Abstract
In eukaryotes, DNA topoisomerase I (Top1) catalyzes the relaxation of supercoiled DNA by a conserved mechanism of transient DNA strand breakage, rotation, and religation. The unusual architecture of the monomeric human enzyme comprises a conserved protein clamp, which is tightly wrapped about duplex DNA, and an extended coiled-coil linker domain that appropriately positions the C-terminal active site tyrosine domain against the Top1 core to form the catalytic pocket. A structurally undefined N-terminal domain, dispensable for enzyme activity, mediates protein-protein interactions. Previously, reversible disulfide bonds were designed to assess whether locking the Top1 clamp around duplex DNA would restrict DNA strand rotation within the covalent Top1-DNA intermediate. The active site proximal disulfide bond in full-length Top1-clamp534 restricted DNA rotation (Woo, M. H., Losasso, C., Guo, H., Pattarello, L., Benedetti, P., and Bjornsti, M. A. (2003) Proc. Natl. Acad. Sci. U. S. A. 100, 13767–13772), whereas the more distal disulfide bond of the N-terminally truncated Topo70-clamp499 did not (Carey, J. F., Schultz, S. J., Sisson, L., Fazzio, T. G., and Champoux, J. J. (2003) Proc. Natl. Acad. Sci. U. S. A. 100, 5640–5645). To assess the contribution of the N-terminal domain to the dynamics of Top1 clamping of DNA, the same disulfide bonds were engineered into full-length Top1 and truncated Topo70, and the activities of these proteins were assessed in vitro and in yeast. Here we report that the N terminus impacts the opening and closing of the Top1 protein clamp. We also show that the architecture of yeast and human Top1 is conserved in so far as cysteine substitutions of the corresponding residues suffice to lock the Top1-clamp. However, the composition of the divergent N-terminal/linker domains impacts Top1-clamp activity and stability in vivo.
Eukaryotic Top1 (DNA topoisomerase I) is a highly conserved enzyme, which plays critical roles in cellular processes, such as DNA replication, transcription, recombination, and chromosome condensation (1–4). Top1 catalyzes changes in DNA topology or the linkage of the individual strands that comprise duplex DNA. The enzyme first binds double-stranded DNA as a protein clamp and then transiently cleaves a single strand forming a 3′-phosphotyrosyl linkage between the active site tyrosine and the nicked DNA. Torsional or flexural strain in the DNA then drives the rewinding or unwinding of the polynucleotide strands. In the subsequent transesterification reaction, the free 5′-OH acts as a nucleophile to resolve the phosphotyrosyl linkage and religate the phosphodiester backbone of the DNA. A random number of supercoils are removed in this process to effect changes in DNA linking number (5, 6).
Top1 is also the cellular target of the camptothecin (CPT)4 class of chemotherapeutic agents (3, 7–9). These drugs convert the enzyme into a cellular poison by reversibly stabilizing the Top1-DNA covalent complex. CPT intercalates into the protein-linked DNA complex, effectively displacing the 5′-OH and preventing DNA religation (10, 11). Although ternary CPT-Top1-DNA complexes form throughout the cell cycle, the interaction of these complexes with the replication machinery during S-phase converts the readily reversible intermediate into irreversible DNA lesions that trigger cell death. Our recent in vivo studies and single molecule nanomanipulations revealed that CPT selectively slows Top1 uncoiling of positive supercoils, thereby inducing the accumulation of positively supercoiled DNA in vivo (6). These findings suggest a novel mechanism of Top1 poisoning by CPT, where the accumulation of positive supercoils in front of advancing replication forks acts as a physical barrier to induce fork collapse and/or DNA lesions (6).
Structural and biochemical data indicate that Top1 comprises four domains: a highly charged N-terminal domain, a conserved protein clamp, a pair of extended α-helices that form a positively charged linker domain, and a conserved C-terminal domain that contains the active site Tyr (12–14). Crystallographic studies of human Topo70 (a C-terminal 70-kDa fragment of the nuclear human enzyme), complexed with DNA, demonstrate that a lone salt bridge between the loops of opposable “lips” completes Top1 clamping of the DNA (12, 14). The tight packing of DNA within the central pore of the enzyme indicated that substantial flexibility of the protein clamp would be necessary for protein binding of DNA and to allow for strand rotation upon covalent complex formation. These predictions were confirmed in studies of the Top1-clamp534 where cysteine residues, substituted for Gly365 and Ser534, formed a reversible disulfide bond (15). Locking the Top1-clamp534 closed prevented DNA binding as well as strand rotation within the covalent complex. Surprisingly, these results contrasted with related studies, where distinct residues (His367 and Ala499) were changed to cysteine in the context of Topo70 (16). In this case, herein referred to as Topo70-clamp499, a disulfide tether between the same upper “lip” as that of Top1-clamp534 and a lower protein loop more distal from the active site did not impede DNA rotation. Thus, seemingly subtle differences in enzyme architecture profoundly influenced enzyme catalysis.
The N-terminal domain, which is missing in Topo70-clamp499, is dispensable for enzyme activity in vitro yet has been implicated in mediating Top1 interactions with other proteins and subcellular localization of the enzyme (1, 17–22). The N terminus has also been shown to bind DNA (19). Recently, we also demonstrated an interaction of the N-terminal and linker domains in regulating enzyme function in vivo.5 Previous studies of Top1 reconstituted from independently expressed protein domains and the Top1A653P mutant indicate that the physical integrity and flexibility of the linker domain are critical determinants of enzyme sensitivity to CPT (20, 23–25). Molecular dynamic simulations and x-ray crystallographic data suggest that restrictions in linker domain flexibility coincide with CPT binding to the Top1-DNA complex (11, 23). Linker flexibility also impacts mutation-induced alterations in enzyme-catalyzed DNA cleavage/religation (24). Indeed, our recent studies suggest that a conserved Gly within the catalytic pocket of Top1 provides a flexible hinge to facilitate linker mobility (26).
Taken together, these observations support a model whereby the orientation of the linker domain impacts enzyme active site architecture to affect enzyme catalyzed DNA cleavage and religation. Without structural information, it remains unclear what role, if any, the N-terminal domain has in coordinating Top1 protein domain movements with DNA transactions. Nevertheless, we wondered if the reported differences between Topo70-clamp499 and Top1-clamp534 (15, 16) were in fact due to differences in global enzyme architecture rather than subtle changes in active site geometry. Taking advantage of these reversible disulfide bonds, we show that the N terminus also impacts the opening and closing of the Top1 protein clamp, whereas the composition of the N-terminal/linker domains affects Top1-clamp stability.
EXPERIMENTAL PROCEDURES
Chemicals, Yeast Strains, and Plasmids—Camptothecin, anti-FLAG M2 affinity-agarose gel, FLAG peptide, and M2 monoclonal antibodies were purchased from Sigma. CPT was dissolved in dimethyl sulfoxide (Me2SO) at 4 mg/ml and stored at -20 °C.
