Abstract
Eukaryotic Okazaki fragment maturation requires complete removal of the initiating RNA primer before ligation occurs. Polymerase δ (Pol δ) extends the upstream Okazaki fragment and displaces the 5′-end of the downstream primer into a single nucleotide flap, which is removed by FEN1 nuclease cleavage. This process is repeated until all RNA is removed. However, a small fraction of flaps escapes cleavage and grows long enough to be coated with RPA and requires the consecutive action of the Dna2 and FEN1 nucleases for processing. Here we tested whether RPA inhibits FEN1 cleavage of long flaps as proposed. Surprisingly, we determined that RPA binding to long flaps made dynamically by polymerase δ only slightly inhibited FEN1 cleavage, apparently obviating the need for Dna2. Therefore, we asked whether other relevant proteins promote long flap cleavage via the Dna2 pathway. The Pif1 helicase, implicated in Okazaki maturation from genetic studies, improved flap displacement and increased RPA inhibition of long flap cleavage by FEN1. These results suggest that Pif1 accelerates long flap growth, allowing RPA to bind before FEN1 can act, thereby inhibiting FEN1 cleavage. Therefore, Pif1 directs long flaps toward the two-nuclease pathway, requiring Dna2 cleavage for primer removal.
During eukaryotic DNA replication, the lagging strand is replicated via synthesis and maturation of Okazaki fragments. These fragments are short stretches of DNA that are joined to generate a continuous strand (1). Each fragment is initiated when DNA polymerase α/primase (Pol α)3 makes an RNA/DNA primer, synthesizing ∼10 nucleotides (nt) of RNA followed by 10–20 nt of DNA (2). The primer is then extended by a complex of DNA polymerase δ (Pol δ), the sliding clamp, proliferating cell nuclear antigen (PCNA), and the clamp loader, replication factor C (RFC). When Pol δ encounters the 5′-end of the downstream Okazaki fragment, it displaces it into a flap. Cleavage of the flap by nucleases generates a nick, which is subsequently sealed by DNA ligase I to form continuous double-stranded DNA (3, 4).
One pathway for cleavage of the flap employs flap endonuclease 1 (FEN1). FEN1 is a single-strand, structure-specific endonuclease that enters the 5′-end of the flap and tracks to the base for cleavage (3, 5, 6). Following displacement of a short flap, less than about 12 nt, by Pol δ, FEN1 cleaves leaving a nick, the substrate for DNA ligase I (7–9). Because the RNA initiating the Okazaki fragments is ∼10 nt in length, short flaps composed entirely of RNA are first displaced by Pol δ. This does not obstruct FEN1, which is active on RNA (10, 11). In addition, displacement and cleavage occurs mostly within the first 25 nt of the downstream fragment, sufficient to remove the entire RNA/DNA primer, which is ∼20–30 nt in length (11). Previous biochemical studies demonstrated that primarily short flaps are cleaved by FEN1. It is likely that in vivo a series of short successive displacements by Pol δ and cleavages by FEN1 are effective for removal of the entire initiating RNA/DNA primer (7–9, 11).
However, although Pol δ and FEN1 appear to be designed to keep flaps short, reconstitutions in vitro demonstrate the creation of some long flaps, ∼20–30 nt in length (11). Biochemical results suggest that once flaps achieve lengths in the range of 30 nt they can be bound by RPA, which inhibits FEN1 cleavage. RPA-coated flaps must then be processed by an alternative pathway.
This second pathway is proposed to require Dna2 (13). Dna2 is an ATP-dependent helicase and a single-stranded DNA-specific endonuclease (14, 15). The endonuclease activity requires tracking from the 5′-end of a flap and is stimulated by RPA (13, 16, 17). Because the specificity of Dna2 does not allow for cleavage at the base of a flap to generate a nick, the terminal Dna2 product is a short flap (17). The remaining flap is too small to be bound by RPA, so it can be cleaved by FEN1 to generate the nick for ligation (13, 15). In this way, the properties of the nucleases and RPA order the reactions with Dna2 cleaving first and FEN1 second.
The proposed coordinated function of Dna2 and FEN1 is supported by other results in which they have been shown to interact physically through co-immunoprecipitation (18) and genetically through deletion studies in Saccharomyces cerevisiae (18, 19). Deletion of DNA2 is lethal in S. cerevisiae (20, 21). Moreover, the dna2-1 mutant, which has impaired nuclease activity, is synthetically lethal with the rad27Δ, which has impaired FEN1 flap-cleavage activity (18). However, dna2-1 nuclease mutants show a temperature-sensitive phenotype that is suppressed by overexpression of FEN1 (18, 19). Likewise, the temperature-sensitive phenotype of rad27Δ mutants is suppressed by overexpression of Dna2 (18).
In addition, in S. cerevisiae, recent deletion studies highlight the importance of Dna2 in replication and implicate the Pif1 helicase in maturation of Okazaki fragments (22). Pif1 is a 5′-3′ helicase conserved from yeast to humans (23, 24). Budd and colleagues (22) found that pif1Δ rescued the lethality of dna2Δ. Also, pif1Δ rescued the temperature-sensitive phenotype of the dna2-1 mutant. They proposed that Pif1 creates a need for Dna2 by promoting formation of long flaps. Although deletion of PIF1 rescued the dna2-1 mutant, the mutant strain had residual phenotypic defects consistent with disruptions in replication. The additional deletion of POL32 in the dna2-1 pif1Δ strain suppressed these defects (22). Pol32 is the subunit of Pol δ that binds to PCNA, and deletion of this subunit has been shown to limit some of the strand-displacement activity of Pol δ (25, 26). The phenotype of the triple mutant dna2-1 pif1Δ pol32Δ further supports the hypothesis that Pif1 promotes long flap formation (22). By deleting both PIF1 and POL32, it is likely that flap displacement lengths are shorter and flaps do not bind RPA, a situation that eliminates the need for Dna2 cleavage.
Similar to Pif1 in S. cerevisiae, it was previously shown that Pfh1, the Pif1 ortholog in Schizosaccharomyces pombe, has an interaction with Dna2 (27). Pfh1 is essential in S. pombe (28). However, mutation of pfh1 suppresses the temperature-sensitive phenotype of dna2 mutants. Moreover, mutations in pfh1 and cdc27, the PCNA-interacting subunit of Pol δ in S. pombe, are synthetically lethal (27). These results suggest that Pfh1, Dna2, and Pol δ participate in similar pathways in yeast, presumably Okazaki fragment maturation.
Together the biochemical and genetic data suggest that both FEN1 and Dna2 are involved in the processing of flaps to remove the RNA/DNA primer initiating Okazaki fragments (3, 4, 29). However, it has not been clearly established how the presence of RPA and Pif1 may affect the directing of flaps into either the FEN1 cleavage pathway or the two-nuclease pathway requiring Dna2. Through reconstitution studies in vitro with purified proteins from S. cerevisiae, we set out to examine the influence of RPA and Pif1 on the length of flap displacement and subsequent cleavage by FEN1.
EXPERIMENTAL PROCEDURES
Materials—Oligonucleotides were synthesized by either Midland Certified Reagents Company (Midland, TX) or Integrated DNA Technologies (Coralville, IA). Radionucleotides [γ-32P]ATP and [α-32P]dCTP were purchased from PerkinElmer Life Sciences. Polynucleotide kinase, the Klenow fragment of Escherichia coli DNA polymerase I, and streptavidin were obtained from Roche Applied Science. All other reagents were the best commercially available.
