are observed in both luminal and basal cells (B (e′) and D (h)). A, a and d-f, cryosections; b and c, paraffin-embedded sections. In B, f and f′ show cryosections, and the rest are paraffin-embedded sections. d′ and e′, enlarged microphotographs of d and e (circled), respectively. C, representative images of TR outgrowths at 6 months after renal capsule transplantation. Five cell types are shown, and individual animal numbers are indicated (Table 3). Note that the large outgrowth in animal 3587 (4; asterisk) was found to be negative for CK8, p63, and hMito (data not shown); therefore, it was probably derived from the contaminated rat epithelium. D, shown are two representative glandular outgrowths (a and b) derived from regular NHP8 cells in animal 428 (Table 3). Note the specific CK8 staining of glandular structures but not mouse kidney cells. The only PSA-positive gland is also positive for CK8 (e and f, arrows). Similar results were observed in glands in animal 426 (Table 3) with also one strongly stained PSA-positive gland (not shown). The prostatic glands in animal 429 did not show any PSA staining (not shown). DAPI, 4′,6-diamidino-2-phenylindole.
The Immortalized NHP Cells Possess Gene Expression Profiles Characteristic of Proliferating Progenitor Cells—Finally, we carried out expression analysis (Fig. 12A) using the 44 K 60-mer oligonucleotide microarrays (Agilent) representing ∼19,000 genes (see “Materials and Methods”). In total, 14,254 genes (all microarray data can be accessed at the NCBI site on the World Wide Web under the accession number GSE10570) showed differential expression in at least one of the seven samples (i.e. 3, 4, 5, 7a, 9, 11, and 13) when compared with young NHP8 cells (Fig. 12A). Unsupervised hierarchical clustering analysis based on Pearson correlation distance measurement and average linkage clustered the two immortalized cell types, presenescent NHP8/p16sh + DNp53 cells, and the young NHP8 cells in one group and the three senescent cell types (types 3-5) with near senescent NHP8/p16sh cells in the other group (not shown). Hierarchical clustering analysis using the 1585 genes that showed changes across all seven comparisons (with sample 1) revealed similar gene expression clustering patterns (Fig. 12B). Analysis of the 100 most highly up-regulated (Fig. 12C, left) and down-regulated (Fig. 12C, right) genes also revealed many commonly altered genes among senescent (Fig. 12C, top) or among presenescent/immortalized (Fig. 12C, bottom) cell types. These results suggest that the immortalized and senescent NHP cells exhibit distinct gene expression profiles.
Among the 100 most highly up-regulated genes, p16 expectedly turned up in multiple probe sets in all three senescent cell types but not in near senescent NHP8/p16sh cells (Fig. 12D). Increased p16 mRNA levels were observed in presenescent NHP8/p16sh + DNp53 and the two immortalized NHP8 cells, although the levels were significantly lower than in senescent NHP8 cells (Fig. 12D), suggesting that p16 mRNA suppression by p16 shRNA in these cells was incomplete. In addition to p16, IGFBP3 (IGF-binding protein 3), tissue plasminogen activator, urokinase plasminogen activator, several integrin subunits, and thrombospondin-1 were uniquely up-regulated in (near) senescent NHP8 cells (Fig. 12D (a)). These genes have all previously been implicated in the senescence of many different cell types (47), including prostate epithelial cells (48-50). Several histone subunits were also commonly up-regulated in senescent NHP cells (Fig. 12D (a)), implicating chromatin remodeling during cell senescence (51). Interestingly, HIF1α, which was recently shown to delay premature senescence in fibroblasts (52), was up-regulated in (near) senescent NHP cells (Fig. 12D (a)). In contrast to senescent cells, presenescent NHP8/p16sh + DNp53 and, in particular, the two immortalized NHP8 cells showed increased mRNA levels of multiple positive cell cycle/division regulators (cyclins, CDCs, survivin, etc.) and components in several signal transduction pathways, including PI3K, NF-κB, Notch, and Stat (Fig. 12D (a)). Of note, β2-microglobulin, a molecule implicated in PCa metastasis (53), was commonly up-regulated in life span-extended NHP cells (Fig. 12D (a)).