Saccharomyces cerevisiae strains EKY3 (MATα, ura3-52, his3Δ200, leu2Δ1, trp1Δ63, top1Δ::TRP1), MMY3 (EKY3, rad9Δ::hisG), MWY9 (EKY3, glr1Δ::his5+), and JCW28 (MATa, ura3-52, his3Δ200, leu2Δ1, trp1Δ63, top1Δ, top2-4) have been described (15, 27–29). For regulated expression of yeast or human Top1, the indicated TOP1 allele was expressed from the galactose-inducible GAL1 promoter on an ARS/CEN, URA3 plasmid. In all cases, the Top1 proteins contained an N-terminal FLAG epitope indicated by the “e” prefix. For clarity, the “h” prefix has been dropped from the discussions of human Top1, with the exception of the chimeric enzymes. However, the “y” prefix has been retained for all discussions of yeast Top1. For the human TOP1, vectors YCpGAL1-eTOP1, YCpGAL1-etop1Y723F, YCpGAL1-etop1-clamp534, and YCpGAL1-etop1-clamp534Y723F were described (15). Individual Top1-clamp499 mutations (H367C and A499C) were generated in human TOP1 cDNA restriction fragments (BamHI/SacII and SacII/ClaI, respectively) excised from YCp-GAL1-eTOP1 or YCpGAL1-etop1Y723F, ligated into vector pRS416, and mutagenized using the QuikChange kit from Stratagene (La Jolla, CA). The DNA fragments were used to replace the corresponding wild-type sequences in the original vectors to yield YCpGAL1-etop1-clamp499 and YCpGAL1-etop1-clamp499Y723F.
Topo70 constructs, which encode a C-terminal 70-kDa fragment of human Top1, were generated by two-step PCR to introduce a FLAG epitope (underlined in MDYKDDDDK) and a canonical nuclear localization signal (NLS) upstream of Met174 in human Top1, designated eTopo70. The NLS sequence used (PKKIKTED) spans human Top1 residues 149–156, which have previously been shown to be sufficient for nuclear localization (21, 22). YCpGAL1-eTOP1 was used as a substrate for the PCRs. The PCR products were then used as self-priming templates for five amplification cycles, followed by successive amplification reactions with flanking primers. A BamHI/SacII restriction fragment was then ligated into the corresponding sites in YCpGAL1-eTOP1 to yield YCp-GAL1-eTopo70. Similar strategies generated YCpGAL1-eTopo70Y723F, YCpGAL1-eTopo70-clamp499, YCpGAL1-eTopo70-clamp499Y723F, YCpGAL1-eTopo70-clamp534, and YCpGAL1-eTopo70-clamp534Y723F.
The yeast TOP1 expression vectors, YCpGAL1eyTOP1 and YCpGAL1eytop1Y727F, were previously described (30). To generate yeast Top1-clamp460 (which corresponds to human Top1-clamp534), G297C and S460C substitutions were introduced into pTP1-4 (17) using the QuikChange mutagenesis kit. A BclI-SpeI DNA fragment was then used to replace the corresponding wild-type sequences in YEpGAL1eyTOP1 and YEpGAL1eytop1Y727F (31) to yield YEpG1eytop1-clamp460 and YEpG1eytop1-clamp460Y727F. BamHI-EagI fragments bearing the eytop1-clamp460 and eytop1-clamp460Y727F sequences were then used to replace the corresponding DNA sequences in YCpGAL1eyTOP1 to generate YCpGAL1-eytop1-clamp460 and YCpGAL1eytop1-clamp460Y727F, respectively.
In plasmids YCpGAL1-(yN,yL)hTop1 and YCpGAL1-(hN,hL)yTop1, chimeric Top1 proteins comprising either the yeast N-terminal/linker domains embedded in the human core/C-terminal domains ((yN,yL)hTop1) or the human N-terminal/linker domains embedded in the yeast core/C-terminal domains ((hN,hL)yTop1) were expressed from the GAL1 promoter.5 A BamHI-ClaI fragment, excised from YCpGAL1-(yN,yL)hTop1, was ligated into pRS416. Following site-directed mutagenesis to introduce G365C and S534C, as previously described (15), the (yN,yL)hTop1-clamp534 sequences in the BamHI-ClaI fragment were ligated into YCpGAL1eTOP1 to generate YCpGAL1-(yN,yL)hTop1-clamp534. The corresponding yTop1-clamp460 sequences were introduced into YCpGAL1-(hN,hL)yTop1 by homologous recombination in yeast (32). PCR-amplified DNA sequences, which encode the eyTop1-clamp460 mutations, were co-transformed into yeast EKY3 (top1Δ) cells with YCpGAL1-(hN,hL)yTop1 DNA linearized with Bsu36I. Following selection for uracil prototrophs on synthetic complete media lacking uracil (SC-uracil) plates, the YCpGAL1-(hN,hL)yTop1-clamp460 plasmid DNA was recovered from individual transformants and amplified in bacteria. Yeast cells were transformed with URA3-marked vectors following treatment with lithium acetate (33), and transformants were maintained on SC-uracil, supplemented with 2% dextrose. In all cases, plasmid constructs were verified by restriction enzyme digests and DNA sequencing. Primer sequences are available upon request.
Yeast Cell Viability and Sensitivity to Camptothecin—To assess the viability and camptothecin sensitivity of cells expressing the indicated yeast/human top1 alleles, exponentially growing cultures of individual transformants were adjusted to an A595 = 0.3 and serially 10-fold diluted, and 5-μl volumes were spotted onto SC-uracil agar supplemented with 25 mm HEPES (pH 7.2), a final 2% dextrose or galactose, and the indicated concentration of CPT in a final 0.125% Me2SO. Cell viability was assessed following incubation at 30 °C. To assess relative steady state levels of Top1 proteins in galactose-induced top1Δ cells, cultures were corrected for A595, lysed with NaOH/trichloroacetic acid as previously described (34), and subjected to immunoblot analysis with the M2 α-FLAG antibody (Sigma).
Top1 Protein Purification—Human enzymes (Top1, Top1Y723F, Top1-clamp534, Top1-clamp534Y723F, Top1-clamp499, and Top1-clamp499Y723F), yeast enzymes (yTop1, yTop1Y727F, and yTop1-clamp460), and chimeras ((hN/hL)y-Top1, (hN/hL)yTop1-clamp460, (yN/yL)hTop1, and (yN/yL)h-Top1-clamp534) were partially purified as described (15) from galactose-induced cultures of top1Δ yeast cells, transformed with relevant YCpGAL1-etop1 vector. For homogenous protein preparations, Top1 proteins were applied to an anti-FLAG M2 affinity gel (Sigma) and eluted with an excess of FLAG peptide in TBS (50 mm Tris, pH 7.4, 150 mm KCl). To remove the peptide, fractions were applied to a phosphocellulose column, and homogeneous Top1 proteins were eluted in TEEG buffer (50 mm Tris (pH 7.4), 1 mm EDTA, 1 mm EGTA, 50% glycerol) plus 1.0 m KCl and protease inhibitors, diluted to a final 40% glycerol, and stored at -20 °C. Protein levels and integrity were assessed in immunoblots with the M2 α-FLAG antibody.
