Abstract
SIRT1, a histone/protein deacetylase, and AMP-activated protein kinase (AMPK) are key enzymes responsible for longevity and energy homeostasis. We examined whether a mechanistic connection exists between these molecules that involves the major AMPK kinase LKB1. Initial studies demonstrated that LKB1 is acetylated in cultured (HEK293T) cells, mouse white adipose tissue, and rat liver. In the 293T cells, SIRT1 overexpression diminished lysine acetylation of LKB1 and concurrently increased its activity, cytoplasmic/nuclear ratio, and association with the LKB1 activator STRAD. In contrast, short hairpin RNA for SIRT1, where studied, had opposite effects on these parameters. Mass spectrometric analysis established that acetylation of LKB1 occurs on multiple, but specific, lysine residues; however, only mutation of lysine 48 to arginine, which mimics deacetylation, reproduced all of the effects of activated SIRT1. SIRT1 also affected downstream targets of LKB1. Thus its overexpression increased AMPK and acetyl-CoA carboxylase phosphorylation, and conversely, RNA interference-mediated SIRT1 knockdown reduced AMPK phosphorylation and that of another LKB1 target MARK1. Consistent with the results in cultured cells, total LKB1 lysine acetylation was decreased by 60% in the liver of 48-h starved rats compared with starved-refed rats, and this was associated with modest but significant increases in both LKB1 and AMPK activities. These results suggest that LKB1 deacetylation is regulated by SIRT1 and that this in turn influences its intracellular localization, association with STRAD, kinase activity, and ability to activate AMPK.
LKB1 is a serine-threonine protein kinase that phosphorylates and activates 13 downstream kinases (1), one of which is AMP-activated protein kinase (AMPK),2 a key enzyme that regulates cellular energy state, growth, inflammation, and mitochondrial function (2). LKB1, when not associated with other proteins, is located predominantly in the nucleus because of its N-terminal nuclear localization signal. However, LKB1 activation takes place predominantly in the cytoplasm, after it complexes with STRAD (STE-related adapter) and MO25 (mouse protein 25) (1, 3). Once activated, LKB1 has been demonstrated to phosphorylate AMPK on Thr-172, an event required for its activation (4). On the other hand, no specific mechanism for regulating the activation and inactivation of the kinase activity of LKB1 has been described. Indeed, it has been suggested that LKB1 may be constitutively active and that its effects on AMPK phosphorylation (e.g. in contracting muscle) may be governed by the action of phosphatases (1, 20).
SIRT1, a class III NAD+-dependent histone/protein deacetylase, has been implicated in the longevity induced by caloric restriction in species ranging from Caenorhabditis elegans to rodents (5). It has been suggested that it may work in part by activating AMPK (5). The expression and deacetylation activities of SIRT1 are enhanced by increases in NAD+ levels or the NAD+/NADH ratio, such as occur during caloric restriction (5, 6). In the investigations described here, we present evidence that SIRT1 deacetylates LKB1 and that this is associated with its movement to the cytoplasm where it is bound to and activated by STRAD. The data also suggest that SIRT1 activates AMPK by this mechanism both in cultured HEK293T cells and in rat liver in vivo.
EXPERIMENTAL PROCEDURES
Cell Cultures—Human embryonic kidney 293T cells (HEK293T) and HepG2 cells were purchased from the ATCC (Manassas, VA). The cells were maintained in either Opti-MEM I reduced serum medium (Invitrogen) for 293T or Dulbecco's modified Eagle's medium for HepG2 cells supplemented with 5–10% fetal bovine serum and antibiotics.
Antibodies—The following primary antibodies were used: total AMPK-α and phospho-Thr-172-AMPK (Cell Signaling Technology, Danvers, MA); glutathione S-transferase (GST), LKB1 (N-19, H-75, M-18, and Ley37D/G6), SIRT1, STRAD, and phospho-Ser-428 LKB1 (Santa Cruz Biotechnologies; Santa Cruz, CA); acetyl-CoA carboxylase (ACC), phospho-Ser-79-ACC, acetyl-lysine (Millipore/Upstate, Charlottesville, VA); β-actin, FLAG tag (Sigma); and phospho-Thr-336-LKB1 (ImmuQuest; Ingleby Barwick, Cleveland, UK). The following secondary antibodies were used: HRP-conjugated donkey anti-rabbit antibody, HRP-conjugated sheep anti-mouse antibody (Amersham Biosciences); HRP-conjugated rabbit anti-goat antibody, and HRP conjugated rabbit anti-sheep antibody (Chemicon, Temecula, CA).
cDNAs and Plasmids—The following is a list of cDNA accession numbers used in this study. Unless stated otherwise, the plasmids were purchased from ATCC. They include LKB1 (STK11), BC007981 (HIP human kinase collection, DF/HCC DNA Resource Core; Boston, MA); STRAD (LYK5 isoform 4), BC081911; MO25 (CAB39, calcium-binding protein 39), BC020570; SirT1, NM_019812 (Upstate). These cDNAs were subcloned by PCR to produce in-frame fusion expression plasmids. The forward cloning primers start at the ATG site, and the reverse cloning primers end at the stop codon. Each primer has a length of ∼20–25 bp and has a calculated Tm of 60 °C. The 5′-end of the forward and reverse primers also contained additional sequences (forward, GGCTTTAAAGGAACC, and reverse, AAGCTGGGTCTAGAT) so that the cloned cDNAs could undergo in vitro homologous ligation with In-Fusion system plasmids (Clontech). After 18 cycles of PCR with proofreading KOD hot start polymerase (Novagen; San Diego), the PCR product was gel-purified (Qiagen; Valencia, CA), cut with XmnI and EcoRV (Invitrogen), and ligated with the pENTR1A vector that had been cut with the same restriction endonucleases. Subsequently, these entry vectors (in which the target gene is flanked by L1 and L2 Gateway sequences) were incubated with the LR enzyme (Invitrogen) and with pDEST27 (N-terminal GST), pDEST26 (N-terminal His), or pDEST53 (N-terminal GFP) to generate fusion proteins tagged with GST, His, or GFP, respectively.