Enzyme Expression and Purification—S. cerevisiae wild-type Pol δ (Pol δ-wt) (25) and 3′-5′ exonuclease-deficient Pol δ (Pol δ-01) (9, 25) were overexpressed and purified from S. cerevisiae as previously described. S. cerevisiae RFC was overexpressed and purified from E. coli as previously described (30). S. cerevisiae Rad27 (31) (referred to as FEN1) and PCNA (11) were cloned into the T7 expression vector pET-24b (Novagen/EMD Biosciences, Madison, WI), expressed in E. coli BL21(DE3) codon plus strain (Stratagene and Novagen/EMD Biosciences, respectively) and purified as previously described, resulting in a recombinant protein with a C-terminal His6 tag. S. cerevisiae Pif1 was cloned into the pET-28b bacterial expression vector (Novagen/EMD Biosciences), expressed in the E. coli Rosetta strain (Novagen/EMD Biosciences), and purified as previously described resulting in recombinant protein with an N-terminal His6 tag (32).
Oligonucleotide Substrates—Substrates were designed to resemble Okazaki fragment processing intermediates using oligonucleotide primers. The primer sequences are listed in Table 1. Primers D1, D2, and D3 were radiolabeled on the 5′-end with [γ-32P]ATP and polynucleotide kinase. Primer D1 was radiolabeled on the 3′-end by annealing a 20-nt labeling template with a 5′-GCTA overhang to the 3′-end of the primer and incorporating dC using [α-32P]dCTP and the Klenow polymerase. Radiolabeled primers were fractionated by electrophoresis on a 15% 7 m urea polyacrylamide gel and then purified. The primers were then annealed into substrates in a 1:2:4 ratio of labeled downstream primer to template to upstream primer. Substrates were annealed by combining corresponding primers in a buffer containing 50 mm Tris-HCl, pH 8.0, 50 mm NaCl, and 1 mm dithiothreitol, heating at 95 °C for 5 min, transferring to 70 °C, and cooling to room temperature.
TABLE 1.
Oligonucleotide sequences
| Primer | Length | Sequence |
|---|---|---|
| nt | ||
| Upstream (5′-3′) | ||
| U1 | 20 | GTCCACCCGCGCCACCTCC |
| U2 | 44 | GTCCACCCGCGCCACCTCCTGCTTCAATGTGCTGGATCCTA |
| Downstream (5′-3′) | ||
| D1 | 60 | AGACGAATTCGGATACGACGGCAGTGCCGACCGTCCAGCCTAAATTCAATCCACCC |
| D2a | 59 | GACGAATTCGGATACGACGGCCGTGCCGACCGTGCAGCCTAAATTCAATCCACCC |
| D3b | 59 | GACGAATTCGGATACGACGGCCGTGCCGACCGTGCAGCCTAAATTCAATCCACCC |
| Template (3′-5′) | ||
| T1 | 42 | CAGGTGGGCGCGGTGGAGGGTCGATTTAAAGTTAGTGGG |
| T2 | 51 | CAGGTGGGCGCGGTGGAGGGCTGCACGGTCGGATTAAAGTTAGGGGG |
| T3 | 60 | CAGGTGGGCGCGGTGGAGGCCGTCACGGCTGGCAGGTCGGATTTAAGTTAGGTGGG |
| T4c | 110 | CAGGTGGGCGCGGTGGAGGACGAAGTTACACGACCTAGGATGTTGTTCTGCTTAAGCCTATGCTCCGGTCACGGCTGCACGGTCGGATTAAAGTTAGGTGG |
Underline indicates the RNA segment.
Underline and italicized indicates the 2′-O-methylated RNA segment.
Template T4 is biotinylated at both the 5′- and 3′-ends.
Three sets of substrates were used in the following experiments. One set was used to examine FEN1 cleavage following strand displacement by Pol δ. These contained a 44-nt upstream primer and a 60-nt downstream primer annealed to the 3′- and 5′-ends, respectively, of a 110-nt template and will be referred to as the standard-44 substrate. A second set of substrates contained the same 44-nt upstream primer and 110-nt template of the standard-44 substrate, except the downstream primer was 59-nt in length and initiated by either a single nucleotide of RNA or a single nucleotide of 2′-O-methylated RNA. The annealed substrates will be referred to as the RNA substrate or methylated RNA substrate, respectively. The third set of substrates were fixed double-flaps, in which an upstream 1-nt 3′ tail complementary to the template overlaps with a 5′ flap on the downstream primer, and was used for examining FEN1 cleavage. The fixed double-flap substrates contained a 20-nt upstream primer and 60-nt downstream primer annealed to the 3′- and 5′-ends, respectively, of varying length templates, 60, 51, or 42 nt, forming 19-, 28-, or 37-nt double-flaps, respectively. The sequence of the fixed double-flap substrates is the same on the flap and at the base of the flap as when flaps of 19, 28, or 37 nt are formed from displacement of the downstream primer on the standard-44 substrates. Specific substrates used are indicated in the figure legends and depicted above the figures.
Enzyme Assays—For strand-displacement reactions, 5 fmol of labeled biotinylated substrate was incubated with 500 fmol of streptavidin for 20 min on ice, prior to starting each reaction. The streptavidin complexes with the biotinylated template strand and blocks the ends of the DNA, providing a substrate on which RFC must load PCNA. In the figures, substrates are depicted without blocked ends for simplicity. Substrate was then incubated with 23 fmol of Pol δ-wt or Pol δ-01, 25 fmol each of PCNA and RFC, 20 fmol of FEN1, 100 fmol of RPA, and 25, 50, or 100 fmol of Pif1 for 10 min at 30 °C in a total volume of 20 μl of reaction buffer. Reaction buffer contained 50 mm Tris-HCl, pH 7.5, 2 mm dithiothreitol, 25 μg/ml bovine serum albumin, 0.5 mm MgCl2, 1 mm ATP, 50 μm dNTPs, and 75 mm NaCl. Reactions were stopped by addition of 10 μl of 2× termination dye (90% formamide (v/v), 10 mm EDTA with 0.01% bromphenol blue and xylene cyanole) and heating at 95 °C for 5 min. Products were separated by electrophoresis on a 15% 7 m urea polyacrylamide gel. The gel was dried, scanned using a GE Healthcare PhosphorImager, and analyzed using Image Quant version 1.2 software. All assays were repeated at least in triplicate with a representative gel shown in the figure.
For the fixed double-flap cleavage reactions, 5 fmol of substrate was incubated with 500 fmol of streptavidin for 20 min on ice, prior to the start of each reaction. Although the substrates were not conjugated with biotin, streptavidin was included to control for any effects of free streptavidin in the reaction mixture. Substrate was then incubated with 20 fmol of FEN1 and 50, 100, or 200 fmol of RPA for 10 min at 30 °C in a total volume of 20 μl of reaction buffer (same as above). Reactions were stopped, electrophoresed, and analyzed as directed above.
Cleavage Product Distribution Analysis—For each lane of interest, the cleavage product bands were identified and correlated with flap length. Using ImageQuant software, peak pixel intensities of each gel lane were obtained and matched with a corresponding cleavage product band and flap length. Pixel intensities of the cleavage bands for FEN1 cleavage alone were subtracted from the intensities of bands in reaction lanes containing Pol δ, PCNA/RFC, and FEN1. The pixel intensity versus the size of the flap cleaved was graphed. Then, a moving average trendline (period of three) was added to the graph to smooth and more clearly indicate the trend of the data.
RESULTS
Because a fraction of the flaps made in our Okazaki fragment processing system was found to become long before cleavage by FEN1 (11), we questioned whether RPA binding on these long flaps is sufficient to effectively inhibit FEN1 cleavage, necessitating cleavage by Dna2. First, we examined RPA inhibition on flaps generated dynamically through strand displacement. Additionally, based on the genetic evidence of a role for Pif1 in Okazaki fragment maturation through its interaction with Dna2 (22), we asked whether Pif1 stimulates formation of long flaps, creating an additional need for the two-nuclease pathway.