Top1 Enzyme Activity Assays—Purified Top1 proteins or crude cell extracts, corrected for protein concentration, were serially 10-fold diluted and incubated in plasmid DNA relaxation assays in the presence or absence of 10 mm DTT and the final KCl concentration indicated as described in Ref. 15. Following a 1-h incubation at 37 °C for human proteins or 30 °C for yeast Top1 proteins, the reactions were terminated by adding 1% SDS, and the extent of plasmid DNA relaxation was assessed following agarose gel electrophoresis and staining with ethidium bromide.
DNA Cleavage Assays—Top1 enzyme sensitivity to CPT was assessed in DNA cleavage assays with or without 10 mm DTT or 5 mm diamide, essentially as described (15). After 30 min at 30 °C (yTop1) or 37 °C (human Top1), the reactions were terminated with 1% SDS at 75 °C, treated with proteinase K, and resolved in 8% polyacrylamide, 7 m urea gels. Cleavage products were visualized with a PhosphorImager (Amersham Biosciences).
RESULTS
The Position of the Disulfide Bond That Locks the Top1-clamp Induces Distinct Effects on Enzyme Function—We previously reported that cysteine substitutions of Gly365 and Ser534 in full-length human Top1 form a reversible disulfide bond that locks the Top1-clamp534 closed and prevents DNA rotation within the covalent enzyme-DNA complex (15). In contrast, cysteine residues substituted for His367 and Ala499 form a disulfide bond more distal to the active site tyrosine (see Fig. 1) that did not affect DNA rotation in the context of Topo70 (16). Although subtle changes in enzyme active site architecture could impact enzyme catalysis, differences in the N-terminal composition of these enzymes might also alter the dynamics of Top1-clamp formation and DNA strand rotation.
FIGURE 1.
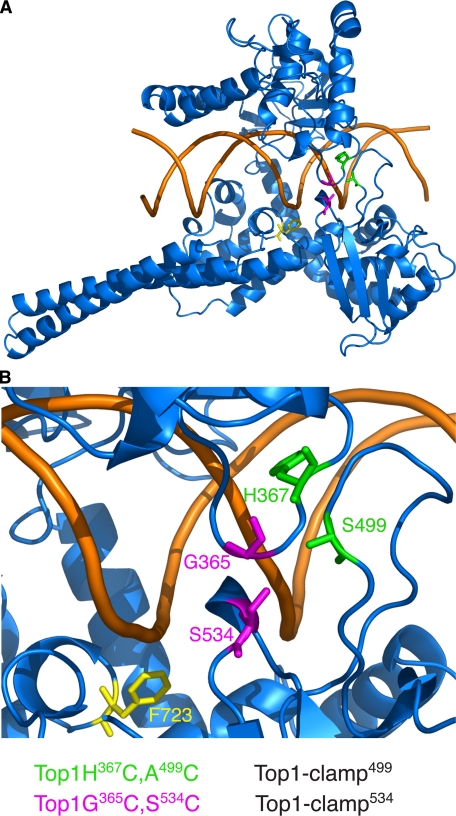
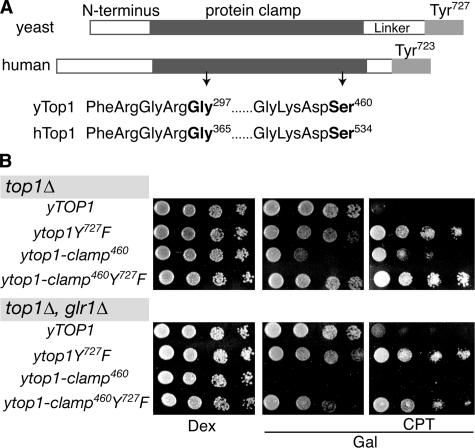
DNA topoisomerase I clamp structures. A, ribbon diagram of the structure of a C-terminal 70-kDa fragment of human Top1 (Topo70) in blue noncovalently bound to duplex DNA (brown) (Protein Data Bank file 1A36) (14). B, close up view of the lip domain residues mutated to cysteines in the Top1-clamp mutants. The active site tyrosine, changed to phenylalanine (F723), is colored yellow. Amino acid residues Gly365 and Ser534 (G365 and S534, shown in magenta) are substituted with cysteines in Top1-clamp534; residues His367 and Ala499 (H367 and A499) (colored green) are substituted with cysteines in Top1-clamp499. All figures were made with PyMol (DeLano Scientific, San Carlos, CA).
As defined by crystallographic data, the clamp domain spans residues 215–635 of nuclear human Top1 (12, 14). However, the function of specific enzyme domains has largely been described by biochemical studies of Topo70, which begins at residue 175 (20). Several studies suggest that residues 175–190 have no discernible effect on enzyme catalysis; however, residues 191–206 play important roles in Top1 binding of DNA and in enzyme processivity (17, 19, 25, 35, 36). Structural data further demonstrate the interaction of hydrophobic residues spanning 205 with the flexible hinge presumed to regulate Top1 protein clamp dynamics (37). Thus, in considering the potential contribution of the N-terminal domain to the function of the Top1 clamp, we restricted our investigations to residues 1–174, which are deleted from Topo70. We began by engineering both sets of cysteine substitutions (comprising clamp534 and clamp499 as shown in Fig. 1) into full-length human Top1 and Topo70 and then assessing the activity of these clamps in vitro and in yeast cells deleted for the endogenous TOP1 gene. An NLS (human residues 149–156) (21, 22) was also included to ensure the appropriate subcellular localization of the Top1 proteins.
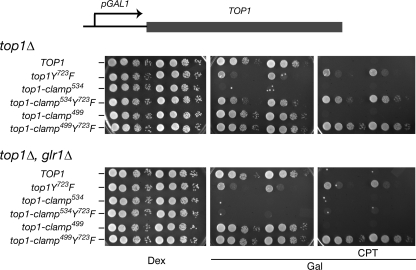
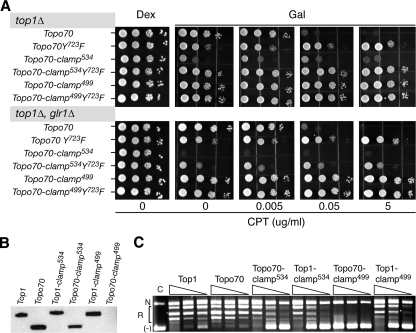
As previously reported (15), expression of Top1-clamp534 from the galactose-inducible GAL1 promoter induced yeast cell lethality, independent of camptothecin (Fig. 2). In a top1Δ, glr1Δ strain, where increased levels of oxidized glutathione favor the formation of disulfide bonds, the catalytically inactive Top1-clamp534Y723F clamp was also cytotoxic. Thus, locking the Top1-clamp534Y723F on DNA, in the absence of DNA cleavage, induced cell death. These results contrast with data obtained with the full-length Top1-clamp499 and the catalytically inactive Top1-clamp499Y723F. In the absence CPT, expression of either clamp had no effect on top1Δ or top1Δ, glr1Δ cell growth (Fig. 2). Although cells expressing Top1-clamp499 were sensitive to CPT (Fig. 2), 100-fold higher concentrations of drug were required to achieve the same level of cytotoxicity as that observed with wild-type Top1 (Fig. S1). Yet, under the same conditions, deletion of the Rad9 DNA damage checkpoint increased the CPT sensitivity of Top1-clamp499-expressing cells (Fig. S1). These data suggest that the Top1-clamp499 may be defective in binding DNA, either as a consequence of efficient disulfide bond formation that prevents DNA binding or impaired mutant Top1-DNA interactions, such that cross-linking of a DNA-bound Top1-clamp499 does not readily occur in vivo.