Among the 100 most highly down-regulated genes in genetically modified NHP8 cells, the most noticeable ones were the genes encoding multiple mitochondrial and intermediate metabolic enzymes as well as ribosomal subunits, which were all specifically down-regulated in senescent NHP cells (Fig. 12D (b)), consistent with senescent cells having lower protein synthesis and slower metabolic rates. By contrast, several genes, including BAMBI (BMP and activin membrane-bound inhibitor homolog), a transmembrane glycoprotein related to TGFβR-I that functions as a negative regulator of TGFβ signaling, and SPRR1A (small proline-rich protein 1A), a member of the small proline-rich family of cell envelope precursor proteins linked to cell differentiation, were specifically down-regulated in the immortalized NHP cells (Fig. 12D (b)). Remarkably, these two classes of genes that demonstrated opposite associations with senescent versus immortalized NHP cells both were down-regulated in near senescent NHP8/p16sh (i.e. sample 7a) cells (Fig. 12D (b)), thus indicating the functional relevancy of these gene changes in determining the NHP cell proliferative properties.
Altogether, the microarray studies indicate that the immortalized NHP cells possess gene expression profiles distinct from senescent NHP cells and characteristic of proliferating progenitor cells.
DISCUSSION
Phenotypic, Proliferative, and Differentiation Properties Classify Primary NHP Cells as Basal-like Progenitor Cells—In this study, we have thoroughly characterized multiple strains of primary NHP cells, most of which are derived from organ donors as young as 14 years of age without prior history of prostatic diseases. These normal epithelial cells are probably different from the “benign” epithelial cells prepared from “uninvolved” prostate tissues of PCa patients, as reported in most published works (29-31). The early passage (i.e. P1-P2) NHP cells have been immunophenotyped as CK5+/CK18+CD44+α2β1+p63+hTERT+ cells that do not express any luminal or NE cell markers. This marker expression profile is typical of the reported intermediate basal-like prostate epithelial stem/progenitor cells. When cultured in PrEBM(EGF + Ins) containing one mitogen (i.e. EGF) and one survival factor (i.e. insulin), NHP cells can undergo extensive proliferation to 20-26 cumulative PDs. Importantly, primary NHP cells can functionally differentiate into prostatic glands in vivo. Altogether, the marker expression profile and proliferative and differentiation capacities of primary NHP cells suggest that they are intermediate basal-like progenitor cells.
The PrEBM(EGF + Ins) medium is apparently insufficient to maintain NHP cells as proliferating progenitor cells, and, like most human epithelial cells, NHP cells gradually lose their proliferative potential. Of note, as NHP cells lose their proliferative capacity, they also down-regulate or lose most of the progenitor markers, suggesting that some of these markers may play a functional role. In support, telomerase is well known to be critical for the long term expansion of most stem/progenitor cells, p63 plays an essential role in regulating epithelial SC life span (3), and p63 deficiency promotes cell senescence and animal aging (54). The proliferative life span of NHP cells in PrEBM(EGF + Ins) cannot be extended by basic fibroblast growth factor, Wnt-3a, or LIF, individually or in combination, suggesting that the signaling pathways initiated by these factors may not be critical in regulating the proliferative potential of NHP cells under the specified conditions. Even when cultured on fibroblast feeders with enforced expression of hTERT, NHP cell life span cannot be extended. This was initially surprising to us, since it has been reported that human keratinocytes and mammary epithelial cells cultured on the fibroblast feeders become immortalized with hTERT (39). However, later studies demonstrate that keratinocyte proliferation is limited by p16 irrespective of culture conditions or telomerase expression (55).
p16 Is the Primary Determinant of the NHP Progenitor Cell Life Span, and Telomerase Is Required for NHP Cell Immortalization—Different human cells may utilize divergent mechanisms to execute senescence. In human fibroblasts, telomere shortening leads to p53 activation, which triggers senescence. By contrast, human keratinocytes and mammary epithelial cells appear to have two senescence programs running simultaneously: progressive telomere shortening and a telomere-independent cell aging process mediated by p16 and p53 (55, 56). Mesothelial cells, retinal pigmented epithelial cells, and endothelial cells behave like fibroblasts in that p53/p21 activation caused by telomere shortening initiates senescence. Nevertheless, p16-deficient human fibroblasts are resistant to Ets- and RAS-induced senescence, and p16 also determines the sensitivity of fibroblast transformation by oncogenes (40), highlighting a critical role of p16 also in fibroblast senescence.