Site-directed Mutagenesis—Site-directed LKB1 mutagenesis was performed by PCR. Essentially, the PCR mixture containing MgCl2, dNTP, and 10× PCR buffer was given 125 ng each of the respective forward and reverse mutation generating primers, 10 ng of template wtLKB1 pENTR vector, and 2 units of KOD proofreading DNA polymerase. The PCR was performed with the following parameters: 95 °C for 30 s, 60 °C for1 min, and 72 °C for 2 min for 18 cycles. After PCR, 20 units of DpnI (New England Biolabs, Ipswich, MA) were added to the mix and incubated at 37 °C for 1 h. The DpnI treatment digests only the original methylated template DNA leaving the newly synthesized unmethylated mutated DNA. The digested PCR mix was transformed into Top10 Escherichia coli. The plasmids were purified, and the mutations were confirmed by sequencing. The following is a list of primers used to make LKB1 mutations: K44R forward, 5′-CGC CGC AAG CGG GCC AGG CTC ATC GGC AAG TAC-3′, and reverse, 5′-GTA CTT GCC GAT GAG CCT GGC CCG CTT GCG GCG-3′; K48R forward, 5′-GCC AAG CTC ATC GGC AGG TAC CTG ATG GGG GAC-3′, and reverse, 5′-GTC CCC CAT CAG GTA CCT GCC GAT GAG CTT GGC-3′; K96R forward, 5′-AAC GGG GAG GCC AAC GTG AGG AAG GAA ATT CAA CTA CTG-3′, and reverse, 5′-CAG TAG TTG AAT TTC CTT CCT CAC GTT GGC CTC CCC GTT-3′; K97R forward, 5′-GGG GAG GCC AAC GTG AAG AGG GAA ATT CAA CTA CTG AGG-3′, and reverse, 5′-CCT CAG TAG TTG AAT TTC CCT CTT CAC GTT GGC CTC CCC-3′.
The SirT1 H335Y mutation was made in a similar way by using following primers: forward, 5′-C CAA AGG ATC CTT CAG TGT TAT GGT TCC TTT GCA ACA GCA TC-3′, and reverse, 5′-GA TGC TGT TGC AAA GGA ACC ATA ACA CTG AAG GAT CCT TTG G-3′.
Creation of Plasmids Expressing GST Fusion LKB1 Fragment Proteins—Indicated fragments of LKB1 were made by PCR with LKB1 (BC007981) as the template and with the following primers: LKB1-(1–44) forward, 5′-CACC ATG GAG GTG GTG GAC CCG-3′, and reverse, 5′-T TCG ACC CAG ATC TA CTA CT TGG CCC GCTT GCG-3′; LKB1-(45–90) forward, 5′-CACC CTC ATC GGC AAG TAC CTG-3′, and reverse 5′-CTA GTT GGG GAT CCT TCG-3′; LKB1-(189–318) forward, 5′-CACC CTC AAA ATC TCC GAC CTG-3′, and reverse, 5′-T TCG ACC CAG ATC TA CTA TGC TTC AGC CGG AGG-3′; and LKB1-(189–433) forward, 5′-CACC CTC AAA ATC TCC GAC CTG-3′, and reverse, 5′-T TCG ACC CAG ATC TA CTA CAA CTG CTG CTT GCA GGC-3′. PCR products were gel-purified and ligated directly into pENTR/D-TOPO vector (Invitrogen). After confirming sequences, GST fusion protein expression vectors were created by LR reaction with pDEST27 vector.
Short Hairpin RNA Expressing Lentivirus—The sequence GTATTGCTGAACAGATGGAA was chosen as a potential target sequence for shRNA-mediated RNAi of human sirt1. The pSilencer 2.0 vector (Ambion; Austin, TX) was used as the template for the human U6 promoter that was cloned using the following PCR primers: forward primer (5′-GAATTCCCCAGTGGAAAGACGC-3′) and reverse primer (5′-GGTGTTTCGTCCTTTCCACAAGATATATAAAGGG-3′). An shRNA expression cassette was created by tandem polymerase reaction of the U6 promoter template with one forward and two reverse primers as follows: forward, 5′-CACCGCGCGCCAAGGTCGGGCA-3′, and reverse 1, 5′-CTACACAAACTCCACCTGTTCAGCAATACGGTGTTTCGTCC-3′ and reverse 2 5′-CCAAAAAAGTATTGCTGAACAGATGGAACTACACAAACTC-3′, which contains two GU pairing mutations in the sense strand. The resulting PCR product was inserted into pCR8-GW-TOPO (Invitrogen) and then transferred by LR reaction to pDSL_hpUGIP shRNA lentivirus expression plasmid (ATCC). Six μg of the lentivirus plasmid was calcium phosphate-transfected along with 10 μg of pLp1(gag-pol), 3 μg of pLp2 (reverse), and 2 μg of pLP/VSVG (VSVG) helper vectors into 293T cells grown in a 100-mm dish. 24 h post-transfection the media were changed; 48 h post-transfection 10 ml of supernatant containing lentivirus was collected. The lentivirus was then purified and concentrated by centrifuging at 10,000 × g for 90 min after adding 5 μl of 100 mg/ml poly-l-lysine solution. It was reconstituted with 500 μl of PBS, and 100-μl aliquots were frozen at -80 °C until use. The 293T cells or HepG2 cells grown in 6-well plates were infected by incubating each well with 100 μl of lentivirus vector and 8 μg/ml Polybrene for 8 h. And the cells were harvested 72 h later.