FIGURE 2.
Expression of Top1-clamp534 but not Top1-clamp499 induces yeast cell lethality. Congenic top1Δ yeast strains, wild-type for GLR1 or glr1Δ, were transformed with the indicated YCpGAL1-human top1 constructs. Individual transformants, grown in SC-uracil dextrose medium, were serially diluted and spotted onto SC-uracil plates supplemented with 25 mm HEPES, pH 7.2, dextrose (Dex) or galactose (Gal), Me2SO, and, as indicated, CPT. Cell viability was assessed after 3 days at 30 °C.
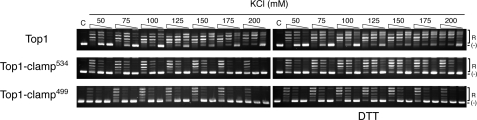
To distinguish between these possibilities, wild type Top1, Top1-clamp534, and Top1-clamp499 proteins were purified to homogeneity, and the specific activities of the enzymes were assessed in plasmid DNA relaxation assays. As seen in Fig. 3, wild-type Top1 activity was unaffected by DTT and was optimal at 150–175 mm KCl. In the absence of DTT, Top1-clamp534 and Top1-clamp499 were about 10- and 15-fold less active, respectively, whereas the addition of DTT enhanced the activity of both clamps by about 5-fold. However, even under these conditions, the apparent decrease in salt optimum to 125–150 mm KCl and 100–125 mm for Top1-clamp534 and Top1-clamp499, respectively, is consistent with a more severe defect in DNA binding for the Top1-clamp499.
FIGURE 3.
The reduced activities of Top1-clamp534 and Top1-clamp499 are partially suppressed by DTT. Equal concentrations of homogenous preparations of Top1, Top1-clamp534, and Top1-clamp499 were serially 10-fold diluted and incubated in plasmid DNA relaxation assays in the presence or absence of 10 mm DTT and the indicated concentration of KCl. After 1 h at 37 °C, the extent of plasmid DNA relaxation was assessed by agarose gel electrophoresis followed by EtBr staining. Lane C, plasmid DNA control. R and (-), the positions of relaxed and supercoiled DNA, respectively.
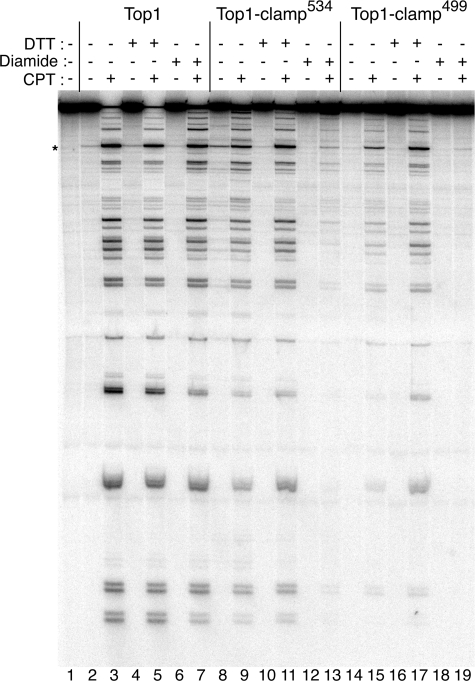
Top1-clamp499 Is Sensitive to CPT in Vitro—To assess the intrinsic CPT sensitivity of the purified proteins, drug-induced stabilization of enzyme-DNA covalent complexes was assayed in DNA cleavage reactions (Fig. 4). In these assays, steady state levels of Top1-linked DNA were assessed at 50 mm KCl, conditions under which the specific activities of the proteins were similar when treated with DTT. As previously reported (15), Top1-clamp534 exhibited slightly higher levels of covalent complexes in absence of the CPT, relative to that observed with wild-type Top1 (compare lanes 8 and 2). However, this effect was reversed with DTT (compare lanes 10 and 4). In the presence of CPT, the slightly lower levels of Top1-clamp534-DNA complexes (Fig. 4, lane 9) were enhanced by DTT treatment to levels achieved with CPT-treated wild-type Top1 (compare lanes 11 and 5). However, when Top1-clamp534 was treated with the oxidizing agent diamide, to lock the clamp prior to the addition of DNA and CPT, DNA cleavage was largely suppressed (Fig. 4, compare lanes 13 and 9). If, on the other hand, DNA was added prior to diamide and CPT, the levels of DNA cleavage mirrored that observed in lane 9 (data not shown). These data suggest that the Top1-clamp534 is predominantly “unlocked” in the absence of DNA.
FIGURE 4.
Top1-clamp499 and Top1-clamp534 are sensitive to CPT in vitro. Equal concentrations of purified Top1 (lanes 2–7), Top1-clamp534 (lanes 8–13), and Top1-clamp499 (lanes 14–19) were incubated in DNA cleavage reactions with a single 3′ 32P-labeled DNA fragment as substrate. As indicated, reactions contained 50 μm CPT (lanes 3, 5, 7, 9, 11, 13, 15, 17, and 19), 10 mm DTT (lanes 4, 5, 10, 11, 16, and 17), and 5 mm diamide (lanes 6, 7, 12, 13, 18, and 19). After 10 min at 37 °C, covalent complexes were trapped by the addition of SDS at 75 °C and treated with proteinase K. The covalent complexes were resolved in 8% polyacrylamide, 7 m urea gels and visualized using a PhosphorImager. The asterisk indicates a high affinity Top1 cleavage site.
These findings contrast with those observed with Top1-clamp499. In this case, the relatively low levels of CPT-induced DNA cleavage were significantly enhanced in the presence of DTT (Fig. 4, compare lanes 15 and 17). Moreover, whether Top1-clamp499 was preincubated with diamide prior to or following the addition of DNA, CPT induced the same low levels of DNA cleavage (Fig. 4, lane 19) (data not shown). Taken together, these data suggest that in the context of the Top1-clamp499, disulfide bond formation occurs more readily in the absence of DNA than with Top1-clamp534. However, the decrease in Top1-clamp499 activity observed at higher salt concentrations in Fig. 3 also indicates a defect in DNA binding under reducing conditions (DTT). In vivo, an increased propensity to form a locked clamp prior to DNA binding would also be manifest as reduced toxicity in response to CPT or expression in a glr1Δ strain.