NHP cell senescence overall resembles keratinocyte and mammary epithelial cell senescence involving p16, p53, and telomere shortening but with important differences (Fig. 13). In NHP cells, p16 up-regulation represents the driving force that impedes unlimited proliferation. Thus, enforced expression of p16 is sufficient to restrict NHP cell expansion, and prevention of p16 accumulation, by itself, significantly extends the cell life span (Fig. 13, B and C). Interestingly, microarray studies suggest that in long term immortalized NHP cells, p16 mRNA is not completely ablated, but immunofluorescence and Western blot analysis reveal little p16 protein, suggesting that the p16 shRNA-mediated suppression of p16 mRNA is sufficient to lead to significantly reduced p16 protein. In NHP cells, p16 seems to function exclusively through pRb-dependent mechanisms. That p16 is the primary determinant of NHP progenitor cell proliferative life span is consistent with recent observations that this CKI also plays a paramount role in restricting the proliferative capacity of many somatic SCs (reviewed in Ref. 57). In stark contrast to the prominent role of p16sh, enforced hTERT or DNp53 expression fails to extend NHP cell life span (Fig. 13B), suggesting that neither loss of telomerase nor early p53 activation (Fig. 13C) constitutes the initiating event in NHP cell senescence. Intriguingly, hTERT has been used to immortalize prostate tumor-derived cells (e.g. see Refs. 29-31). Since p16 is under complex transcriptional regulation by multiple molecules, including Ets and Id proteins and chromatin remodeling proteins like Bmi, CBX-7, and EZH2 (40, 57), these tumor cells may have sustained physical or functional compromises in the p16/pRb pathway, such as p16 deletion or promoter hypermethylation (58).
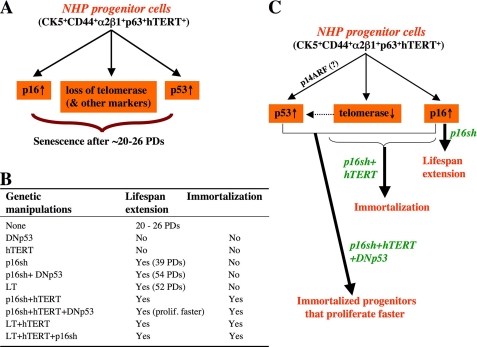
FIGURE 13.
Critical and distinct roles of p16, hTERT, and p53 in NHP cell replicative life span. A, NHP cell senescence involving changes in p16, telomerase, and p53. B, summary of genetic manipulations in NHP cells in this study. C, NHP cell immortalization. Cultured NHP cells up-regulate p16, lose telomerase, and activate p53. p53 activation results possibly from both early p14ARF up-regulation and late telomere erosion (due to p16sh-conferred, continued cell proliferation in the absence of telomerase). p16sh extends NHP cell-proliferative life span, but both p16sh and hTERT are required to immortalize NHP cells.
Telomerase, although insufficient by itself to extend the NHP cell life span, is nevertheless required for immortalization of NHP cells. Conceivably, continued NHP cell proliferation conferred by p16 inhibition but in the absence of telomerase would result in telomere erosion and deprotection, which in turn may lead to p53 activation, thus further contributing to NHP cell senescence (Fig. 13C). In support, NHP8/p16sh cells accumulate p53, p16sh combined with DNp53 extends NHP cell life span beyond that conferred by p16sh alone, and senescent NHP/p16sh and NHP/p16sh + DNp53 cells show signs of DDR. Importantly, p16sh or p16sh plus DNp53 is insufficient and telomerase is required to immortalize NHP cells. Curiously, NHP8/p16sh + hTERT + DNp53 cells start to proliferate faster, from ∼60 PDs on, than NHP8/p16sh + hTERT cells (Fig. 5A (a)), suggesting that in extended cultures, activated p53 caused by both telomere attrition and telomere erosion-independent mechanism(s), such as p14ARF, also slows down NHP cell proliferation (Fig. 13C).