RNAi Experiments—RNAi-mediated down-regulation of SIRT1 was performed by either siRNA (SIRT1 Smartpool siRNA and control siRNA purchased from Dharmacon (Lafayette, CO)) or lentiviral infection of house-made shRNA targeting human sirt1. The effects were assessed 72 h after induction of RNAi.
Expression of Tagged Fusion Proteins in HEK293 Cells and Purification of GST-tagged Proteins—Transient transfection of plasmids containing tagged genes was performed with PerFectin transfection reagent (Genlantis, San Diego, CA) or the calcium phosphate method in 293T cells. The indicated quantity of plasmid DNA was transfected per well of a 6-well plate. Post-transfection (48–72 h), the cells were lysed with lysis buffer containing 20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EGTA, 1 mm EDTA, 1% Triton X-100, 2.5 mm sodium pyrophosphate, 1 mm β-glycerophosphate, 1 mm Na3VO4, 1 μg/ml leupeptin, 10 μg/ml PMSF, 1 mm dithiothreitol, and 10 mm sodium butyrate. To purify GST-tagged protein, the lysates were clarified by centrifugation at 14,000 × g for 15 min and then incubated overnight at 4 °C with 50% glutathione-Sepharose 4 B beads (Amersham Biosciences). The beads were washed four times with NETN buffer containing 300 mm NaCl, 0.1 mm EDTA, 20 mm Tris, pH 7.4, and 0.5% Nonidet P-40. The proteins were eluted with 50 μl of 2× LDS, including 10% 0.5 m dithiothreitol.
LKB1 Activity Assay—Endogenous LKB1 was immunoprecipitated with the N-19 LKB1 antibody and protein G beads. The reaction was initiated by adding 100 μl of kinase buffer containing 1 mm ATP, 10 μCi of [32P]ATP (PerkinElmer Life Sciences), and 300 μm LKBtide (Upstate) into tubes containing the immunoprecipitated sample. After incubation at 30 °C for 10 or 20 min, supernatant was applied onto P81 paper (Whatman), and 32P incorporation was determined by liquid scintillation counting. GST-tagged LKB1 activities were assessed similar way on GSH beads.
SIRT1 Deacetylation of LKB1 in Vitro—GST-LKB1 resuspended in 100 μl of 1× Sir2 assay buffer that came comes with the Cyclex SIRT1 deacetylase kit (CY-1151) was incubated for 30 min at 30 °C with recombinant SIRT1 1 μl with/without 0.2 mm NAD+. After washing with NETN buffer three times, LKB1 was eluted with 2× LDS-PAGE sample.
LKB1 Immunoprecipitation to Detect Acetylation—Because the molecular weight of LKB1 is very close to that of the reduced IgG heavy chain (∼50 kDa), covalent cross-linking of the LKB1 antibody with protein A or G beads (7) was performed to eliminate the IgG heavy chain from the Western blot. Briefly, 100 μg of the goat N-19 LKB1 antibody (for 293 cells), rabbit H-75 LKB1 antibody (for mouse adipose tissue), or goat M-18 LKB1 (for rat liver) antibody was incubated with 400 μl of 50% protein G or A beads in PBS pH8.2 at 4 °C for 2 h. After washing with 1 ml of PBS three times and 1 ml of 0.2 m triethanolamine, pH 8.2, the beads were resuspended with 1 ml of 20 mm dimethylpimelimidate-HCl in 0.2 m triethanolamine, pH 8.2, and incubated with mixing for 30 min at room temperature. After aspirating the solution, the beads were resuspended in 1 ml of 50 mm Tris buffer, pH 7.5, for 15 min. The cross-linked beads/antibody were washed with TBS-T three times and then resuspended with 2× diluted blocking solution (the same used for immunoblotting) at 4 °C until use. For liver, 500 μl (100 μg) of lysate was first cleared by incubating with protein A beads for 2 h and then incubated with the cross-linked LKB1 antibody overnight. For 293T cells, 300–500 μl of lysate were incubated with cross-linked LKB1 beads overnight. The immunoprecipitated beads were washed four times with lysis buffer and then eluted with 30 μl of elution buffer (Pierce) or 2× LDS sample buffer.
GFP-LKB1 Localization Analysis— 0.5 μg of pDEST53-LKB1-WT (or K-mutant GFP-LKB1) alone or in combination with 0.5 μg of SIRT1 (or pDsRed-SIRT1) was transfected into 293T cells grown on coverslips in 6-well plates. Sixteen hours post-transfection, the cells were fixed with 2% paraformaldehyde/PBS. Cells (20–30) were randomly chosen, and their fluorescent images were captured for localization analysis of LKB1 using a CCD image camera. The average brightness of GFP of nucleus, cytoplasm, and background were assessed in NIH-image, and the cytoplasmic/nuclear ratio was calculated.
Mass Spectrometric Analysis—GST-LKB1 fusion proteins (∼80 kDa) were cut into small pieces and subjected to “in gel” digestion with trypsin as described previously (8). MALDI-TOF and LC-MS/MS analyses of the digested samples were performed at the Dana-Farber Cancer Institute Molecular Biology Core Facilities (Boston).