The N-terminal Domain of Top1 Affects the in Vivo Activities of the Top1-clamps—To determine if the presence of N-terminal residues 1–174 contributes to the phenotypes induced by Top1-clamp534 and/or Top1-clamp499, the same cysteine substitutions were engineered into Topo70 constructs that also contain an NLS. Expression of Topo70, but not the catalytically inactive Topo70Y723F, induced cell sensitivity to CPT (Fig. 5A), albeit to slightly higher concentrations of drug than cells expressing full-length Top1 (data not shown). However, deletion of the N-terminal domain partially suppressed the lethal phenotype and CPT sensitivity of Top1-clamp534. In top1Δ cells, expression of Topo70-clamp534 induced a slow growth phenotype, which was unaffected by CPT (Fig. 5A). However, the levels of Topo70-clamp534 protein expression and catalytic activity in DNA relaxation assays mirrored that of the full-length Top1-clamp534 (Fig. 5, B and C). The increased levels of oxidized glutathione in a top1Δ, glr1Δ strain induced a Topo70-clamp534 lethal phenotype. However, in contrast to full-length Top1-clamp534Y723F, the Topo70-clamp534Y723F only exhibited a slow growth phenotype in top1Δ, glr1Δ cells. These data suggest that the N-terminal domain enhances the formation of the Top1-clamp534 on duplex DNA.
FIGURE 5.
The N-terminal domain of Top1 enhances Top1-clamp534 and Top1-clamp499-dependent toxicity. A, congenic top1Δ yeast strains, wild-type for GLR1 or glr1Δ, were transformed with the indicated YCpGAL1-Topo70 constructs bearing an NLS. As in Fig. 2, cultures of individual transformants were serially diluted and spotted onto SC-uracil supplemented with dextrose (Dex) or galactose (Gal) and the indicated concentration of CPT in a final 0.125% Me2SO. Cell viability was assessed after incubation at 30 °C. B, trichloroacetic acid extracts of equivalent numbers of galactose-induced top1Δ cells expressing the indicated Top1 proteins were resolved in a 4–12% SDS-polyacrylamide gel and immunoblotted with the M2 α-FLAG antibody. C, crude cell extracts of the galactose-induced cultures were corrected for protein concentration, serially 10-fold diluted, and incubated in a plasmid DNA relaxation assay in the presence of 150 mm KCl and 10 mm DTT at 37 °C. Reaction products were resolved in an agarose gel and visualized by ethidium bromide staining. N, R, and (-), relative positions of nicked, relaxed, and supercoiled DNAs, respectively.
Deleting the N terminus of Top1-clamp499 was even more deleterious. Cells expressing Topo70-clamp499 and Topo70-clamp499Y723F were perfectly viable, even in the presence of high concentrations of CPT (Fig. 5A). In fact, this phenotype coincided with our inability to detect any Topo70-clamp499 protein in galactose-induced cells (Fig. 5, B and C). Thus, in yeast cells, the N terminus of Top1-clamp499 is apparently required for protein stability. Whether this reflects the inappropriate folding of the nascent polypeptide or the proteolytic degradation of Topo70-clamp499 has yet to be determined.
The yTop1-clamp460 Is Toxic—As shown in Fig. 6A, the core and C-terminal domains of human and yeast Top1 are highly conserved, whereas the N-terminal and linker domains differ in sequence and length. Recent studies of yeast/human Top1 chimeras demonstrate the functional interaction of these divergent protein domains to regulate enzyme function in vivo.5 These findings further suggest that relative to yeast Top1, the longer N-terminal domain of human Top1 compensates for a shorter linker domain. To investigate whether conserved core and C-terminal domains of yeast Top1 would also form a functional Top1-clamp, we substituted cysteines for Gly297 and Ser460 in yTop1-clamp460 (Fig. 6A). Based on sequence alignments, these changes correspond to the G365C and S534C substituents in human Top1-clamp534. Interestingly, expression of yTop1-clamp460 induced similar effects on cell viability and CPT sensitivity as those induced by the human Topo70-clamp534; a slow growth phenotype in top1Δ cells that was unaffected by CPT and a lethal phenotype in top1Δ, glr1Δ cells (Fig. 6B). Moreover, mutation of the catalytic Tyr (yTop1-clamp460Y727F) only impaired cell growth in a glr1Δ background.
FIGURE 6.
The yeast Top1-clamp460 induces a glr1Δ-dependent lethal phenotype. A, schematic representation of the domain structure of human and yeast Top1 (hTop1 and yTop1, respectively). Cysteine substitutions of conserved Gly and Ser residues (in boldface type) constitute the corresponding hTop1-clamp534 and yTop1-clamp460. B, congenic top1Δ yeast strains (GLR1 and glr1Δ), transformed with the indicated YCpGAL1-ytop1 constructs, were serially diluted and spotted onto SC-uracil plates supplemented with dextrose (Dex) or galactose (Gal), 25 mm HEPES, pH 7.2, Me2SO and as indicated, CPT. As in Fig. 5, cell viability was assessed following incubation at 30 °C.
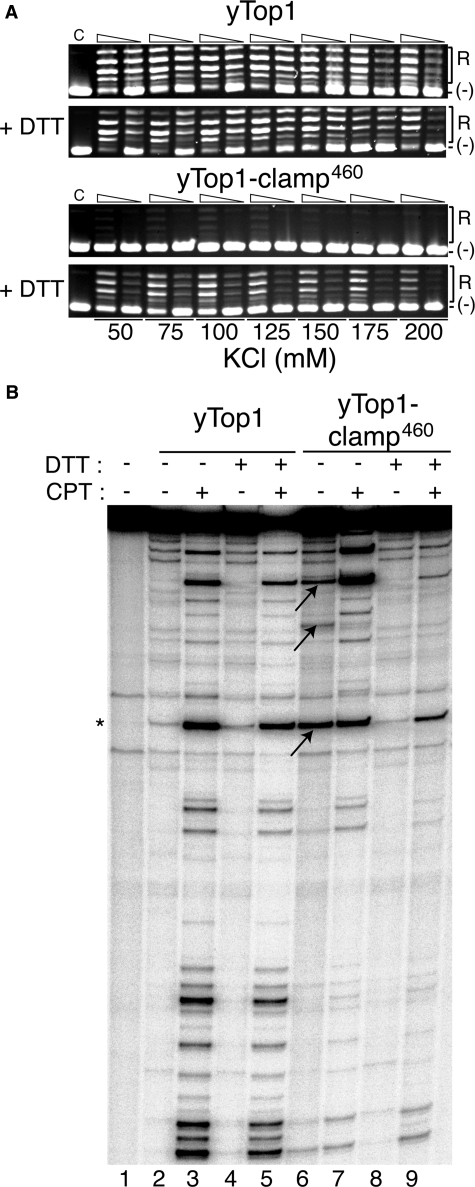
As with human Top1-clamp534 and Topo70-clamp534, the specific activity of the purified yTop1-clamp460 was about 10-fold lower than that of wild-type yTop1 in the presence of DTT, with a slightly lower salt optimum (100 mm versus 150 mm KCl) (Fig. 7A). In the absence of DTT, yTop1-clamp460 activity was barely detectable (Fig. 7A). However, the patterns of DNA cleavage obtained with yTop1-clamp460 (Fig. 7B) were quite distinct from those shown for human Top1-clamp534 in Fig. 2. First, in the absence of DTT and CPT, yTop1-clamp460 exhibited enhanced covalent complex formation at several sites along the DNA duplex substrate, over that observed with wild-type yTop1 (Fig. 7B, compare arrows in lane 6 with corresponding bands in lane 2). CPT enhanced only a subset of these complexes (lane 7), yet all were resolved by DTT (lane 8). Moreover, in contrast to the CPT sensitivity of human Top1-clamp534 (Fig. 4, lane 11), yTop1-clamp460 was much less sensitive to CPT in the presence of DTT than yTop1 (Fig. 7B, compare lanes 9 and 5). These data suggest that the architecture of the Top1-clamp is conserved in so far as the formation of a disulfide bond to lock the protein clamp closed. However, in the context of the shorter N-terminal domain of yTop1, locking of the yTop1-clamp460 increases the stability of the covalent enzyme-DNA complex, which is only marginally enhanced by CPT. This is consistent with the increased toxicity induced by yTop1-clamp460 in glr1Δ versus wild-type GLR1 cells, seen in Fig. 6B.