Cleavage Product Distribution Indicates Both Short and Long Products—During the maturation of Okazaki fragments in vivo, the flaps form through the dynamic process of strand-displacement synthesis. Accordingly, FEN1 cleavage was investigated during strand displacement on a substrate that simulates an intermediate of Okazaki fragment maturation, the standard-44 substrate. This substrate contains an upstream primer and downstream primer annealed to the 3′- and 5′-ends, respectively, of an oligonucleotide template strand. Between the two primers is a 6-nt gap allowing space for the polymerase to synthesize, extending the upstream primer. FEN1 cleavage product size was measured during strand-displacement synthesis by either Pol δ-wt or the 3′-5′ exonuclease-deficient Pol δ-01, PCNA, and RFC on the standard-44 substrate (Fig. 1). For visualization of the initial FEN1 cleavage products, the substrate was radiolabeled on the 5′-end of the downstream primer.
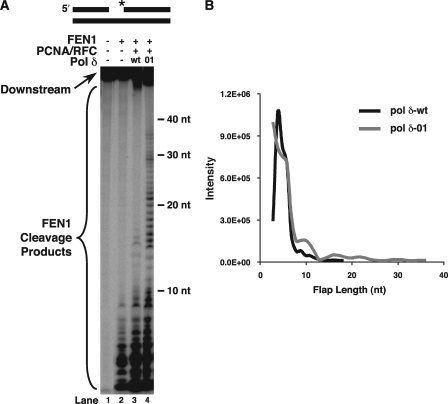
FIGURE 1.
Cleavage product distribution indicates both short and long products. A, initial FEN1 cleavage was assayed on the standard-44 substrate (U2:T4:D1) in the presence of PCNA/RFC and either Pol δ-wt, denoted wt (lane 3), or Pol δ-01, denoted 01 (lane 4), as described under “Experimental Procedures.” The substrate is depicted above the figure. The asterisk indicates location of radiolabel. B, the size of flap cleavage products versus the intensity of the product band was graphed and moving average trendline added, as described under “Experimental Procedures.” The black line corresponds to the cleavage products for reconstitutions with Pol δ-wt. The gray line corresponds to the products for reconstitutions with Pol δ-01.
A 6-nt single-stranded DNA gap between the upstream and downstream primers was specifically chosen for use in this study. This 6-nt gap was expected to be too small for RPA binding (33). We also performed band shift experiments comparing the level of RPA binding to a 6-nt gap substrate, a 30-nt gap substrate, and a 60-nt length of single-stranded DNA. Results of these experiments demonstrated that RPA is unable to bind the 6-nt gap substrate even at a concentration 2-fold higher than used in the strand-displacement experiments (data not shown). However, RPA did bind well to both the 30-nt gap substrate and the single-stranded DNA. Consequently, the effects of RPA binding are exclusively confined to the displaced flaps created dynamically by Pol δ.
In the presence of PCNA, RFC, and either Pol δ-wt or Pol δ-01, the maximum lengths of FEN1 cleavage products were ∼18 and 36 nt, respectively, for the two polymerases (Fig. 1A, lanes 3 and 4, respectively). This is consistent with the product lengths observed in our prior studies using a 30-nt gap-containing substrate, indicating that the length of the gap does not influence the results, because these gaps are filled in efficiently by Pol δ prior to strand-displacement synthesis (11). The longer flaps created and cleaved in the presence of Pol δ-01 are understood to derive from the increased strand-displacement activity of 3′-5′ exonuclease-deficient Pol δ mutants (8, 9, 11).
Distribution of the FEN1 cleavage products accompanying either Pol δ-wt or Pol δ-01 displacement synthesis was evaluated. After quantitating the pixel intensities of the product bands, band intensity versus the size of the flap cleaved was graphed (Fig. 1B). The distribution revealed both a large population of flaps cleaved at the short stage, up to ∼10 nt, and a second small population of products cleaved from long flaps, up to ∼18 or 36 nt, following displacement by either the Pol δ-wt or Pol δ-01, respectively. Similar to what was observed before on the standard 30-nt gap-containing substrate (11), there was a sharp peak distribution for the short cleavage products, and the long cleavage product distribution was broad and flat (Fig. 1B). Similarity of the two-population distributions with different substrates suggests that such distributions are generally characteristic of FEN1 cleavage of Pol δ-displaced flaps.
FEN1 Cleavage during Flap Displacement Is Only Moderately Inhibited by RPA—Assessing the effects of RPA on FEN1 cleavage of long flaps is important for understanding the pathways of flap removal. This is because the anticipated inhibition of FEN1 by RPA is expected to create the need for Dna2 and the two-nuclease pathway. Cleavage by FEN1 was measured in the presence of Pol δ-01, PCNA, RFC, and increasing amounts of RPA on the standard-44 substrate radiolabeled at the 5′-end of the downstream primer (Fig. 2). RPA did not noticeably affect strand displacement and cleavage of flaps made in the presence of Pol δ-wt, which were less than ∼20 nt (data not shown). Because of its ability to generate longer flaps than the Pol δ-wt (8, 9, 11), we anticipated that RPA inhibition of FEN1 would be more pronounced with Pol δ-01. Therefore, in this experiment, the exonuclease-deficient Pol δ-01 was used to examine RPA inhibition of FEN1 cleavage following displacement.
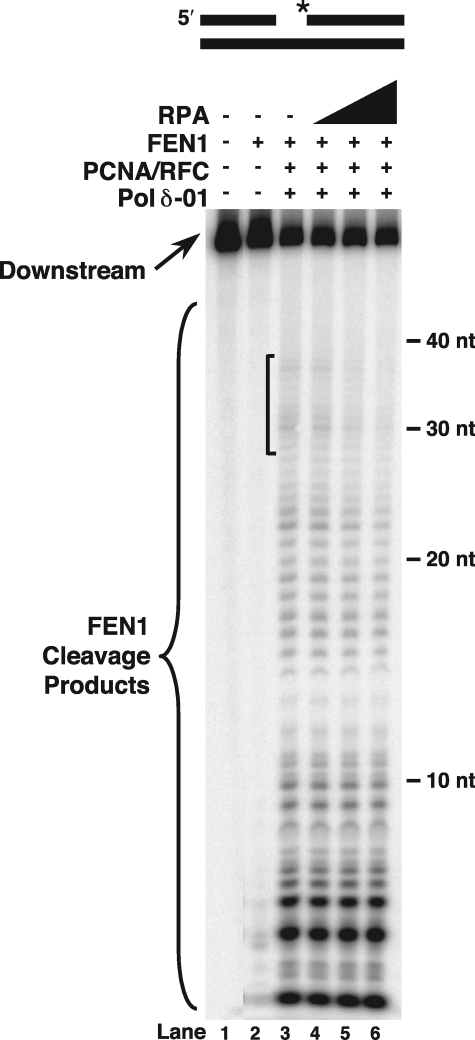
FIGURE 2.
FEN1 cleavage during flap displacement is moderately inhibited by RPA. Initial cleavage by FEN1 was assayed on the standard-44 substrate (U2:T4:D1) in the presence of PCNA/RFC, Pol δ-01, and increasing amounts of RPA (50, 100, or 200 fmol) as described under “Experimental Procedures” (lanes 4–6). The substrate is depicted above the figure. Asterisk indicates location of radiolabel.