Experiments with Rats—Sprague-Dawley rats weighing 150–170 g were either starved for 48 h or starved for 48 h and refed standard chow for 24 h. Under pentobarbital anesthesia, the liver was freeze-clamped in liquid nitrogen and stored at -80 °C until used for assays. Immunoprecipitation, Western blotting, and LKB1 activity measurements were performed as described. All studies were done in accordance with protocols approved by the Boston University Institutional Animal Use and Care Committee.
Statistical Analysis—Statistical analyses were done by the GLM procedure using SAS (Cary, NC). All results are expressed as means ± S.E. p < 0.05 was taken as significant.
RESULTS
Endogenous and Exogenously Expressed LKB1 Are Acetylated, and the Levels of Acetylation Can Be Modulated by SIRT1 Function—We immunoprecipitated LKB1 from various cells and blotted it for total acetylated lysines. To eliminate the heavily acetylated 50-kDa IgG heavy chain band that runs in close proximity to LKB1, we covalently cross-linked our antibodies to protein A/G beads. When this was done, acetylation of LKB1 was clearly demonstrable in HEK293T cells (Fig. 1A) in which Myc-tagged LKB1 was expressed by transient transfection and immunoprecipitated with covalently cross-linked Myc antibody. Using the same procedure, but with an LKB1 antibody (Santa Cruz Biotechnology M-18), we also observed acetylation of LKB1 in white adipose tissue of the mouse (supplemental Fig. S1) and in mouse testis. Thus, LKB1 was acetylated both in cultured 293T cells and in vivo.
FIGURE 1.
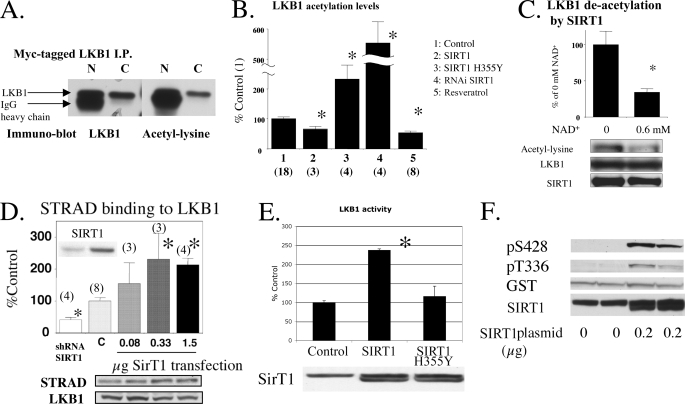
LKB1 acetylation and the effects of SIRT1. A, lysine acetylation of an exogenously expressed LKB1. Myc-tagged LKB1 expressed in HEK293T cells was immunoprecipitated with native (N) and covalently linked (C) anti-Myc antibody-protein A beads. B, modulation of LKB1 acetylation by SIRT1 in 293T cells. Plasmids expressing GST-LKB1 were transfected into HEK293T cells together with pcDNA or shRNA lentivirus for control, SIRT1 WT, or H355Y plasmid (0.5 μg) or shRNA for SIRT1 lentivirus. In addition, some cells were treated with 50–100 μm resveratrol for 0.5–1 h. The cells were harvested for Western blotting to quantify LKB1 acetylation. *, p <0.05 versus control. Number of samples are shown in parentheses. C, direct deacetylation of GST-LKB1 by SIRT1 in vitro. Purified GST-LKB1 was incubated in a SIRT1 reaction buffer containing 1 μl of recombinant SIRT1 solution with/without 0.6 mm NAD+ at 30 °C for 30 min (n = 6). D, SIRT1 expression levels affect STRAD association with LKB1. The 293T cells were transfected with SIRT1 plasmid or shRNA for sirt1. STRAD binding to LKB1 was assessed by calculating the STRAD to LKB1 ratio and is expressed as percent of the ratio in control cells (*, p < 0.05 versus C. Number of samples are in parenthesis). E and F, effects of SIRT1 overexpression on LKB1 activity (E) and phosphorylation (F). E, 293T cells were transfected with the 0.2 μg of SIRT1 plasmid. LKB1 activity was assessed in LKB1-immunoprecipitated samples (*, p < 0.005 versus control (pcDNA) plasmid, n = 3). The H355Y mutant did not increase LKB1 activity. F, 293T cells were co-transfected with 0.2 μg of SIRT1 and 0.5 μg of GSTl-LKB1 plasmids and blotted with the indicated antibodies.
To investigate the effect of SIRT1 on LKB1 acetylation status, GST-tagged LKB1 was expressed together with wild type SIRT1 and catalytically inactive H355Y SIRT1 in 293T cells. Wild type SIRT1 diminished LKB1 acetylation by 50%, whereas H355Y SIRT1 caused a 3-fold increase in its acetylation (Fig. 1B). Similarly, SIRT1 knockdown by shRNA caused a 5.5-fold increase in LKB1 acetylation, and incubation with 50–100 μm resveratrol, a putative Sir2 activator (5), for 0.5–1 h decreased its acetylation by 60% (Fig. 1B). Direct deacetylation of LKB1 by SIRT1 was also observed in a cell-free system in which isolated GST tagged LKB1 was co-incubated with SIRT1 and its substrate NAD+ (Fig. 1C). Acetylation was not observed with GST tag alone or with the control protein, GST-glucuronidase, suggesting that acetylation was exclusively on LKB1 protein. These results strongly suggest that SIRT1 can modulate LKB1 acetylation.