FIGURE 7.
yTop1-clamp460 exhibits increased levels of covalent complexes in the absence of CPT. A, equal concentrations of purified yTop1 and yTop1-clamp460 were serially 10-fold diluted and incubated in a plasmid DNA relaxation assay in the presence or absence of 10 mm DTT and the indicated salt concentrations. Following incubation at 30 °C for 1 h, the extent of plasmid DNA relaxation was assessed by agarose gel electrophoresis and ethidium bromide staining. Lane C, plasmid DNA control. R and (-), the positions of relaxed and supercoiled DNA, respectively. B, equal concentrations of yTop1 (lanes 2–5) and Top1-clamp460 (lanes 6–9) were incubated in DNA cleavage reactions with a single 3′-32P-labeled DNA substrate as in Fig. 4. Where indicated, reactions contained 50 μm CPT (lanes 3, 5, 7, and 9) and 10 mm DTT (lanes 4, 5, 8, and 9). After 10 min at 30 °C, covalent complexes were resolved in polyacrylamide/urea gels and visualized by PhosphorImager analysis. *, position of a Top1 high affinity cleavage site. The arrows indicate yTop1-clamp460 complexes stabilized in the absence of DTT and CPT.
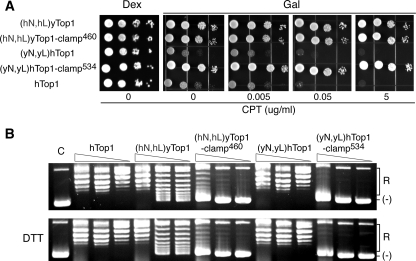
In our yeast/human chimera studies, replacing the shorter human linker domain of hTop1 with the longer linker domain of yeast Top1 (in (yL)hTop1) was toxic to yeast top1Δ cells.5 However, when the corresponding yeast N-terminal domain was used to replace the human N terminus (in (yN,yL)hTop1), yeast cell sensitivity to CPT mirrored that induced by expression of wild-type hTop1 (Fig. 8A). Based on these findings, we asked what effect reciprocal swaps of the paired yeast and human N-terminal/linker domains might have on Top1-clamp activity. As shown in Fig. 8A, when human Top1-clamp534 was engineered to contain the yeast N-terminal and linker domains ((yN,yL)hTop1-clamp534) or yTop1-clamp460 contained the corresponding human Top1 domains ((hN,hL)yTop1-clamp460), expression of these chimeric Top1-clamps failed to induce any adverse effects on yeast cell viability, either in the presence or absence of CPT. In the context of wild-type yTop1, the presence of human N-terminal/linker domains ((hN,hL)y-Top1) failed to induce CPT sensitivity, despite the fact that the catalytic activity of this mutant enzyme mirrored that of wild-type yTop1 (Fig. 8, A and B) (data not shown). The specific activity of (hN,hL)yTop1-clamp460 (Fig. 8B) also resembled that of the yTop1-clamp460 in the presence of DDT (Fig. 7A). However, the activity of chimera ((hN,hL)yTop1-clamp460) was unaffected by the addition of DTT. These data suggest that swapping the paired N-terminal/linker domains impaired the formation of the disulfide bond to lock the protein clamp, thereby suppressing the toxicity of the yTop1-clamp460 in yeast. In contrast, introducing yeast N-terminal/linker domains into hTop1-clamp534 rendered the chimera unstable, since the lack of enzyme activity corresponded to the absence of detectable protein in Western blots (Fig. 8B) (data not shown). Taken together, our findings indicate that the presence of a functional N-terminal domain and its interaction with the appropriate linker impact the dynamics of Top1 protein clamp closure and the toxicity of the Top1-clamp in vivo.
FIGURE 8.
The N-terminal and linker domains modulate locking of the yTop1-clamp460. A, exponential cultures of top1Δ cells transformed with the indicated YCpGAL1-yeast and human top1 chimeras were serially diluted and spotted onto SC-uracil plates supplemented with dextrose (Dex) or galactose (Gal), 25 mm HEPES (pH 7.2), 0.125% Me2SO, and the indicated concentrations of CPT. Cell viability was assessed after incubation at 30 °C. B, crude cell extracts of galactose-induced cultures were corrected for protein concentration, serially 10-fold diluted, and incubated in plasmid DNA relaxation assays as described under “Experimental Procedures.” Lane C, plasmid DNA control. (R) and (-), the positions of relaxed and supercoiled DNA, respectively.
DISCUSSION
A DNA-bound clamp is a common structure adopted by proteins or complexes that function in DNA replication, repair, and transcription (2, 38–40). However, the clamping of eukaryotic Top1 around duplex DNA presents some unique architectural features. First, the DNA duplex is tightly packed within the central pore of the Topo70-clamp (Fig. 1) (2, 12, 14, 15, 41). Second, the extended α-helices of the flexible linker domain and the shorter α-helices of the top portion of the clamp impose asymmetry on the complex. Both pairs of helices flank the DNA that rotates in response to the torsional and/or flexural strain induced by the movement of replication forks or the transcription machinery along the DNA.
One strategy to investigate enzyme catalysis is the design of a reversible disulfide bond to restrict the relative movement of protein domains. For instance, Roca et al. (42) engineered a disulfide bond across the DNA topoisomerase II dimer interface. Similarly, disulfide bonds were engineered in human Top1 to assess whether locking the Top1-clamp would impede DNA rotation within the covalent Top1-DNA complex (15, 16). We demonstrated that locking the full-length Top1-clamp534 prevented DNA rotation (15), whereas Carey et al. (16) reported that DNA rotation was not affected by the locked Topo70-clamp499.
Considering these distinct experimental outcomes, we reasoned that subtle alterations in enzyme architecture might profoundly affect enzyme catalysis (15). The disulfide bond of Top1-clamp534 tethers the upper lip to a more active site proximal loop of the lower lip, whereas a more distal lower loop is cross-linked in the Topo70-clamp499 (Fig. 1). However, these reports did not address the in vivo activity of the Topo70-clamp499 or the contribution of the N-terminal domain to enzyme catalysis. Although the spatial organization of the N terminus remains an enigma, recent studies of yeast/human Top1 chimeras revealed a functional interaction between the N-terminal and linker domains in vivo.5 Indeed, our studies indicate that the position of the disulfide linkage and the N-terminal domain affect both the dynamics of Top1 clamping of duplex DNA and the cytotoxicity induced by locking the Top1 protein clamp in vivo. Several aspects of these studies are worth considering.