Immortalized NHP Progenitors Can Functionally Differentiate into Prostatic Glands and Possess Progenitor Cell Gene Expression Profiles—Most immortalization studies utilized viral genes, such as SV40 LT, HPV-18 genome, or HPV-16 E6/E7 (e.g. see Refs. 23 and 29) or used “benign” prostate epithelial cells from PCa samples. Here we have achieved genetically defined, nonviral gene-mediated immortalization of primary normal prostate epithelial cells. Our immortalized NHP cells possess normal DDR and are nontumorigenic. Significantly, the immortalized NHP cells retain expression of most reported progenitor markers and have the ability to differentiate into glandular/ductal structures containing not only luminal and basal cells but also NE-like cells. Many of the glands contain eosinophilic secretions, and some glands even show PSA, suggesting that NHP cell-derived prostatic glands are functional. Importantly, although prostatic organoids have been shown to form glandular structures in TR assays (12), the present study is the first to show that both primary and immortalized NHP cells, as dissociated cells, can differentiate into functional prostatic glands. TR assays using varying numbers of NHP cells indicate that we could obtain glandular outgrowths with 10,000 cells,8 suggesting that at least some of the immortalized NHP cells have regenerative capacity. Since the immortalized NHP cells antigenically resemble basal cells lacking CK18 expression (i.e. CK5+CD44+p63+hTERT+), the present results provide further support to the concept that the basal cell compartment in prostatic glands harbors regenerative stem/progenitors. We are currently performing more TR assays using <10,000 cells aiming to better characterize the common stem/progenitor cell that regenerates the epithelial and NE cells in the outgrowths.
Gene expression profiling has uncovered, in senescent NHP cells, multiple genes known to be involved in cell senescence as well as potentially novel regulators of NHP cell senescence and immortality, including molecules like histone variants, HIF1α, β2-microglobulin, BAMBI, and SPRR1A. Further studies on these molecules will undoubtedly deepen our knowledge on molecular mechanisms regulating the NHP progenitor cell life span. Overall, the microarray studies clearly indicate that the immortalized NHP cells possess gene expression profiles characteristic of proliferating progenitor cells, supporting our functional studies. The availability of the immortalized NHP progenitor cells should greatly facilitate our future studies on how NHP progenitor cells might be involved in PCa development and how the normal mechanisms regulating NHP cell life span might be disrupted in PCa stem/progenitor cells (1).
Supplementary Material
Acknowledgments
We thank D. Chopra, D. Galloway, D. Johnson, A. Lloyd, S. Lowe, G. Peters, and J. Shay for providing cells/reagents, Histology and Animal Facility Cores for technical assistance, the T. C. Hsu Cytogenetics Core for cytogenetic services, and other members of the Tang laboratory for discussion and support. We apologize to those colleagues whose original work could not be cited due to space constraints.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-AG023374, R01-ES015888, and R21-ES015893-01A1. This work was also supported by American Cancer Society Grant RSG MGO-105961, Department of Defense Grants W81XWH-07-1-0616 and PC073751, the Prostate Cancer Foundation, the Elsa Pardee Foundation (to D. G. T.), and two Center Grants, CCSG-5 P30 CA166672 and ES07784. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S12 and Tables S1 and S2.
Footnotes
The abbreviations used are: NE, neuroendocrine; AR, androgen receptor; PSA, prostate-specific antigen; 15-LOX2, 15-lipoxygenase 2; SC, stem cell; NHP, normal human prostate; EGF, epidermal growth factor; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid; FISH, fluorescence in situ hybridization; TR, tissue recombination; rUGM, rat urogenital sinus mesenchyme; Ex, embryonic day x; PD, population doubling; NSE, neuron-specific enolase; CKI, cyclin-dependent kinase inhibitor; p16sh, p16 short hairpin RNA; GFP, green fluorescent protein; DNp53, dominant negative p53; DDR, DNA damage response; TRITC, tetramethylrhodamine isothiocyanate; shRNA, short hairpin RNA; BrdUrd, bromodeoxyuridine.
