Abstract
Membrane skeletons are cytoskeletal elements that have important roles in cell development, shape, and structural integrity. Malaria parasites encode a conserved family of putative membrane skeleton proteins related to articulins. One member, IMC1a, is expressed in sporozoites and localizes to the pellicle, a unique membrane complex believed to form a scaffold onto which the ligands and glideosome are arranged to mediate parasite motility and invasion. IMC1b is a closely related structural paralogue of IMC1a, fostering speculation that it could be functionally homologous but in a different invasive life stage. Here we have generated genetically modified parasites that express IMC1b tagged with green fluorescent protein, and we show that it is targeted exclusively to the pellicle of ookinetes. We also show that IMC1b-deficient ookinetes display abnormal cell shape, reduced gliding motility, decreased mechanical strength, and reduced infectivity. These findings are consistent with a membrane skeletal role of IMC1b and provide strong experimental support for the view that membrane skeletons form an integral part of the pellicle of apicomplexan zoites and function to provide rigidity to the pellicular membrane complex. The similarities observed between the loss-of-function phenotypes of IMC1a and IMC1b show that membrane skeletons of ookinetes and sporozoites function in an overall similar way. However, the fact that ookinetes and sporozoites do not use the same IMC1 protein implies that different mechanical properties are required of their respective membrane skeletons, likely reflecting the distinct environments in which these life stages must operate.
Over 125 years after the discovery of its causative agent by Alphonse Laveran in 1880, malaria remains one of the most devastating infectious diseases in the world. With 300–500 million cases and over a million deaths a year, this apicomplexan parasite represents a huge public health problem and a considerable economic burden (1). Malaria control efforts suffer from widespread resistance to anti-parasitic drugs and insecticides, underpinning the urgent need for novel intervention strategies. Transmission of malaria parasites starts with the ingestion of male and female gametocytes by vector mosquitoes during blood feeding on a parasite-infected host. Rapid gametogenesis and fertilization occur in the mosquito midgut, from which ookinetes develop that invade the midgut epithelium and transform into oocysts. After a two-week period of growth, mature oocysts release thousands of sporozoites into the mosquito hemolymph that invade the salivary glands of the insect and enter the vertebrate host during blood feeding to initiate new malaria infections.
Invasive stages of malaria parasites, as well as related apicomplexan parasites, possess a unique cortical structure called the pellicle. This structure is made up of the plasma membrane, the inner membrane complex (IMC),2 and subpellicular microtubules (2, 3). An additional structure of the pellicle, named the subpellicular network (SPN), was identified in Toxoplasma gondii tachyzoites (4). The SPN consists of a two-dimensional network of intermediate filaments located on the cytoplasmic side of the IMC (4). It displays mechanical strength and takes the shape of the cell, indicating that it acts as a membrane skeleton (4). A protein component of the T. gondii SPN was identified, TgIMC1, which has structural homology to the articulins, the membrane skeleton proteins of free-living protists (4). A conserved family of putative membrane skeleton proteins structurally related to TgIMC1 has been identified in Plasmodium species (5). The first member of this family, IMC1a, was previously characterized and shown to be specifically expressed in sporozoites and to be essential for sporozoite infectivity (5), highlighting malaria membrane skeletons as potential intervention targets. In this study, we characterize a second member of the Plasmodium berghei IMC1 membrane skeleton family, IMC1b, using fluorescent protein tagging and gene disruption. Our findings show that IMC1b is functionally homologous to IMC1a, but operates in a different invasive life stage, the ookinete.
EXPERIMENTAL PROCEDURES
Parasite Maintenance, Culture, and Purification
P. berghei ANKA clone 234 parasites were maintained as cryopreserved stabilates or by mechanical blood passage and regular mosquito transmission. To purify gametocytes, white blood cells were removed from gametocytemic blood by passage through CF11 columns and further purified by centrifugation (300 × g for 30 min) through 48% Nycodenz cushions, followed by phosphate-buffered saline (PBS) washes. Ookinete cultures were set up overnight from unpurified gametocytemic blood as described previously (6). After 18–20 h, ookinetes were purified via ice-cold 0.17 m ammonium chloride lysis and centrifugation at 800 × g for 10 min, followed by PBS washes. Oocyst cultures were performed as described previously (7). Mosquito transmission assays were as described previously (8–11).
Reverse Transcription (RT)-PCR
RT-PCR was carried out as described (12) with the imc1b-specific primers TCCCAAACCCAAAATTATAGATG and TTATGTATTTGTTTCAATTGAGAAATG, and the α-tubulin-specific primers ACACATCAATGACTTCTTTACC and GAAGTAATAAGTATACATGTAGG.