First, in the context of the full-length human enzyme, our biochemical data suggest that the formation of a disulfide bond to lock the Top1-clamp499 occurs more readily in the absence of DNA than with Top1-clamp534. These differences were readily apparent in DNA cleavage assays, where the Top1-clamp534 appeared to be mostly “unlocked” in the absence of DNA, whereas efficient CPT poisoning of the Top1-clamp499 was only observed in the presence of DTT. Mechanistically, these differences could be attributed to a more severe defect in DNA binding induced by the cysteine substitutions in Top1-clamp499 than in Top1-clamp534. In the case of Top1-clamp499, a decreased affinity for DNA would favor disulfide bond formation prior to DNA binding and thereby preclude effective locking of Top1 on the DNA in vivo. Indeed, contrary to Top1-clamp534, Top1-clamp499 was not toxic to glr1Δ cells in the absence of CPT and only induced a low level of CPT sensitivity in wild-type cells.
The N-terminal 190 residues of human Top1 are dispensable for enzyme catalysis in vitro, yet they have been implicated in mediating protein interactions in vivo. Our results suggest that residues 1–174 also affect the stability and dynamics of Top1-clamp locking in vivo. As reported for Top1, deletion of the N-terminal residues of Top1-clamp534 did not alter the specific activity of the enzyme in vitro or the levels of protein expressed in vivo (Fig. 5). Nevertheless, the cytotoxicity of the truncated Topo70-clamp534 and Topo70-clamp534Y723F were diminished relative to that observed with the full-length clamps. The increased toxicity induced in the more oxidizing environment of glr1Δ cells also argues for the impaired formation of a locked Topo70-clamp534 in vivo. Since the specific activity of the full-length and truncated proteins are comparable, the N-terminal domain does not affect Top1 protein binding of DNA. Rather, we think it more likely that alterations in the interaction of Topo70-clamp534 with other proteins or complexes decrease the likelihood of forming a potentially lethal protein clamp on the DNA. Several lines of evidence suggest the association of Top1 with gene transcription and replication machinery, consistent with a role for this enzyme as a swivelase to alleviate the overwinding of DNA generated in advance of these processive complexes (1, 2, 7). The mechanisms by which Top1 is recruited to these complexes in vivo have yet to be elucidated. However, protein interactions with the N-terminal domain of Top1 have been reported to alter the intracellular localization of the enzyme (18). Moreover, the N-terminal domain of Top1 contains consensus sites for covalent modification by the small ubiquitin-related modifier, which may also regulate the formation of multiprotein complexes containing Top1 (34, 43). Thus, it is reasonable to propose that protein-protein interactions would not only function to target Top1 but could also influence the interaction of the N-terminal domain with the rest of the Top1 protein clamp. Similar considerations would hold for the catalytically inactive Topo70-clamp534Y723F.
It is also tempting to posit that alterations in protein-protein interactions adversely impact the stability of the Topo70-clamp499. In this case, specific proteins or complexes may be required to prevent the proteolytic degradation of the locked Top1-clamp499. Indeed, the ubiquitin-dependent down-regulation of CPT-stabilized Top1-DNA complexes has been reported (44, 45), which may be triggered by the accumulation of stabilized Top1 clamps. Alternatively, alterations in N-terminal and linker domain composition of the Top1-clamp constructs could prevent appropriate Top1 protein folding. For instance, replacing the human N-terminal and linker domains with the corresponding yeast domains (in (yN/yL)hTop) did not impair Top1 enzyme function or CPT sensitivity in vivo (Fig. 8). However, the same domain swaps in the context of the Top1-clamp534 suppressed the expression of detectable protein in vivo. Further studies will address these possibilities. Nevertheless, these data suggest that the N-terminal/linker composition of Top1 affects protein clamp dynamics. Frøhlich et al. (46) recently reported the in vitro activity of a human Top1 that contains a 6-amino acid linker from the Cre recombinase. It would be interesting to assess the dynamics of Top1-clamp function in the context of a minimal linker.
It is also worth noting that the overall architecture of yeast and human Top1 and, in particular, the juxtaposition of the lip domains appears to be conserved. Our molecular modeling of human Topo70-DNA complexes identified only two residue pairs with the necessary orientation and proximity to form a disulfide bond when substituted with cysteines (15). Indeed, these predictions were borne out in the characterizations of human Top1-clamp534 and Topo70-clamp499 (15, 16). Remarkably, introducing the Top1-clamp534 cysteine substitutions at the corresponding positions in yeast Top1 also produced a reversible disulfide bond that locked the yeast Top1-clamp460 closed. The enhanced toxicity of yTop1-clamp460 and yTop1-clamp460Y727F in the more oxidizing environment of a glr1Δ strain and the DTT-dependent alterations in enzyme activity argue for the formation of a reversible disulfide bond that locks the yeast protein clamp. Thus, despite extensive differences in the composition of yeast and human Top1 N-terminal/linker domains, the intramolecular interactions critical for the formation and flexibility of the Top1 protein clamp appear to be conserved.
Supplementary Material
Acknowledgments
We thank Padma Thimmaiah and Hong Guo for assistance and members of the Bjornsti and Benedetti laboratories for thoughtful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants CA58755 and CA111542 (to M.-A. B.) and National Institutes of Health, NCI, Cancer Center Core Grant CA21765. This work was also supported by the American Lebanese Syrian Associated Charities and grants from Ministero dell'Istruzione, dell'Università e della Ricerca, Cofinanziamento 2005–2007 and Ministero della Salute (to P. B.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Footnotes
The abbreviations used are: CPT, camptothecin; NLS, nuclear localization signal; DTT, dithiothreitol.