B. Bhatia and D. G. Tang, unpublished observations.
References
- 1.Tang, D. G., Patrawala, L., Calhoun, T., Bhatia, B., Choy, G., Schneider-Broussard, R., and Jeter, C. (2007) Mol. Carcinog. 46 1-14 [DOI] [PubMed] [Google Scholar]
- 2.Oudes, A. J., Campbell, D. S., Sorensen, C. M., Walashek, L. S., True, L. D., and Liu, A. Y. (2006) BMC Genomics 7 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Senoo, M., Pinto, F., Crum, C. P., and McKeon, F. (2007) Cell 129 523-536 [DOI] [PubMed] [Google Scholar]
- 4.Wang, Y., Hayward, S., Cao, M., Thayer, K., and Cunha, G. (2001) Differentiation 68 270-279 [DOI] [PubMed] [Google Scholar]
- 5.Robinson, E. J., Neal, D. E., and Collins, A. T. (1998) Prostate 37 149-160 [DOI] [PubMed] [Google Scholar]
- 6.Lawson, D. A., and Witte, O. N. (2007) J. Clin. Invest. 117 2044-2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goto, K., Salm, S. N., Coetzee, S., Xiong, X., Burger, P. E., Shapiro, E., Lepor, H., Moscatelli, D., and Wilson, E. L. (2006) Stem Cells 24 1859-1868 [DOI] [PubMed] [Google Scholar]
- 8.Wang, S., Garcia, A. J., Wu, M., Lawson, D. A., Witte, O. N., and Wu, H. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 1480-1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhatia, B., Tang, S., Yang, P., Doll, A., Aumueller, G., Newman, R. A., and Tang, D. G. (2005) Oncogene 24 3583-3595 [DOI] [PubMed] [Google Scholar]
- 10.Hudson, D. L., O'Hare, M., Watt, F. M., and Masters, J. R. (2000) Lab. Invest. 80 1243-1250 [DOI] [PubMed] [Google Scholar]
- 11.Uzgare, A. R., Xu, Y., and Isaacs, J. T. (2004) J. Cell. Biochem. 91 196-205 [DOI] [PubMed] [Google Scholar]
- 12.Hayward, S. W., Haughney, P. C., Rosen, M. A., Greulich, K. M., Weier, H. U., Dahiya, R., and Cunha, G. R. (1998) Differentiation 63 131-140 [DOI] [PubMed] [Google Scholar]
- 13.Liu, A. Y., True, L. D., LaTray, L., Nelson, P. S., Ellis, W. J., Vessella, R. L., Lange, P. H., Hood, L., and van den Engh, G. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 10705-10710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collins, A. T., Habib, F. K., Maitland, N. J., and Neal, D. E. (2001) J. Cell Sci. 114 3865-3872 [DOI] [PubMed] [Google Scholar]
- 15.Richardson, G. D., Robson, C. N., Lang, S. H., Neal, D. E., Maitland, N. J., and Collins, A. T. (2004) J. Cell Sci. 117 3539-3545 [DOI] [PubMed] [Google Scholar]
- 16.Heer, R., Robson, C. N., Shenton, B. K., and Leung, H. Y. (2007) J. Cell. Physiol. 212 572-578 [DOI] [PubMed] [Google Scholar]
- 17.Brown, M. D., Gilmore, P. E., Hart, C. A., Samuel, J. D., Ramani, V. A., George, N. J., and Clarke, N. W. (2007) Prostate 67 1384-1396 [DOI] [PubMed] [Google Scholar]
- 18.Patrawala, L., Calhoun, T., Schneider-Broussard, R., Zhou, J., Claypool, K., and Tang, D. G. (2005) Cancer Res. 65 6207-6219 [DOI] [PubMed] [Google Scholar]
- 19.Pascal, L. E., Oudes, A. J., Petersen, T. W., Goo, Y. A., Walashek, L. S., True, L. D., and Liu, A. Y. (2007) BMC Urol. 7 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Leenders, G., Dijkman, H., Hulsbergen-van de Kaa, C., Ruiter, D., and Schalken, J. (2000) Lab. Invest. 80 1251-1258 [DOI] [PubMed] [Google Scholar]