Constructs
pLP-DHFR2—Plasmid pLP-DHFR/SR (13) was digested with NdeI and self-ligated using T4 DNA ligase.
pDNR-IMC1b/EGFP—The coding sequence of IMC1b plus 0.53 kb of the 5′-UTR were PCR-amplified with primers pDNR-IMC1b-F (ACGAAGTTATCAGTCGACGGTACCATTGAGACGTTACGTATTAATTGTG) and pDNR-IMC1b-R (ATGAGGGCCCCTAAGCTTGTATTTGTTTTCAATTGAGAAATGG) and introduced into SalI/HindIII-digested donor plasmid pDNR-EGFP (13) via in-fusion cloning.
pLP-DHFR/IMC1b—A 0.74-kb sequence corresponding to the 3′-UTR of imc1b was PCR-amplified with primers pLP-IMC1b-F (TAACCATTGGTCATAAAAAAGGAACTGAAACAGGATA) and pLP-IMC1b-R (CGGCCGCTCTAGCATACTACTTAAATAATATTTATTTCCTTTAGTGTGAA) and introduced into SacII-digested pLP-DHFR2 via in-fusion cloning.
pLP-IMC1b/EGFP—The IMC1b-specific sequence from pDNR-IMC1b/EGFP was introduced into pLP-DHFR/IMC1b via Cre-loxΨ site-specific recombination (BD Biosciences).
pLP-ΔIMC1b/EGFP—pLP-IMC1b/EGFP was digested with HindIII, removing most of the IMC1b coding sequence, and self-ligated using T4 DNA ligase.
A Dual Plasmid System for the Generation of Constructs Facilitating GFP Tagging
To facilitate the generation of DNA constructs that allowed the GFP tagging of imc1b via double crossover homologous recombination, we designed and constructed a dual plasmid system (supplemental Fig. S1). The first plasmid, pDNR-EGFP, is derived from pDNR-Dual (BD Biosciences) and was modified to contain the coding sequence for enhanced GFP (EGFP) followed by a “generic” 3′-UTR derived from pbdhfr. This plasmid also contains the chloramphenicol resistance gene without a bacterial promoter. These combined sequences are flanked by two loxP sites. The second plasmid, pLP-DHFR2, is derived from pBS-DHFR (8) and was modified to contain a modified tgdhfr gene (conferring resistance to the antimalarial drug pyrimethamine) flanked upstream by the promoter sequence of pbdhfr and downstream by the 3′-UTR of pbsr (12). This plasmid also contains the loxP promoter cassette (BD Biosciences) containing a single loxP site followed by a bacterial promoter (supplemental Fig. S1). The ensuing generation of the DNA construct for GFP tagging of IMC1b involved three steps (supplemental Fig. S1). In the first step, the coding sequence plus 5′-UTR of imc1b was PCR-amplified and introduced into pDNR-EGFP upstream of, and in-frame with, the egfp sequence. A unique KpnI restriction site was introduced upstream of the imc1b sequence during PCR. In the second step, the 3′-UTR of imc1b was PCR-amplified and introduced in pLP-DHFR2 downstream of the tgdhfr cassette. In the third step, the imc1b sequence contained within pDNR-EGFP was transferred by Cre-loxΨ site-specific recombination to the pLP-vector containing the imc1b-specific 3′-UTR. This recombination event places the chloramphenicol resistance gene present in the pDNR vector downstream of the bacterial promoter present in pLP vector, allowing antibiotic selection of desired recombinants.
Generation and Genomic Analysis of Genetically Modified Parasites
Parasite transfection, pyrimethamine selection, and dilution cloning were performed as described previously (14). Prior to performing transfections, plasmid DNA was digested with KpnI and SacII to remove the vector backbone. Genomic DNA extraction and Southern blot were performed as described previously (8). All clonal genetically modified parasite populations were checked for the absence of wild-type parasites by diagnostic PCR.
Western Blot Analysis
Parasite samples were heated directly in SDS-PAGE loading buffer at 70 °C for 10 min. Proteins were fractionated by electrophoresis through NuPAGE 4–12% BisTris precast gels (Invitrogen) and transferred to polyvinylidene difluoride membrane (Invitrogen) according to the manufacturer's instruction. Membranes were blocked for nonspecific binding in PBS supplemented with 0.1% Tween 20 and 5% skimmed milk for 1 h at room temperature. Goat polyclonal antibody to GFP horseradish peroxidase-conjugated (Abcam ab6663) diluted 1:5000 was applied to the membrane for 1 h at room temperature. After washing, the signal was detected by chemiluminescence (Pierce ECL western blotting substrate) according to the manufacturer's instructions.
Confocal Microscopy
For assessment of fluorescence, live or paraformaldehyde-fixed parasite samples were mounted in Vectashield containing DAPI. Images were captured on a Zeiss Axiovert 200 M inverted microscope using Zeiss LSM 510 software.
Assessment of Ookinete Shape and Motility
Images of Giemsa-stained ookinetes were captured by microscopy and their length and width measured. For motility assays, ookinetes were purified on magnetic columns (MACS, Miltenyi Biotec) and resuspended in ookinete medium. Images of live ookinetes were captured at 2-min intervals by microscopy. These time points were used to measure ookinete movement over a 20-min period.