RPA did not significantly alter the strand-displacement synthesis by Pol δ-01 (data not shown). In examining FEN1 cleavage products formed in the presence of Pol δ-01, some moderate inhibitory effects were discernible when increasing amounts of RPA were added to the reconstitution (Fig. 2, lanes 4–6 compared with lane 3). Percent cleavage inhibition was determined by comparing the amount of cleavage for reactions in the presence versus absence of RPA. The amount of cleavage is defined as the sum intensities of the cleavage product bands divided by the sum intensities of the downstream primer and cleavage product bands. To calculate percent inhibition, the value for cleavage amount in the presence of RPA was divided by the value for cleavage amount by FEN1 alone, subtracted from 1, and then multiplied by 100. Relative to the total amount of cleavage, all cleavage products greater than 28 nt long were diminished by only 36% with addition of the highest concentration of RPA (Fig. 2, lane 6). Likewise, all cleavage products of greater than 19 nt were decreased by only 38% (Fig. 2, lane 6). Cleavage products in the 28–37-nt size range were most sensitive with 48% inhibition by RPA (Fig. 2, lane 6 compared with lane 3, bracketed region). Similarly, products in the 19–28-nt size range were inhibited by 41% (Fig. 2, lane 6). This contrasts with products in the 10–19-nt size range, which were less susceptible to RPA inhibition, displaying only 17% reduction at the highest concentration of RPA (Fig. 2, lane 6). Greater inhibition of cleavage that gave products greater than 19 nt is consistent with RPA binding more stably on single-stranded DNA greater than 30 nt long (33). However, based on the decrease of starting substrate, comparing the overall amount of cleavage in the absence and presence of RPA revealed similar cleavage levels, indicating that RPA has only a moderate effect on overall rate of displacement and cleavage.
RPA Inhibits FEN1 Cleavage of Fixed Flap Substrates—Previous studies demonstrated that RPA binding, which blocks FEN1 tracking to its cleavage site, was effective at inhibiting FEN1 cleavage of long fixed flaps (13, 17). However, our results showed only moderate inhibition of FEN1 cleavage by RPA on the strand-displaced flaps. Consequently, we sought to verify earlier results, and confirm the proper functionality of our RPA, by examining the influence of RPA on FEN1 cleavage of fixed flaps (Fig. 3). Fixed flap cleavage experiments with RPA and FEN1 utilized a set of double-flap substrates that contained a complementary 1-nt 3′ tail overlapping with the 5′ flap, creating a double flap. It was previously demonstrated through studies in vitro that this type of double flap is the preferred substrate for FEN1 (31, 34, 35). During polymerase extension of the 3′-end of the upstream Okazaki fragment and displacement of the 5′-end of the downstream primer, reversible unannealing of the 3′-end and realignment of the 5′- and 3′-ends may occur, resulting in formation of a double flap (8, 12, 31). For this experiment, the substrates contained 5′ flaps 19, 28, and 37 nt in length, all of which are long enough to bind RPA (33) and are representative of the lengths of long flaps seen in strand-displacement reconstitutions (11). Each substrate, depicted above the figure, was radiolabeled at the 5′-end of the downstream primer to allow visualization of the flap cleavage product.
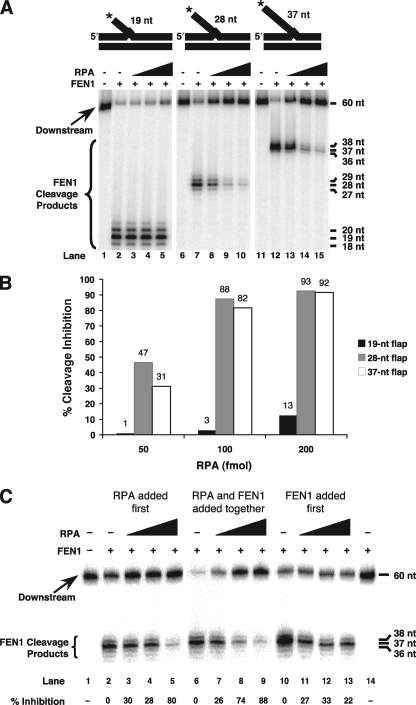
FIGURE 3.
RPA inhibits FEN1 cleavage of fixed flap substrates. A, cleavage by FEN1 was assayed in the presence of increasing amounts of RPA (50, 100, or 200 fmol) as described under “Experimental Procedures.” The 19-nt fixed double-flap (U1:T3:D1)(lanes 1–5), 28-nt fixed double-flap (U1:T2:D1)(lanes 6–10), and 37-nt fixed double-flap (U1:T1:D1)(lanes 11–15) are depicted above the figure. The asterisk indicates location of radiolabel. B, the amount of RPA versus the percent cleavage inhibition was graphed for the 19-nt fixed double-flap (black boxes), 28-nt fixed double-flap (gray boxes), and 37-nt fixed double-flap (white boxes) for the gel in A. Percentages are indicated above the bar for each substrate at each concentration of RPA. C, cleavage by FEN1 was assayed in the presence of increasing amounts of RPA (50, 100, or 200 fmol) using the 37-nt fixed double-flap, as in A. In lanes 2–5, RPA was added at the start of the reaction and FEN1 was added after 5 min. In lanes 6–9, both FEN1 and RPA were added at the start of the reaction, as in lanes 12–15 of A. In lanes 10–13, FEN1 was added at the start of the reaction in the absence of Mg+2, followed by addition of both RPA and Mg+2 after 5 min. Lane 14 contains FEN1 alone in the absence of Mg+2. The percent cleavage inhibition is shown under each lane.
There were three significant FEN1 cleavage products seen with each flap substrate, the major one representing cleavage at the base of the 5′ flap in the double-flap configuration (Fig. 3A). A minor product is one nucleotide longer and derives from cleavage at the base of the 5′ flap when the substrate has equilibrated to a nick-flap configuration. A faint product is 1 nucleotide shorter, possibly representing a transient flap configuration.
When increasing amounts of RPA were titrated into a reaction with FEN1, the amount of FEN1 cleavage decreased substantially with the longer flaps (Fig. 3A, lanes 7–10 and 12–15), but only marginal inhibition was observed with the 19-nt flap that does not readily bind RPA (Fig. 3A, lanes 2–5). To quantitate the level of this inhibition, we compared the cleavage for reactions in the presence of RPA to the cleavage by FEN1 alone, as described above. In the presence of the highest concentration of RPA, the amount of FEN1 cleavage was inhibited to the greatest extent on the long 28- and 37-nt flaps, with a maximum inhibition of 93 and 92%, respectively (Fig. 3B). However, the 19-nt flap was markedly less susceptible to the effects of RPA, with a maximum inhibition of only 13% (Fig. 3B). The extent of inhibition seen here is consistent with previous biochemical results (13, 17).
Evidently, FEN1 cleavage of the flaps created by the dynamic process of strand displacement is less sensitive than cleavage of fixed flaps to the inhibitory activity of RPA. To simulate a dynamic competition, FEN1 cleavage of the 37-nt flap substrate was assessed under conditions in which RPA was added before, with, or after the FEN1 (Fig. 3C). When FEN1 was added first, the reaction lacked magnesium, which was added later with the RPA (Fig. 3C, lanes 10–13). This allowed the FEN1 to bind the flap, but not cleave until the RPA was added, because FEN1 did not cleave in the absence of magnesium (Fig. 3C, lane 14). Inhibition by RPA was similar when it was added before or with FEN1 (Fig. 3C, lanes 2–5 and 6–9, respectively), but considerably less when FEN1 was added first (Fig. 3C, lanes 10–13). Such an outcome is consistent with the interpretation that a growing flap can acquire FEN1 before it is long enough for stable binding by RPA.