SIRT1 Affects LKB1 Binding to STRAD and Its Activity and Phosphorylation—Transient transfection of wild type (WT) SIRT1 in 293T cells increases both STRAD binding to LKB1 (Fig. 1D) and endogenous LKB1 activity, neither of which were observed when the catalytically inactive SIRT1 H355Y mutant was used (Fig. 1E). Conversely, STRAD binding to LKB1 was diminished in cells infected with SIRT1 shRNA (Fig. 1D). Overexpression of WT SIRT1 also increased LKB1 phosphorylation at both Ser-428 and Thr-336 (Fig. 1F), indicative of LKB1 activation as will be discussed later. Changes in the protein abundance of LKB1, STRAD, MO25, and AMPK-α were not observed in these experiments (data not shown).
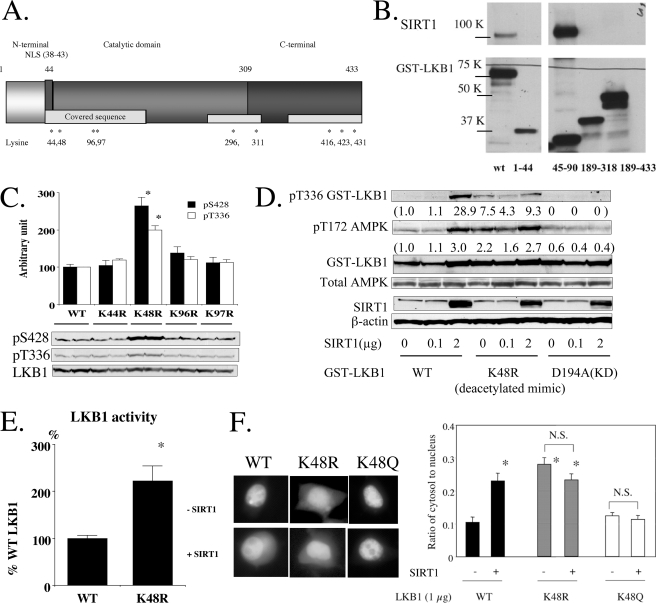
Lys-48 Is a Key Lysine at Which SIRT1 Modulates LKB1 Functions—To determine which of the acetylated lysine residues (AcK) on LKB1 play a role in the regulation of its activity, GST-tagged LKB1 was expressed in 293T cells and subjected to MALDI-TOF and LC-MS/mass spectrometry to determine its acetylation sites. As shown in Fig. 2A and supplemental Fig. S2, at least 9 lysines are acetylated over the three domains (N- and C-terminal regulatory and catalytic) of LKB1. Interaction analysis between a series of LKB1 fragments and SIRT1 revealed that SIRT1 associates with LKB1 on the N-terminal side of its catalytic domain (amino acids 45–90) (Fig. 2B). This association was specific, because no binding was observed between SIRT1 and a negative control protein (GST-glucuronidase). In separate experiments, we also observed an association of endogenous LKB1 and SIRT1 in 293T cells by co-immunoprecipitation. Based on these findings, we created a series of LKB1 point mutants in which specific lysines in the N-terminal catalytic domain were converted to arginine, a change that mimics to some extent the effects of a deacetylated lysine (9). Mutations were created at Lys-44, Lys-48, Lys-96, and Lys-97 in GST fusion protein expression plasmids. As shown in Fig. 2C, in the 293T cells, only the K48R mutant demonstrated increased phosphorylation of LKB1 at Ser-428 and Thr-336, as would be predicted if Lys-48 deacetylation was an early event in LKB1 activation. The K48R mutant of LKB1 also caused more AMPK phosphorylation under basal conditions and, as expected, because it lacked lysine at position 48, it was less sensitive to SIRT1 overexpression (Fig. 2D). Furthermore, LKB1 activity of the Lys-48 mutant was 2-fold higher than that of WT LKB1 when not stimulated by SIRT1 (Fig. 2E). In keeping with the importance of Lys-48 deacetylation in mediating the action of SIRT1, we also found that it was deacetylated in cells incubated with resveratrol (supplemental Fig. S2B).
FIGURE 2.
Identification of key lysines regulated by deacetylation in LKB1. A, LC-MS/MS and MALDI-TOF analyses revealed that LKB1 is acetylated on a number of lysine residues in each of its three domains. The asterisks indicate the positions of the acetylated lysines. The analysis was repeated six times, and the results for acetylation modification were compiled (see supplemental Fig. S2 for detail). B, identification of the domain on LKB1 that interacts with endogenous SIRT1. Plasmids expressing the indicated GST-tagged fragments of LKB1 and SIRT1 were co-transfected into HEK293T cells. GST-LKB1 fragments were pulled down with GSH beads and blotted with GST and SIRT1 antibodies. A strong association was observed between the N-terminal catalytic domain (amino acids 45–90) of LKB1 and SIRT1. C, effects of point mutations of lysine to arginine at various sites between 44 and 97. N-terminal lysine residues were systematically mutated to the deacetylation mimic arginine and expressed in HEK293T cells. Only Lys-48 showed enhanced phosphorylation of Ser-428 and Thr-336 (n = 4). D and E, effects of the arginine point mutation at Lys-48 on SIRT1-induced phosphorylation of AMPK, LKB1 Thr-336, and LKB1 activity. SIRT1 overexpression (2 μg) markedly increased the phosphorylation of Thr-336 on wild type (WT) LKB1. In contrast, phosphorylation of Thr-336 was increased basally but was insensitive to SIRT1 overexpression in the K48R mutant. A similar trend was observed in AMPK phosphorylation. No changes were observed in KD-expressing cells. Numbers refer to fold-changes in density of a band compared with that in control cells (D). E, activity of GST-K48R LKB1 expressed in 293T cells were 2-fold higher than wild type (*, p < 0.05, n = 4). F, effects of SIRT1 overexpression on cellular distribution of K48R LKB1. Plasmids expressing GFP-tagged LKB1 harboring the indicated mutations were transfected with or without SIRT1 into HEK293T cells. SIRT1 overexpression increased the cytosol/nuclear ratio of WT LKB1, but failed to do so in the K48Q LKB1 mutant. When K48R was used, the cytosolic distribution of LKB1 was already increased (*, p < 0.05 versus WT LKB1 not infected with SIRT1, n = 15–20), and it was not increased further by SIRT1 overexpression.