- 21.Sharpless, N. E., and DePinho, R. A. (2007) Nat. Rev. Mol. Cell. Biol. 8 703-713 [DOI] [PubMed] [Google Scholar]
- 22.Yaswen, P., and Campisi, J. (2007) Cell 128 233-234 [DOI] [PubMed] [Google Scholar]
- 23.Jarrard, D. F., Sarkar, S., Shi, Y., Yeager, T. R., Magrane, G., Kinoshita, H., Nassif, N., Meisner, L., Newton, M. A., Waldman, F. M., and Reznikoff, C. A. (1999) Cancer Res. 59 2957-2964 [PubMed] [Google Scholar]
- 24.Sandhu, C., Peehl, D. M., and Slingerland, J. (2000) Cancer Res. 60 2616-2622 [PubMed] [Google Scholar]
- 25.Castro, P., Giri, D., Lamb, D., and Ittmann, M. (2003) Prostate 55 30-38 [DOI] [PubMed] [Google Scholar]
- 26.Choi, J., Shendrik, I., Peacocke, M., Peehl, D., Buttyan, R., Ikeguchi, E. F., Katz, A. E., and Benson, M. C. (2000) Urol. 56 160-166 [DOI] [PubMed] [Google Scholar]
- 27.Chen, Z., Trotman, L. C., Shaffer, D., Lin, H. K., Dotan, Z. A., Niki, M., Koutcher, J. A., Scher, H. I., Ludwig, T., Gerald, W., Cordon-Cardo, C., and Pandolfi, P. P. (2005) Nature 436 725-730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhatia, B., Multani, A. S., Patrawala, L., Chen, X., Calhoun-Davis, T., Zhou, J., Schroeder, L., Schneider-Broussard, R., Shen, J., Pathak, S., Chang, S., and Tang, D. G. (2008) Int. J. Cancer 122 1483-1495 [DOI] [PubMed] [Google Scholar]
- 29.Bright, R. K., Vocke, C. D., Emmert-Buck, M. R., Duray, P. H., Solomon, D., Fetsch, P., Rhim, J. S., Linehan, W. M., and Topalian, S. L. (1997) Cancer Res. 57 995-1002 [PubMed] [Google Scholar]
- 30.Yasunaga, Y., Nakamura, K., Ewing, C. M., Isaacs, W. B., Hukku, B., and Rhim, J. S. (2001) Cancer Res. 61 5969-5973 [PubMed] [Google Scholar]
- 31.Kogan, I., Goldfinger, N., Milyavsky, M., Cohen, M., Shats, I., Dobler, G., Klocker, H., Wasylyk, B., Voller, M., Aalders, T., Schalken, J. A., Oren, M., and Rotter, V. (2006) Cancer Res. 66 3531-3540 [DOI] [PubMed] [Google Scholar]
- 32.Tang, S., Bhatia, B., Maldonado, C. J., Yang, P., Newman, R. A., Liu, J., Chandra, D., Traag, J., Klein, R. D., Fischer, S. M., Chopra, D., Shen, J., Zhau, H. E., Chung, L. W., and Tang, D. G. (2002) J. Biol. Chem. 277 16189-16201 [DOI] [PubMed] [Google Scholar]
- 33.Patrawala, L., Calhoun, T., Schneider-Broussard, R., Li, H., Bhatia, B., Tang, S., Reilly, J. G., Chandra, D., Zhou, J., Claypool, K., Coghlan, L., and Tang, D. G. (2006) Oncogene 25 1696-1708 [DOI] [PubMed] [Google Scholar]
- 34.Peehl, D. M., and Stamey, T. A. (1986) In Vitro Cell. Dev. Biol. 22 82-90 [DOI] [PubMed] [Google Scholar]
- 35.Chopra, D. P., Grignon, D. J., Joiakim, A., Mathieu, P. A., Mohamed, A., Sakr, W. A., Powell, I. J., and Sarkar, F. H. (1996) J. Cell. Physiol. 169 269-280 [DOI] [PubMed] [Google Scholar]
- 36.Patrawala, L., Calhoun-Davis, T., Schneider-Broussard, R., and Tang, D. G. (2007) Cancer Res. 67 6796-6805 [DOI] [PubMed] [Google Scholar]
- 37.Pounds, S., and Morris, S. W. (2003) Bioinformatics 19 1236-1242 [DOI] [PubMed] [Google Scholar]
- 38.Bhatia, B., Maldonado, C. J., Tang, S., Chandra, D., Klein, R. D., Chopra, D., Shappell, S. B., Yang, P., Newman, R. A., and Tang, D. G. (2003) J. Biol. Chem. 278 25091-25100 [DOI] [PubMed] [Google Scholar]