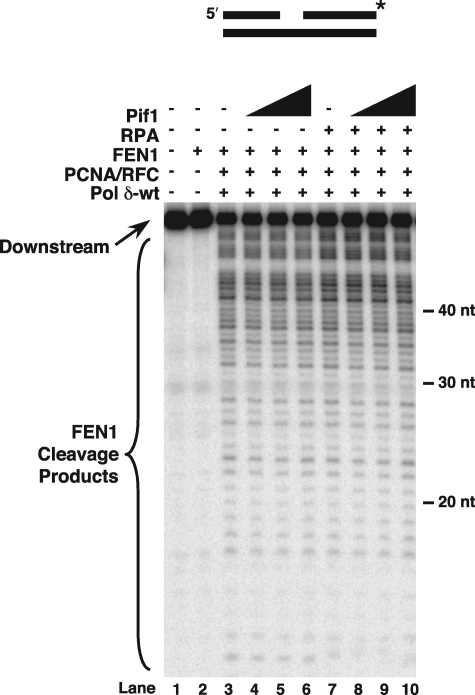
Pif1 Improves Flap Displacement and Promotes RPA Inhibition of FEN1 Cleavage—In S. cerevisiae, pif1Δ rescues the lethality of dna2Δ suggesting that Pif1 creates a need for Dna2, possibly by promoting the formation of long flaps (22). These genetic studies support a role for Pif1 in Okazaki fragment maturation, specifically in the two-nuclease primer removal pathway. To investigate this potential role, we asked whether Pif1 stimulates strand displacement and subsequent FEN1 cleavage of long flaps. Additionally, because RPA was inefficient at inhibiting FEN1 cleavage on long displaced flaps, we questioned whether the action of Pif1 would exacerbate RPA inhibition of FEN1 cleavage, augmenting the need for flap cleavage by Dna2. Again using the standard-44 substrate radiolabeled at the 5′-end of the downstream primer in reconstitution assays, the effects of Pif1 and RPA on initial FEN1 cleavages following displacement by Pol δ-wt were examined (Fig. 4). With Pol δ-wt, PCNA, and RFC, the maximum length of FEN1 cleavage products was ∼18 nt (Fig. 4, lane 3), consistent with previous results (11). When increasing amounts of Pif1 were added, the maximum cleavage product length increased to ∼33 nt (Fig. 4, lanes 4–6). Moreover, the density of longer products, ∼28–33 nt in length notably increased (Fig. 4, lanes 4–6 compared with lane 3, bracket region). These results show that addition of Pif1 improves the displacement and cleavage of longer flaps. The levels of Pol δ synthesis were mildly stimulated by the presence of Pif1 (data not shown), suggestive that the polymerase and helicase cooperate in strand displacement.
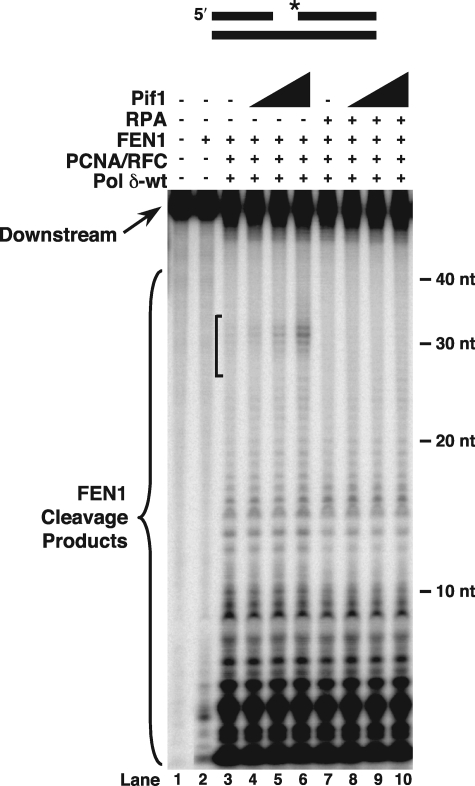
FIGURE 4.
Pif1 improves flap displacement and promotes RPA inhibition of FEN1 cleavage. Initial cleavage by FEN1 was assayed on the standard-44 substrate (U2:T4:D1) in the presence of PCNA/RFC, Pol δ-wt, and increasing amounts of Pif1 (25, 50, or 100 fmol) as described under “Experimental Procedures” (lanes 4–6). Cleavage was also assayed in the presence of RPA (100 fmol) and increasing amounts of Pif1 (25, 50, or 100 fmol) as described under “Experimental Procedures” (lanes 8–10). The substrate is depicted above the figure. Asterisk indicates location of radiolabel.
Strand lengths of 28–33 nt had previously been reported to allow stable RPA binding (33). Because of this, we wanted to know whether RPA would inhibit the cleavage of the longer flaps formed in the presence of Pif1. Synthesis by Pol δ-wt was not significantly altered by the addition of both RPA and Pif1 (data not shown). However, in examining the cleavage of displaced flaps in the presence of RPA and Pif1, there was a marked decrease in long cleavage products, particularly those in the 28–33-nt size range that had been augmented by addition of Pif1 alone (Fig. 4, lanes 4–6 compared with lanes 8–10, bracketed region). These results indicate that Pif1 promotes RPA inhibition of FEN1.
Evidence that Pif1-stimulated Long Flaps Are a Small Subset of Total Flaps—Because Pif1 stimulated the creation and cleavage of long flaps, we anticipated that Pif1 would also have increased the overall distance of displacement and cleavage into the downstream primer region. To test this, reconstitution assays were performed using the standard-44 substrate, radiolabeled at the 3′-end of the downstream primer to allow visualization of the products remaining after cleavage (Fig. 5). As strand-displacement proceeds, the 3′-labeled cleavage products are expected to become progressively shorter. Following synthesis by Pol δ-wt, PCNA, and RFC, there was a distribution of cleavage product sizes (Fig. 5, lane 3) up to 48 nt into the downstream primer, at which point the primers should spontaneously dissociate. This distribution was consistent with previous results (11). When increasing amounts of Pif1 were added, there was no significant change in the distribution of cleavage products (Fig. 5, lanes 4–6 compared with lane 3). Moreover, addition of RPA and Pif1 together slightly increased the amount of cleavage products greater than 20 nt while decreasing the smallest cleavage products ∼12–13 nt in length (Fig. 5, lanes 8–10 compared with lanes 4–6). This slight shift in distribution toward longer products indicates that RPA limits some cleavage by FEN1, as expected from our other results, and that this limitation is evident in the presence of Pif1. Overall, the effects of Pif1 on the cleavage product distribution were minor, consistent with the conclusion that long flaps promoted by Pif1 are a small subset of the total population of cleavage products.
FIGURE 5.
Pif1-stimulated products are a small subset of cleavage products. The extent of FEN1 cleavage into the downstream primer was assayed on the standard-44 substrate (U2:T4:D1) in the presence of PCNA/RFC, Pol δ-wt, and increasing amounts of Pif1 (25, 50, or 100 fmol) as described under “Experimental Procedures” (lanes 4–6). Cleavage was also assayed in the presence of RPA (100 fmol) and increasing amounts of Pif1 (25, 50, or 100 fmol) as described under “Experimental Procedures” (lanes 8–10). The substrate is depicted above the figure. Asterisk indicates location of radiolabel.