Cytosolic Localization of LKB1 Is Increased by Both SIRT1 Overexpression and K48R Mutation of LKB1—To examine further the role of Lys-48, we transfected K48R GFP fusion proteins and compared their localization to that of WT LKB1. The studies were done both under basal conditions and after overexpressing SIRT1. Overexpression of SIRT1 significantly increased the cytoplasmic/nuclear ratio of WT LKB1. In contrast, the K48R LKB1 mutant was initially more prominent in the cytoplasm, and this was not further altered by exogenous SIRT1 (Fig. 2F and supplemental Fig. S3). Interestingly, increased cytoplasmic localization of both WT LKB1 and especially the K48R mutant was observed when STRAD was exogenously expressed, suggesting that STRAD levels maybe rate-limiting in these cells (supplemental Fig. S4). The distribution of the negative control K48Q mutant was similar to that of control cells under basal conditions, but, like the latter, it was not affected by exogenous SIRT1. Taken together, these results indicate that deacetylation at Lys-48 is a key event in SIRT1-mediated LKB1 activation, phosphorylation, and cytosolic relocation.
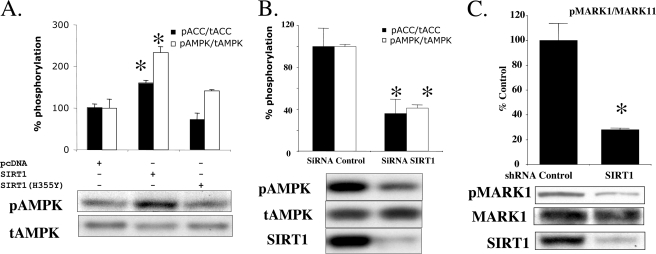
SIRT1 Modulates LKB1-induced Phosphorylation of Its Downstream Targets—Phosphorylation of AMPK at Thr-172 was increased by overexpression of WT SIRT1 (Fig. 3A), and conversely, transfection of siRNA for SIRT1 down-regulated the phosphorylation of AMPK and its substrate acetyl-CoA carboxylase (Fig. 3B). In addition, we found that the phosphorylation of another LKB1 target, MARK1, was down-regulated by SIRT1 knockdown in HepG2 cells (Fig. 3C).
FIGURE 3.
A, effects of overexpression of SIRT1 on AMPK and ACC phosphorylation. Cells grown in 6-well plates were transfected with 2 μg of wild type SIRT1 or the inactive SIRT1 H355Y mutant. The ratios of phosphorylated/total AMPK and ACC were increased by SIRT1 overexpression, whereas SIRT1 H355Y had no effects (*, p < 0.05 versus pcDNA, n = 3). A similar increase in AMPK phosphorylation was observed in other experiments. B, effects of SIRT1 knockdown on AMPK and ACC phosphorylation in HEK293T cells. Transfection of HEK293T cells with siRNA targeting SIRT1 decreased a ratio of phosphorylated/total of AMPK and ACC (*, p < 0.05 versus siRNA control, n = 6). C, effects of SIRT1 knockdown on MARK1 phosphorylation in HepG2 cells. Lentiviral infection of shRNA targeting sirt1 in HepG2 cells decreased the ratio of phosphorylated to total MARK1 by 69% (*, p <0.05, n = 6).
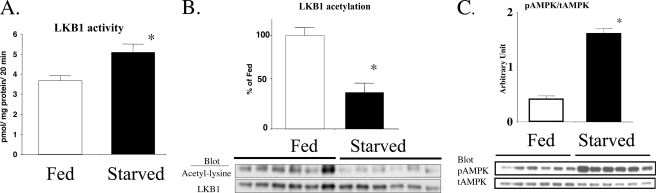
LKB1 Deacetylation and Activity in Rat Liver Are Increased by Starvation—LKB1 activity was 33% higher (Fig. 4A), and its total acetylation was 60% lower (Fig. 4B) in the liver of rats starved for 48 h than in the liver of starved-refed rats. Furthermore, this increase in LKB1 activity was associated with a greater than 2-fold increase in pAMPK (Fig. 4C) and pACC (not shown) as described previously (10).
FIGURE 4.
A–C, effects of starvation on hepatic LKB1 activity (A), acetylation (B), and AMPK phosphorylation (C). Sprague-Dawley rats were starved for 48 h or starved and then refed standard chow for 24 h (Fed). Under pentobarbital anesthesia, the livers were removed and then freeze-clamped for the indicated measurements. After 48 h of starvation, LKB1 activity and the phosphorylation of AMPK were higher, and LKB1 acetylation was lower that in the starved-refed rats (*, p < 0.05 versus fed, n = 6).
DISCUSSION
The principal new findings of this study are that LKB1 can be deacetylated by SIRT1 and that in HEK293T cells such deacetylation correlates with increases in LKB1 activity, cytoplasmic localization, and binding to STRAD, and AMPK and ACC phosphorylation. In contrast, down-regulation of SIRT1 with RNAi diminished LKB1 binding to STRAD and the phosphorylation (activity) of both AMPK and another LKB1 target, MARK1. We also found that similar increases in LKB1 deacetylation and activity and AMPK activation occur in rat liver in vivo during starvation, a state in which activation of SIRT1 has been described previously (6). Finally, we observed LKB1 acetylation in adipose tissue and testis (not shown) suggesting that acetylation and deacetylation are likely regulated in many organs. To our knowledge, these observations represent the first evidence that changes in the acetylation and deacetylation of LKB1 are mediated by SIRT1 and that they can affect its activity and secondarily that of AMPK. A schema describing the proposed mechanism by which SIRT1 activates LKB1 and secondarily AMPK and MARK1 is depicted in Fig. 5.