- 39.Ramirez, R. D., Morales, C. P., Herbert, B. S., Rohde, J. M., Passons, C., Shay, J. W., and Wright, W. E. (2001) Genes Dev. 15 398-403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gil, J., and Peters, G. (2006) Nat. Rev. Mol. Cell. Biol. 7 667-677 [DOI] [PubMed] [Google Scholar]
- 41.Tang, D. G., Tokumoto, Y. M., Apperly, J. A., Lloyd, A. C., and Raff, M. C. (2001) Science 291 868-871 [DOI] [PubMed] [Google Scholar]
- 42.Benanti, J. A., and Galloway, D. A. (2004) Mol. Cell. Biol. 24 2842-2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hemann, M. T., Fridman, J. S., Zilfou, J. T., Hernando, E., Paddison, P. J., Cordon-Cardo, C., Hannon, G. J., and Lowe, S. W. (2003) Nat. Genet. 33 396-400 [DOI] [PubMed] [Google Scholar]
- 44.Mitchell, P. J., Perez-Nadales, E., Malcolm, D. S., and Lloyd, A. C. (2003) Mol. Cell. Biol. 23 2530-2542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berger, R., Febbo, P. G., Majumder, P. K., Zhao, J. J., Mukherjee, S., Signoretti, S., Campbell, K. T., Sellers, W. R., Roberts, T. M., Loda, M., Golub, T. R., and Hahn, W. C. (2004) Cancer Res. 64 8867-8875 [DOI] [PubMed] [Google Scholar]
- 46.Taylor, R. A., Cowin, P. A., Cunha, G. R., Pera, M., Trounson, A. O., Pedersen, J., and Risbridger, G. P. (2006) Nat. Methods 3 179-181 [DOI] [PubMed] [Google Scholar]
- 47.Campisi, J. (2005) Cell 120 513-522 [DOI] [PubMed] [Google Scholar]
- 48.Cho, K. A., Ryu, S. J., Oh, Y. S., Park, J. H., Lee, J. W., Kim, H. P., Kim, K. T., Jang, I. S., and Park, S. C. (2004) J. Biol. Chem. 279 42270-42278 [DOI] [PubMed] [Google Scholar]
- 49.Schwarze, S. R., DePrimo, S. E., Grabert, L. M., Fu, V. X., Brooks, J. D., and Jarrard, D. F. (2002) J. Biol. Chem. 277 14877-14883 [DOI] [PubMed] [Google Scholar]
- 50.Stern, M., Savill, J., and Haslett, C. (1996) Am. J. Pathol. 149 911-921 [PMC free article] [PubMed] [Google Scholar]
- 51.Gevry, N., Chan, H. M., Laflamme, L., Livingston, D. M., and Gaudreau, L. (2007) Genes Dev. 21 1869-1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Welford, S. M., Bedogni, B., Gradin, K., Poellinger, L., Broome Powell, M., and Giaccia, A. J. (2006) Genes Dev. 20 3366-3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang, W. C., Wu, D., Xie, Z., Zhau, H. E., Nomura, T., Zayzafoon, M., Pohl, J., Hsieh, C. L., Weitzmann, M. N., Farach-Carson, M. C., and Chung, L. W. (2006) Cancer Res. 66 9108-9116 [DOI] [PubMed] [Google Scholar]
- 54.Keyes, W. M., Wu, Y., Vogel, H., Guo, X., Lowe, S. W., and Mills, A. A. (2005) Genes Dev. 19 1986-1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rheinwald, J. G., Hahn, W. C., Ramsey, M. R., Wu, J. Y., Guo, Z., Tsao, H., De Luca, M., Catricala, C., and O'Toole, K. M. (2002) Mol. Cell. Biol. 22 5157-5172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kiyono, T., Foster, S. A., Koop, J. I., McDougall, J. K., Galloway, D. A., and Klingelhutz, A. J. (1998) Nature 396 84-88 [DOI] [PubMed] [Google Scholar]
- 57.Kim, W. Y., and Sharpless, N. E. (2006) Cell 127 265-275 [DOI] [PubMed] [Google Scholar]
- 58.Konishi, N., Nakamura, M., Kishi, M., Nishimine, M., Ishida, E., and Shimada, K. (2002) Am. J. Pathol. 160 1207-1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.