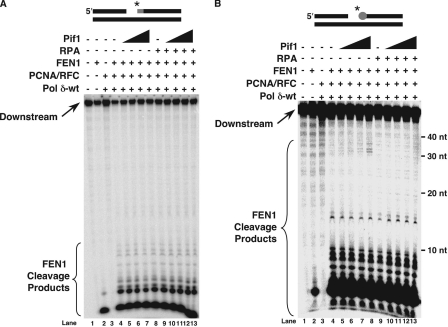
The Presence of RNA on the Downstream Primer Does Not Inhibit Lengthening of Flaps by Pif1—We considered the possibility that the initiator RNA of every Okazaki fragment would influence the action of Pif1. We investigated the effects of Pif1 on FEN1 cleavage of a strand-displaced flap when a single ribonucleotide was present at the 5′-end of the downstream Okazaki fragment. Although Pif1 does not bind RNA (32, 40), we expected no change in FEN1 cleavage when RNA initiated the downstream primer compared with when RNA was absent, as a single ribonucleotide is unlikely to significantly affect Pif1 binding to the emerging flap. We designed a substrate, the RNA substrate, which is identical in sequence to the standard-44 substrate, except it contains a ribonucleotide at the 5′-end of the downstream primer. We performed the same reactions with the RNA substrate as with the substrate composed only of DNA (Fig. 6A). Surprisingly, no long products were observed as increasing amounts of Pif1 were titrated into the reaction with Pol δ, PCNA, RFC, and FEN1 (Fig. 6A, lanes 5–8). Examination of the reaction in which only FEN1 was present, representing synthesis-independent FEN1 cleavage (Fig. 6A, lane 2), revealed that the background level of FEN1 cleavage of the ribonucleotide was very high. This is consistent with previous findings from our laboratory demonstrating efficient FEN1 cleavage of 5′-terminal ribonucleotides (41). FEN1 cleavage occurred in the absence and presence of an upstream primer, and also when the upstream primer formed either a nick or a gap with the downstream primer (41). Therefore, we hypothesize that in the current experiment, FEN1 was very efficient at cleaving off the ribonucleotide before Pol δ could begin displacing a flap. Because the downstream primer was radiolabeled at the 5′-end, the majority of FEN1 cleavage removing the ribonucleotide resulted in loss of the radiolabel. Therefore, additional cleavage of Pol δ-displaced flaps was not observed. The radiolabel would be preserved in any flaps that escape FEN1 cleavage. However, in this system, the combination of initial low abundance and loss of most radiolabel reduced long cleavage products to below the level of detection.
FIGURE 6.
RNA does not inhibit lengthening of flaps by Pif1 during synthesis of Okazaki fragments. A, initial cleavage by FEN1 was assayed on the RNA substrate (U2:T4:D2) in the presence of PCNA/RFC, Pol δ-wt, and increasing amounts of Pif1 (25, 50, 100, or 200 fmol) as described under “Experimental Procedures” (lanes 5–8). Cleavage was also assayed in the presence of RPA (100 fmol) and increasing amounts of Pif1 (25, 50, 100, or 200 fmol) as described under “Experimental Procedures” (lanes 10–13). The substrate is depicted above the figure. The gray segment represents the ribonucleotide. The asterisk indicates location of radiolabel. B, the same reactions as in A except the substrate used was the methylated substrate (U2:T4:D3). The substrate is depicted above the figure. The gray circle represents the 2′-O-methylated ribonucleotide. The asterisk indicates the location of radiolabel.
Although they may be rare, we wanted to examine the fate of any RNA-initiated flaps that might escape FEN1 cleavage of the RNA. To address the problem of cleavage of the radiolabeled RNA, we utilized a substrate with a terminal 2′-O-methylated ribonucleotide, a modification that inhibits RNA cleavage by ribonucleases. We hypothesized that the modification would also inhibit the ability of FEN1 to efficiently cleave off the single ribonucleotide prior to flap production. Inhibition of FEN1 cleavage preserves the radiolabeled RNA, allowing us to observe the effects on flap production and cleavage by FEN1 in the presence of Pif1 and RPA following synthesis. Our substrate for this experiment was identical to the RNA substrate except that the single ribonucleotide on the downstream primer was 2′-O-methylated. We performed the same experiment as with the RNA substrate, titrating in increasing amounts of Pif1 to a reaction with Pol δ, PCNA, RFC, and FEN1 first without RPA and then in the presence of RPA (Fig. 6B). As expected, the amount of synthesis-independent FEN1 cleavage was reduced with the methylated RNA substrate (Fig. 6B, lane 2). In the absence of RPA, a population of long flap cleavage products appeared as the amount of Pif1 increased (Fig. 6B, lanes 5–8). When RPA was present, these products disappeared (Fig. 6B, lanes 10–13). We conclude that the ribonucleotide does not affect the ability of Pif1 to lengthen a flap created by Pol δ, thus resulting in formation of long flaps cleavable by FEN1. When RPA is present, it binds these long flaps and inhibits FEN1 cleavage.
DISCUSSION
We developed a system to reconstitute the reactions of eukaryotic Okazaki fragment processing in vitro. We have used it to evaluate two proposed processing pathways of flap removal, one involving FEN1 as the only nuclease (7–9), and the other involving the ordered action of Dna2 and FEN1 (13, 15). The two nuclease pathways would be required if flaps become long enough to bind RPA. The presence of RPA is expected to inhibit FEN1, necessitating initial cleavage by Dna2.
Previous biochemical studies indicated that primarily short flaps 1–2 nt in length are displaced by Pol δ during RNA primer removal and cut by FEN1, and the iterative process of displacement and cutting is likely to facilitate primer removal on the majority of Okazaki fragments (7–9, 11). Application of our reconstituted system in subsequent work (11), as well as current observations (Fig. 1), revealed two populations of FEN1 cleavage products following displacement by Pol δ. One is a sharp peak of products displaced and cleaved by FEN1 while short, and the other is a broad, flat distribution of products that reeled out and became long flaps, 10–36 nt in length, before cleavage by FEN1. We hypothesized that in vivo, these long displaced flaps would be bound by RPA, effectively preventing FEN1 cleavage. Once bound by RPA, the long flaps would require Dna2 for cleavage before FEN1 could cleave, generating a nick that can be ligated.
In the reconstitution assays, we predicted that inclusion of RPA with Pol δ and PCNA/RFC would prevent FEN1 cleavage of the observed long flaps. Following displacement by Pol δ-wt, there was no apparent effect of RPA on FEN1 cleavage products (data not shown). This absence of inhibition could have been anticipated because observed long flaps cleaved in the presence of Pol δ-wt were 10–20 nt, too small for stable RPA binding that occurs at a length of ∼30 nt (33). This result is also consistent with the small levels of RPA inhibition of FEN1 cleavage on a 19-nt fixed double-flap (Fig. 3).
The exonuclease-deficient Pol δ-01 generated longer flap cleavage products (8, 9, 11), up to ∼36 nt (Fig. 1). Flaps of this length, which are readily observable with Pol δ-01, should stably bind RPA, thus inhibiting FEN1 cleavage. Surprisingly, however, in the presence of Pol δ-01, PCNA/RFC, and FEN1, RPA only moderately inhibited the formation of the long cleavage products (Fig. 2). The longest cleavage products in the 28–37-nt size range were most sensitive to RPA inhibition (Fig. 2), which is consistent with the fixed flap results showing greater levels of inhibition with the 28- and 37-nt double-flaps compared with the 19-nt flap (Fig. 3).
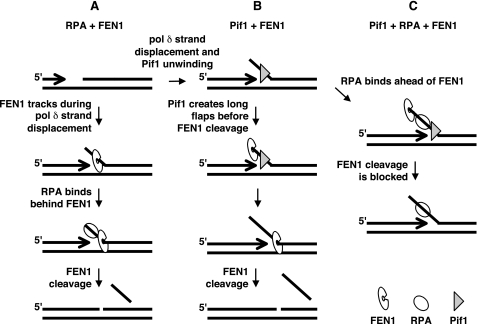
Because RPA was a highly effective inhibitor of FEN1 cleavage on long fixed flaps, we expected inhibition to be similar on long strand-displaced flaps. An advantage of the system that we have employed is that it allowed us to make this important comparison. In actuality, moderate inhibition of FEN1 cleavage by RPA in the context of dynamically lengthening flaps is in obvious contrast with RPA inhibition on fixed flaps. FEN1 cleavage assays on fixed double-flaps revealed more than 90% inhibition on 28- and 37-nt flaps (Fig. 3A and B) compared with only 36% inhibition on cleavage products greater than 28 nt when cleavage occurs during displacement (Fig. 2). These results indicate an inherent difference in the ability of RPA to inhibit FEN1 cleavage when the flap is preformed and fixed versus when flaps are lengthening during strand-displacement. In the displacement assays, the moderate RPA inhibition of long flap cleavage suggests that when flaps are created dynamically, there is a competition between RPA and FEN1 for binding to the flap favoring FEN1. Moreover, assessment of competition on a fixed flap when the RPA was added before, with, or after the FEN1, indicates that initial binding of FEN1 thwarts the inhibitory ability of RPA (Fig. 3C). We propose that by the time most flaps become long enough to bind RPA, FEN1 has already recognized their 5′-ends and begun tracking (Fig. 7A). We envision FEN1 molecules tracking near the base of the elongating flap, racing the displacement process to eventually reach the cleavage site. FEN1 may achieve this binding advantage because it is already a stable part of the lagging strand maturation complex. This would position it on the flap at the earliest time of strand displacement. Alternatively, FEN1 may simply have the ability to bind and track on nearly every elongating flap before it becomes long enough to form a stable complex with the RPA. In either case, RPA molecules would then bind behind the FEN1, too late to inhibit cleavage.