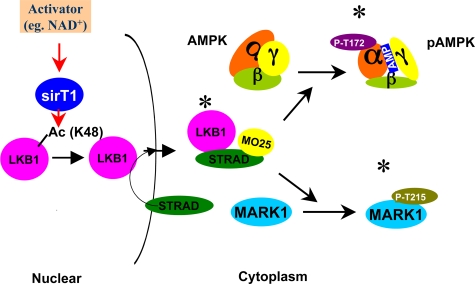
FIGURE 5.
Proposed mechanism of LKB1 activation by SIRT1. Activation of LKB1 and LKB1 target molecules by SIRT1, proposed mechanism. Factors that activate SIRT1 lead to deacetylation of one (Lys-48) or more key lysine residues on LKB1. This in turn enhances LKB1 binding to STRAD and MO25, which increases its kinase activity, and leads to the phosphorylation and activation of AMPK, MARK1, and possibly other LKB1 targets. The scheme assumes that LKB1 acetylation by SIRT1 occurs in the nucleus and that this facilitates LKB1 movement to the cytoplasm by binding to STRAD. Not shown is that the activation of LKB1 by STRAD also leads to LKB1 phosphorylation at Ser-428 and Thr-336 (see text). (* = activated enzyme.)
How LKB1 deacetylation leads to its activation is incompletely understood. LKB1 exists in both the nucleus and cytosol of various cells, and its activation and movement to the cytosol take place when it binds to STRAD and/or MO25 and forms a complex that appears to be constitutively active (1). We found that the deacetylation of LKB1 is associated with 2–3-fold increases in its activity, cytoplasmic/nuclear ratio, and its binding to endogenous STRAD. Experiments with 293T cells containing point mutations of lysine to arginine (which mimic the effects of deacetylation) strongly suggested that Lys-48 is a major site that mediates these events. However, the fact that SIRT1 overexpression or mutation of Lys-48 to arginine did not result in total movement of LKB1 to the cytoplasm suggests that Lys-48 deacetylation is not the sole factor governing LKB1 distribution. Because expression of exogenous STRAD enhanced the cytoplasmic localization of WT LKB1 and especially the K48R LKB1 mutant, STRAD levels appear to be rate-limiting for this process. Studies to determine the relationship between Lys-48 acetylation and the STRAD shuttling mechanism that has been reported to control LKB1 movement into and out of the nucleus (11) will clearly be needed.
Phosphorylation at Ser-428 (Ser-431 in the mouse) on LKB1 by protein kinases such as cAMP-dependent protein kinase and protein kinase Cζ and autophosphorylation at Thr-336, both of which occur after LKB1 is activated (1), were used as markers of LKB1 activation in the present study. Phosphorylation at these sites is not needed for LKB1 kinase activation (1); indeed, the splice variant of mouse LKB1 (accession number EU730638) that lacks the Ser-431 sequence is catalytically active and able to phosphorylate AMPK.3 Finally, the physiological significance of each of these phosphorylation events is incompletely understood, although Ser-428 phosphorylation has been linked to such events as tumor suppression (12), axon distribution (13), and the cellular localization and activation of AMPK (14).
The results presented here suggest that SIRT1 knockdown diminishes AMPK phosphorylation (Fig. 3, A and B); however, it has been reported that cells from SIRT1 knock-out mice show either no change (15) or an increase in AMPK activity (16). This discrepancy is unlikely because of an off-target effect associated with RNAi, because we employed two different RNAi preparations, and both gave the same result. A more likely explanation is that an overall low energy state observed in SIRT1 knock-out cells (16) overcomes the effect of LKB1 acetylation. In support of this possibility, it has been reported that SIRT1 knock-out is associated with constitutively high UCP2 expression and a low rate of ATP production by mitochondria (17). In this study, we employed RNAi-mediated down-regulation of SIRT1, and no change in ATP and ADP levels was observed within 72 h, suggesting that the cells were less energy-stressed.
This study focused on SIRT1 and its ability to deacetylate LKB1. Whether other sirtuins or class I or II histone deacetylases have a similar effect is not known. In support of this notion, these deacetylases and SIRT1 appear to target some proteins as common substrates. For example, FOXO1 is deacetylated by both SIRT1 (5) and SIRT2 (18). Thus, it is conceivable that in some cells sirtuins other than SIRT1 produce the changes in LKB1 described here. In this regard, Dasgupta and Milbrandt (15) have demonstrated that resveratrol can activate AMPK in neuronal cells and that LKB1 is required for this to occur even when SIRT1 is absent. The possibility that another sirtuin replaces SIRT1 in activating LKB1 in these cells has not been specifically examined; however, resveratrol has been reported to activate SIRT2 in neuronal cells (19).