M. van der Merwe, C.M. Wright, and M.-A. Bjornsti, unpublished results.
References
- 1.Champoux, J. J. (2001) Annu. Rev. Biochem. 70 369-413 [DOI] [PubMed] [Google Scholar]
- 2.Corbett, K. D., and Berger, J. M. (2004) Annu. Rev. Biophys. Biomol. Struct. 33 95-118 [DOI] [PubMed] [Google Scholar]
- 3.Pommier, Y. (2006) Nat. Rev. Cancer 6 789-802 [DOI] [PubMed] [Google Scholar]
- 4.Wang, J. C. (2002) Nat. Rev. Mol. Cell. Biol. 3 430-440 [DOI] [PubMed] [Google Scholar]
- 5.Koster, D. A., Croquette, V., Dekker, C., Shuman, S., and Dekker, N. H. (2005) Nature 434 671-674 [DOI] [PubMed] [Google Scholar]
- 6.Koster, D. A., Palle, K., Bot, E. S., Bjornsti, M. A., and Dekker, N. H. (2007) Nature 448 213-217 [DOI] [PubMed] [Google Scholar]
- 7.Bjornsti, M. A. (2002) Cancer Cell 2 267-273 [DOI] [PubMed] [Google Scholar]
- 8.Leppard, J. B., and Champoux, J. J. (2005) Chromosoma 114 75-85 [DOI] [PubMed] [Google Scholar]
- 9.Liu, L. F., Desai, S. D., Li, T. K., Mao, Y., Sun, M., and Sim, S. P. (2000) Ann. N. Y. Acad. Sci. 922 1-10 [DOI] [PubMed] [Google Scholar]
- 10.Chrencik, J. E., Staker, B. L., Burgin, A. B., Pourquier, P., Pommier, Y., Stewart, L., and Redinbo, M. R. (2004) J. Mol. Biol. 339 773-784 [DOI] [PubMed] [Google Scholar]
- 11.Staker, B. L., Hjerrild, K., Feese, M. D., Behnke, C. A., Burgin, A. B., Jr., and Stewart, L. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 15387-15392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Redinbo, M. R., Stewart, L., Kuhn, P., Champoux, J. J., and Hol, W. G. J. (1998) Science 279 1504-1513 [DOI] [PubMed] [Google Scholar]
- 13.Stewart, L., Ireton, G. C., and Champoux, J. J. (1996) J. Biol. Chem. 271 7602-7608 [DOI] [PubMed] [Google Scholar]
- 14.Stewart, L., Redinbo, M. R., Qiu, X., Hol, W. G. J., and Champoux, J. J. (1998) Science 279 1534-1541 [DOI] [PubMed] [Google Scholar]
- 15.Woo, M. H., Losasso, C., Guo, H., Pattarello, L., Benedetti, P., and Bjornsti, M. A. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 13767-13772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carey, J. F., Schultz, S. J., Sisson, L., Fazzio, T. G., and Champoux, J. J. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 5640-5645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bjornsti, M.-A., and Wang, J. C. (1987) Proc. Natl. Acad. Sci. U. S. A. 84 8971-8975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haluska, P. J., and Rubin, E. H. (1998) Adv. Enzyme Regul. 38 253-262 [DOI] [PubMed] [Google Scholar]
- 19.Lisby, M., Olesen, J. R., Skouboe, C., Krogh, B. O., Straub, T., Boege, F., Velmurugan, S., Martensen, P. M., Andersen, A. H., Jayaram, M., Westergaard, O., and Knudsen, B. R. (2001) J. Biol. Chem. 276 20220-20227 [DOI] [PubMed] [Google Scholar]
- 20.Stewart, L., Ireton, G., and Champoux, J. (1997) J. Mol. Biol. 269 355-372 [DOI] [PubMed] [Google Scholar]
- 21.Alsner, J., Svestrup, J. Q., Kjeldsen, E., Sorensen, B. S., and Westergaard, O. (1992) J. Biol. Chem. 267 12408-12411 [PubMed] [Google Scholar]
- 22.Mo, Y. Y., Wang, C., and Beck, W. T. (2000) J. Biol. Chem. 275 41107-41113 [DOI] [PubMed] [Google Scholar]
- 23.Fiorani, P., Bruselles, A., Falconi, M., Chillemi, G., Desideri, A., and Benedetti, P. (2003) J. Biol. Chem. 278 43268-43275 [DOI] [PubMed] [Google Scholar]
- 24.Losasso, C., Cretaio, E., Palle, K., Pattarello, L., Bjornsti, M. A., and Benedetti, P. (2007) J. Biol. Chem. 282 9855-9864 [DOI] [PubMed] [Google Scholar]
- 25.Stewart, L., Ireton, G. C., and Champoux, J. J. (1999) J. Biol. Chem. 274 32950-32960 [DOI] [PubMed] [Google Scholar]
- 26.van der Merwe, M., and Bjornsti, M. A. (2008) J. Biol. Chem. 283 3305-3315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fiorani, P., Reid, R. J., Schepis, A., Jacquiau, H. R., Guo, H., Thimmaiah, P., Benedetti, P., and Bjornsti, M. A. (2004) J. Biol. Chem. 279 21271-21281 [DOI] [PubMed] [Google Scholar]
- 28.Kauh, E. A., and Bjornsti, M. A. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 6299-6303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Megonigal, M. D., Fertala, J., and Bjornsti, M. A. (1997) J. Biol. Chem. 272 12801-12808 [DOI] [PubMed] [Google Scholar]
- 30.Colley, W. C., van der Merwe, M., Vance, J. R., Burgin, A. B., Jr., and Bjornsti, M. A. (2004) J. Biol. Chem. 279 54069-54078 [DOI] [PubMed] [Google Scholar]
- 31.Woo, M. H., Vance, J. R., Marcos, A. R., Bailly, C., and Bjornsti, M. A. (2002) J. Biol. Chem. 277 3813-3822 [DOI] [PubMed] [Google Scholar]
- 32.van Waardenburg, R. C., Duda, D. M., Lancaster, C. S., Schulman, B. A., and Bjornsti, M. A. (2006) Mol. Cell. Biol. 26 4958-4969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaiser, C., Michaelis, S., and Mitchell, A. (1994) Methods in Yeast Genetics, 1994 Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- 34.Jacquiau, H. R., van Waardenburg, R. C., Reid, R. J., Woo, M. H., Guo, H., Johnson, E. S., and Bjornsti, M. A. (2005) J. Biol. Chem. 280 23566-23575 [DOI] [PubMed] [Google Scholar]
- 35.Frohlich, R. F., Andersen, F. F., Westergaard, O., Andersen, A. H., and Knudsen, B. R. (2004) J. Mol. Biol. 336 93-103 [DOI] [PubMed] [Google Scholar]
- 36.Frohlich, R. F., Veigaard, C., Andersen, F. F., McClendon, A. K., Gentry, A. C., Andersen, A. H., Osheroff, N., Stevnsner, T., and Knudsen, B. R. (2007) Nucleic Acids Res. 35 6170-6180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Redinbo, M. R., Champoux, J. J., and Hol, W. G. (2000) Biochemistry 39 6832-6840 [DOI] [PubMed] [Google Scholar]
- 38.Dionne, I., Brown, N. J., Woodgate, R., and Bell, S. D. (2008) Mol. Microbiol. 68 216-222 [DOI] [PubMed] [Google Scholar]
- 39.Georgescu, R. E., Kim, S. S., Yurieva, O., Kuriyan, J., Kong, X. P., and O'Donnell, M. (2008) Cell 132 43-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jansen, J. G., Fousteri, M. I., and de Wind, N. (2007) Mol. Cell 28 522-529 [DOI] [PubMed] [Google Scholar]
- 41.Cheng, C., Kussie, P., Pavletich, N., and Shuman, S. (1998) Cell 92 841-850 [DOI] [PubMed] [Google Scholar]
- 42.Roca, J., Berger, J. M., Harrison, S. C., and Wang, J. C. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 4057-4062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geiss-Friedlander, R., and Melchior, F. (2007) Nat. Rev. Mol. Cell. Biol. 8 947-956 [DOI] [PubMed] [Google Scholar]
- 44.Desai, S. D., Liu, l. F., Vazquez-Abad, D., and D'Arpa, P. (1997) J. Biol. Chem. 272 24159-24164 [DOI] [PubMed] [Google Scholar]
- 45.Lin, C. P., Ban, Y., Lyu, Y. L., Desai, S. D., and Liu, L. F. (2008) J. Biol. Chem. 283 21074-21083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frøhlich, R. F., Juul, S., Nielsen, M. B., Vinther, M., Veigaard, C., Hede, M. S., and Andersen, F. F. (2008) Biochemistry 47 7127-7136 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.