FIGURE 7.
Pif1 directs long flaps toward the two-nuclease pathway for primer removal. A, in most cases FEN1 is tracking near the base of the elongating flap, racing the displacement process to eventually reach the site of cleavage. Most RPA molecules would then bind behind the FEN1, too late to prevent cleavage (refer to Fig. 2, lanes 3–6). B, Pif1 promotes flap displacement, allowing some flaps to become long before FEN1 cleavage occurs (refer to Fig. 4, lanes 3–6). C, Pif1 increases the rate of flap formation such that FEN1 molecules take longer to track to the flap base for cleavage. In the presence of RPA, the flap can bind RPA ahead of the tracking FEN1. RPA bound in this position can prevent FEN1 cleavage (refer to Fig. 4, lanes 7–10).
This current biochemical data prompted a re-evaluation of the pathways of primer removal during Okazaki fragment maturation, specifically of the role of RPA in directing flaps into the two-nuclease pathway. Inhibition of FEN1 cleavage by RPA binding of long flaps was proposed to be the key reason for use of the two-nuclease pathway (13, 15). Yet, the results presented here demonstrate that even in the presence of RPA, FEN1 cleavage of displaced flaps is only moderately inhibited by RPA (Fig. 2). Is it possible that the two-nuclease pathway is either completely redundant or virtually never used?
Recent genetic results in S. cerevisiae suggest the involvement of another factor, the Pif1 helicase, in directing flaps to the two-nuclease pathway. The lethality of dna2Δ mutation is suppressed by pif1Δ (22). Studies in S. pombe indicate a similar relationship between the Pif1 ortholog Pfh1 and Dna2, because mutations in pfh1 suppress the temperature-sensitive phenotype of dna2 mutations (27). These genetic results imply that the Pif1 helicase creates a need for Dna2, possibly by promoting long flap formation (22, 27), and indicate a role for Pif1 in Okazaki fragment processing. Results presented here support this hypothesis. The addition of Pif1 to the Okazaki fragment processing system promoted the appearance of long FEN1 cleavage products (Fig. 4). Pif1 notably increased products in the 28–33-nt size range. It appears that Pif1 accelerated flap displacement, allowing flaps to become longer before FEN1 cleavage could occur (Fig. 7B). Yet, the Pif1-stimulated products were a small subset of FEN1 products, because addition of Pif1 only mildly stimulated the amount of synthesis (data not shown) and did not notably affect the extent of cleavage into the downstream primer (Fig. 5). Although these long Pif1-stimulated products were not abundant in our system, the actual numbers in vivo would be significant because millions of Okazaki fragments are processed during synthesis in a replication cycle.
Our observations that Pif1 stimulates long flap cleavage by FEN1 (Fig. 4) and that RPA only moderately inhibits FEN1 cleavage of long flaps (Fig. 2) prompted the question of whether Pif1 is the critical additional component that necessitates the Dna2-requiring pathway. Pif1-stimulated flaps in the 28–33-nt size range are long enough to bind RPA (33). Correspondingly, RPA did effectively inhibit FEN1 cleavage of this size range of flaps created in the presence of Pif1 (Fig. 4). This result suggests that Pif1 increases the rate of flap displacement, such that FEN1 takes longer to track to the base of the flap for cleavage. Significantly, Pif1 appears to promote such rapid flap formation that RPA is able to bind to the flap ahead of the tracking FEN1 (Fig. 7C). With Pif1, RPA binding prevents cleavage of a significant number of flaps by FEN1 alone and necessitates cleavage via the two-nuclease pathway (13).
Okazaki fragments in vivo are initiated by RNA, made by Pol α, which must be removed. Removal of only the ribonucleotides is not sufficient to maintain high replication fidelity because Pol α lacks a 3′-5′ proofreading activity, and some of the DNA nucleotides incorporated by Pol α just beyond the RNA may be erroneous (42). As a result, additional flap displacement and cleavage of the downstream Okazaki fragment is necessary to remove the DNA synthesized by Pol α (29, 43). Consequently, we were interested in the effect of Pif1 on flap formation with substrates that more closely resemble the natural RNA-primed Okazaki fragment (2, 29). Therefore, we felt it was reasonable to test a model fragment with a single initiator ribonucleotide.
In the first experiments, FEN1 was so efficient at removing the labeled ribonucleotide of the downstream primer that long FEN1 cleavage products were virtually undetectable (Fig. 6A). This result supports the validity of our results with full DNA fragments, because the RNA of nearly all fragments is likely to have been removed in vivo before the flaps could achieve significant length. To determine the effects of any fragments that escaped complete RNA removal by FEN1, we designed and used a downstream primer with a 2′ O-methylated 5′-ribonucleotide (Fig. 6B). As with the substrates containing only DNA, we found that Pif1 promoted RPA inhibition of FEN1 cleavage. This result further verifies that the DNA-initiated substrates served as a sufficient representative substrate for the RNA-initiated Okazaki fragments, consistent with previous work (11). Moreover, in helicase assays, Pfh1 from S. pombe was equally active on RNA- and DNA-initiated flaps, suggesting that Pif1 helicases are active in vitro on substrates that are relevant in vivo (27).
Supporting previous genetic data (22), our biochemical results reveal a role for Pif1 in eukaryotic cellular Okazaki fragment maturation to remove the initiator RNA primer, specifically in the processing of long flaps. Results with our model reconstitution system show that once the Pol δ strand displaces the downstream primer, FEN1 can track onto the flap and cleave at its base (Fig. 7A). The dynamics of flap formation do not allow RPA to bind quickly enough to inhibit FEN1. Pif1 accelerates flap formation allowing the flaps to achieve greater lengths before FEN1 cleavage (Fig. 7B). However, when RPA is present with Pif1 and FEN1, flap growth is sufficiently rapid that RPA can bind and block FEN1 cleavage (Fig. 7C). Because Pif1 is unable to bind RNA (32, 40), Pif1 involvement in unwinding the downstream Okazaki fragment necessitates that Pol δ displaces whatever remains of the RNA primer (2) before Pif1 can bind to the flap and aid displacement. However, our results indicate that the presence of the initiator RNA does not alter the fundamental role of Pif1. The biological implication is that by promoting RPA inhibition of FEN1 cleavage, Pif1 is directing flaps toward the two-nuclease cleavage pathway, requiring Dna2 cleavage for primer removal. Examining the influence of Pif1 on flap cleavage by Dna2 in the context of strand-displacement will be an important next step to further elucidate the role of Pif1 in replication. Additional experiments will be necessary to expand our understanding of the conditions that determine which of the cleavage pathways is followed for primer removal during Okazaki fragment maturation.