To date, studies to determine whether LKB1 activation mediates increases in AMPK activity have principally been carried out in cardiac or skeletal muscle during ischemia, contraction, or chemical stimulation with phenformin or 5-aminomidazole-4-carboxamido-1-β-d-ribofuranoside (AICAR). The observation that LKB1 activity was not changed in these situations led to the view that it is constitutively active (20). The results of this study suggest that these effects may be tissue-specific. Thus, we found that in rat liver, LKB1 activity and acetylation status could be modulated by physiological stimuli (i.e. starvation and starvation-refeeding). Although a causal role of SIRT1 in mediating these events needs to be evaluated in mice with an acute knockdown of SIRT1, our findings suggest that during starvation, a condition associated with increased hepatic SIRT1 activity (6), deacetylation of LKB1 may be a key regulator of the LKB1-AMPK cascade. The reason for the apparently differing mechanisms for LKB1 activation in muscle and liver is unclear.
Recently, it has been reported that AMPK activation by polyphenols requires both SIRT1 and LKB1 in HepG2 cells and mouse liver (21). The results of this study provide a possible molecular mechanism for this observation, namely that SIRT1, by causing the deacetylation of LKB1, leads to its activation and that of AMPK. As already noted whether this mechanism is tissue-specific will require further studies as will the identification of the situations in which it operates in vivo.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants P01 HL08758 and DK19514 (to N. R.). This work was also supported by grants from the Kilo Diabetes Foundation (to Y. I.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
Footnotes
The abbreviations used are: AMPK, AMP-activated protein kinase; GST, glutathione S-transferase; GFP, green fluorescent protein; MALDI-TOF, matrix-assisted laser desorption ionization time-of-flight; LC-MS/MS, liquid chromatography/tandem mass spectrometry; LDS, lithium dodecyl sulfate; HRP, horseradish peroxidase; shRNA, short hairpin RNA; ACC, acetyl-CoA carboxylase; RNAi, RNA interference; WT, wild type.
F. Lan, J. M. Cacicedo, N. Ruderman, and Y. Ido, unpublished data.
References
- 1.Alessi, D. R., Sakamoto, K., and Bayascas, J. R. (2006) Annu. Rev. Biochem. 75 137-163 [DOI] [PubMed] [Google Scholar]
- 2.Ruderman, N., and Prentki, M. (2004) Nat. Rev. Drug Discov. 3 340-351 [DOI] [PubMed] [Google Scholar]
- 3.Boudeau, J., Baas, A. F., Deak, M., Morrice, N. A., Kieloch, A., Schutkowski, M., Prescott, A. R., Clevers, H. C., and Alessi, D. R. (2003) EMBO J. 22 5102-5114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanders, M. J., Grondin, P. O., Hegarty, B. D., Snowden, M. A., and Carling, D. (2007) Biochem. J. 403 139-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sinclair, D. A., and Guarente, L. (2006) Sci. Am. 294 48-51, 54-47 [DOI] [PubMed] [Google Scholar]
- 6.Rodgers, J. T., Lerin, C., Haas, W., Gygi, S. P., Spiegelman, B. M., and Puigserver, P. (2005) Nature 434 113-118 [DOI] [PubMed] [Google Scholar]
- 7.Schneider, C., Newman, R. A., Sutherland, D. R., Asser, U., and Greaves, M. F. (1982) J. Biol. Chem. 257 10766-10769 [PubMed] [Google Scholar]
- 8.Adachi, T., Pimentel, D. R., Heibeck, T., Hou, X., Lee, Y. J., Jiang, B., Ido, Y., and Cohen, R. A. (2004) J. Biol. Chem. 279 29857-29862 [DOI] [PubMed] [Google Scholar]
- 9.Megee, P. C., Morgan, B. A., Mittman, B. A., and Smith, M. M. (1990) Science 247 841-845 [DOI] [PubMed] [Google Scholar]
- 10.Assifi, M. M., Suchankova, G., Constant, S., Prentki, M., Saha, A. K., and Ruderman, N. B. (2005) Am. J. Physiol. Metab. 289 E794-E800 [DOI] [PubMed] [Google Scholar]
- 11.Dorfman, J., and Macara, I. G. (2008) Mol. Biol. Cell 19 1614-1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sapkota, G. P., Kieloch, A., Lizcano, J. M., Lain, S., Arthur, J. S., Williams, M. R., Morrice, N., Deak, M., and Alessi, D. R. (2001) J. Biol. Chem. 276 19469-19482 [DOI] [PubMed] [Google Scholar]
- 13.Shelly, M., Cancedda, L., Heilshorn, S., Sumbre, G., and Poo, M. M. (2007) Cell 129 565-577 [DOI] [PubMed] [Google Scholar]
- 14.Xie, Z., Dong, Y., Scholz, R., Neumann, D., and Zou, M. H. (2008) Circulation 117 952-962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dasgupta, B., and Milbrandt, J. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 7217-7222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, I. H., Cao, L., Mostoslavsky, R., Lombard, D. B., Liu, J., Bruns, N. E., Tsokos, M., Alt, F. W., and Finkel, T. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 3374-3379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bordone, L., Motta, M. C., Picard, F., Robinson, A., Jhala, U. S., Apfeld, J., McDonagh, T., Lemieux, M., McBurney, M., Szilvasi, A., Easlon, E. J., Lin, S. J., and Guarente, L. (2006) Plos Biol. 4 e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jing, E., Gesta, S., and Kahn, C. R. (2007) Cell Metab. 6 105-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki, K., and Koike, T. (2007) Biochem. Biophys. Res. Commun. 359 665-671 [DOI] [PubMed] [Google Scholar]
- 20.Hardie, D. G., Hawley, S. A., and Scott, J. W. (2006) J. Physiol. (Lond.) 574 7-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou, X., Xu, S., Maitland-Toolan, K. A., Sato, K., Jiang, B., Ido, Y., Lan, F., Walsh, K., Wierzbicki, M., Verbeuren, T. J., and Cohen, R. A. (2008) J. Biol. Chem. 283 20015-20026 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.