Acknowledgments
We thank the members of the Bambara laboratory for helpful discussions and suggestions.
This work was supported by National Institutes of Health (NIH) Grant GM024441 (to R. A. B.) with additional support from NIH Grants GM087666 (to J. L. C.) and GM32431 (to P. M. J. B.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: Pol α, DNA polymerase α/primase; Pol δ, DNA polymerase δ; nt, nucleotide(s); PCNA, proliferating cell nuclear antigen; RFC, replication factor C; FEN1, flap endonuclease 1; RPA, replication protein A; wt, wild type; Pol δ-01, 3′-5′ exonuclease-deficient polymerase δ.
References
- 1.Kornberg, A., and Baker, T. A. (1992) in DNA Replication, 2nd Ed., pp. 113–195, W. H. Freeman, New York
- 2.Bambara, R. A., Murante, R. S., and Henricksen, L. A. (1997) J. Biol. Chem. 272 4647–4650 [DOI] [PubMed] [Google Scholar]
- 3.Liu, Y., Kao, H. I., and Bambara, R. A. (2004) Annu. Rev. Biochem. 73 589–615 [DOI] [PubMed] [Google Scholar]
- 4.Rossi, M. L., Purohit, V., Brandt, P. D., and Bambara, R. A. (2006) Chem. Rev. 106 453–473 [DOI] [PubMed] [Google Scholar]
- 5.Harrington, J. J., and Lieber, M. R. (1994) EMBO J. 13 1235–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murante, R. S., Rust, L., and Bambara, R. A. (1995) J. Biol. Chem. 270 30377–30383 [DOI] [PubMed] [Google Scholar]
- 7.Ayyagari, R., Gomes, X. V., Gordenin, D. A., and Burgers, P. M. (2003) J. Biol. Chem. 278 1618–1625 [DOI] [PubMed] [Google Scholar]
- 8.Garg, P., Stith, C. M., Sabouri, N., Johansson, E., and Burgers, P. M. (2004) Genes Dev. 18 2764–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin, Y. H., Ayyagari, R., Resnick, M. A., Gordenin, D. A., and Burgers, P. M. (2003) J. Biol. Chem. 278 1626–1633 [DOI] [PubMed] [Google Scholar]
- 10.Murante, R. S., Rumbaugh, J. A., Barnes, C. J., Norton, J. R., and Bambara, R. A. (1996) J. Biol. Chem. 271 25888–25897 [DOI] [PubMed] [Google Scholar]
- 11.Rossi, M. L., and Bambara, R. A. (2006) J. Biol. Chem. 281 26051–26061 [DOI] [PubMed] [Google Scholar]
- 12.Jin, Y. H., Obert, R., Burgers, P. M., Kunkel, T. A., Resnick, M. A., and Gordenin, D. A. (2001) Proc. Natl. Acad. Sci. U. S. A 98 5122–5127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bae, S. H., Bae, K. H., Kim, J. A., and Seo, Y. S. (2001) Nature 412 456–461 [DOI] [PubMed] [Google Scholar]
- 14.Bae, S. H., Choi, E., Lee, K. H., Park, J. S., Lee, S. H., and Seo, Y. S. (1998) J. Biol. Chem. 273 26880–26890 [DOI] [PubMed] [Google Scholar]
- 15.Bae, S. H., and Seo, Y. S. (2000) J. Biol. Chem. 275 38022–38031 [DOI] [PubMed] [Google Scholar]
- 16.Kao, H. I., Campbell, J. L., and Bambara, R. A. (2004) J. Biol. Chem. 279 50840–50849 [DOI] [PubMed] [Google Scholar]
- 17.Kao, H. I., Veeraraghavan, J., Polaczek, P., Campbell, J. L., and Bambara, R. A. (2004) J. Biol. Chem. 279 15014–15024 [DOI] [PubMed] [Google Scholar]
- 18.Budd, M. E., and Campbell, J. L. (1997) Mol. Cell. Biol. 17 2136–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Budd, M. E., and Campbell, J. L. (2000) Mutat. Res. 459 173–186 [DOI] [PubMed] [Google Scholar]
- 20.Budd, M. E., Choe, W., and Campbell, J. L. (2000) J. Biol. Chem. 275 16518–16529 [DOI] [PubMed] [Google Scholar]
- 21.Lee, K. H., Kim, D. W., Bae, S. H., Kim, J. A., Ryu, G. H., Kwon, Y. N., Kim, K. A., Koo, H. S., and Seo, Y. S. (2000) Nucleic Acids Res. 28 2873–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Budd, M. E., Reis, C. C., Smith, S., Myung, K., and Campbell, J. L. (2006) Mol. Cell. Biol. 26 2490–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bessler, J. B., Torredagger, J. Z., and Zakian, V. A. (2001) Trends Cell Biol. 11 60–65 [DOI] [PubMed] [Google Scholar]
- 24.Boule, J. B., and Zakian, V. A. (2006) Nucleic Acids Res. 34 4147–4153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burgers, P. M., and Gerik, K. J. (1998) J. Biol. Chem. 273 19756–19762 [DOI] [PubMed] [Google Scholar]
- 26.Johansson, E., Garg, P., and Burgers, P. M. (2004) J. Biol. Chem. 279 1907–1915 [DOI] [PubMed] [Google Scholar]
- 27.Ryu, G. H., Tanaka, H., Kim, D. H., Kim, J. H., Bae, S. H., Kwon, Y. N., Rhee, J. S., MacNeill, S. A., and Seo, Y. S. (2004) Nucleic Acids Res. 32 4205–4216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou, J. Q., Qi, H., Schulz, V. P., Mateyak, M. K., Monson, E. K., and Zakian, V. A. (2002) Mol. Biol. Cell 13 2180–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kao, H. I., and Bambara, R. A. (2003) Crit. Rev. Biochem. Mol. Biol. 38 433–452 [DOI] [PubMed] [Google Scholar]
- 30.Gerik, K. J., Gary, S. L., and Burgers, P. M. (1997) J. Biol. Chem. 272 1256–1262 [DOI] [PubMed] [Google Scholar]
- 31.Kao, H. I., Henricksen, L. A., Liu, Y., and Bambara, R. A. (2002) J. Biol. Chem. 277 14379–14389 [DOI] [PubMed] [Google Scholar]
- 32.Boule, J. B., Vega, L. R., and Zakian, V. A. (2005) Nature 438 57–61 [DOI] [PubMed] [Google Scholar]
- 33.Wold, M. S. (1997) Annu. Rev. Biochem. 66 61–92 [DOI] [PubMed] [Google Scholar]
- 34.Friedrich-Heineken, E., Henneke, G., Ferrari, E., and Hubscher, U. (2003) J. Mol. Biol. 328 73–84 [DOI] [PubMed] [Google Scholar]
- 35.Storici, F., Henneke, G., Ferrari, E., Gordenin, D. A., Hubscher, U., and Resnick, M. A. (2002) EMBO J. 21 5930–5942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deleted in proof
- 37.Deleted in proof
- 38.Deleted in proof
- 39.Deleted in proof
- 40.Zhang, D. H., Zhou, B., Huang, Y., Xu, L. X., and Zhou, J. Q. (2006) Nucleic Acids Res. 34 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang, L., Rumbaugh, J. A., Murante, R. S., Lin, R. J., Rust, L., and Bambara, R. A. (1996) Biochemistry 35 9266–9277 [DOI] [PubMed] [Google Scholar]
- 42.Hubscher, U., Maga, G., and Spadari, S. (2002) Annu. Rev. Biochem. 71 133–163 [DOI] [PubMed] [Google Scholar]
- 43.Waga, S., and Stillman, B. (1998) Annu. Rev. Biochem. 67 721–751 [DOI] [PubMed] [Google Scholar]