Osmotic Shock and Viability Assays
Ookinetes in ookinete medium were subjected to hypo-osmotic shock of 0.5× normal osmotic strength by adding an equal volume of water. After 5 min, normal osmotic conditions were restored by adding an appropriate amount of 10× PBS. Cell viability was scored by fluorescence microscopy in the presence of 5 ml/liter propidium iodide and 1% Hoechst 33258. Ookinetes whose nucleus stained positive for both propidium iodide and Hoechst were scored as nonviable, whereas ookinetes whose nucleus only stained positive for Hoechst were scored as viable.
RESULTS
IMC1b Sequence and Structure—Among the paralogues of Plasmodium IMC1a, IMC1b is structurally the most closely related as it shares three conserved domains with IMC1a (5). The P. berghei IMC1b gene was identified from BLAST searches of P. berghei genomic sequences with the IMC1b sequence from Plasmodium yoelii (GenBank™/EMBL/DDJB accession number EAA15257). Three overlapping sequences were identified (PB_RP1359, berg-2278d04.qlk, and PB_RP2104) from which the entire P. berghei IMC1b coding sequence could be assembled. IMC1b is encoded by a single exon. The predicted full-length protein is composed of 535 amino acids with a predicted Mr of 62,590 sharing 93% amino acid sequence identity with its orthologue in P. yoelii. BLAST searches identified orthologues in all other Plasmodium species examined, including Plasmodium falciparum (AAN36013), Plasmodium vivax (EDL45805), Plasmodium chabaudi (CAH84924), Plasmodium knowlesi (PKH_093930), and Plasmodium gallinaceum (gal28a.d000013616.Contig1). A multiple alignment of the IMC1b orthologues reveals the presence of three conserved domains, separated by regions of variable length and sequence (Fig. 1A). These three domains, the first two of which are structurally related to articulins, are conserved in the T. gondii membrane skeleton protein TgIMC1 as well as in IMC1a (Fig. 1B) (4, 5). None of the Plasmodium IMC1b proteins identified possess the cysteine motifs at amino and carboxyl termini described in TgIMC1 and PbIMC1a (Fig. 1B) (5, 15).
FIGURE 1.
Sequence and structure of Plasmodium IMC1b. A, multiple amino acid sequence alignment of the predicted IMC1b proteins from P. berghei (Pb), P. yoelii (Py), P. chabaudi (Pc), P. knowlesi (Pk), P. vivax (Pv), P. falciparum (Pf), and P. gallinaceum (Pg). Indicated are three conserved domains (shaded), and gaps were introduced to allow optimal alignment (hyphens). Conserved amino acid identities (asterisks) and similarities (colons and points) are indicated below. The alignment was made with ClustalW with default parameters. B, schematic diagram of the protein structures of TgIMC1 and its structural relatives PxIMC1a and PxIMC1b in Plasmodium. Boxes mark domains corresponding to the amino-terminal (hatched), central (gray), and carboxyl-terminal (open) conserved domains highlighted in A. Conserved terminal cysteine motifs are indicated by open circles.
Generation of Genetically Modified Parasites—To study the expression and localization of IMC1b, we generated a genetically modified P. berghei parasite stably expressing IMC1b containing a carboxyl-terminal GFP tag. This was achieved by replacing the native imc1b allele by double homologous crossover recombination with a recombinant gene linked to the egfp coding sequence. Concomitantly, a modified T. gondii dihydrofolate reductase (tgdhfr) gene cassette, which confers resistance to the antimalarial drug pyrimethamine, was introduced (supplemental Fig. S2A). To study the function of IMC1b and its contribution to parasite development, a genetically modified P. berghei parasite was constructed in which all of the imc1b coding sequence was removed except for the first 30 residues. In this parasite line GFP is expressed under control of the native imc1b gene promoter (supplemental Fig. S2A). After transfection of purified schizont preparations, pyrimethamine-resistant parasites were selected. Diagnostic PCR across the predicted integration sites showed correct integration of the tgdhfr cassette into the imc1b locus (data not shown). This was confirmed by assessing the integrity of clonal populations of the genetically modified parasite lines (named IMC1b/GFP and IMC1b-KO, respectively) by Southern blot analysis of HindIII-digested genomic DNA (supplemental Fig. S2B). Two DNA probes were used as follows: one specific to part of the 5′-UTR plus complete imc1b coding sequence, and one specific to the tgdhfr gene. The imc1b-specific probe gave rise to a 1.1-kb band in IMC1b-KO parasites, 1.1- and 1.6-kb bands in IMC1b/GFP parasites, and 1.1- and 3-kb bands in the parental, wild-type (WT) parasites, as predicted (supplemental Fig. S2B; the doublet at 3 kb in the WT sample is the result of a small amount of partially digested DNA). The tgdhfr-specific probe gave rise to a single band in the IMC1b/GFP and IMC1b-KO parasites but no signal in WT parasites as expected (supplemental Fig. S2B). These combined results confirmed correct integration of the recombinant imc1b and tgdhfr alleles into the P. berghei imc1b locus. All genetically modified clonal parasite lines developed normally in mice and were morphologically indistinguishable from WT parasites in Giemsa-stained blood films (data not shown).
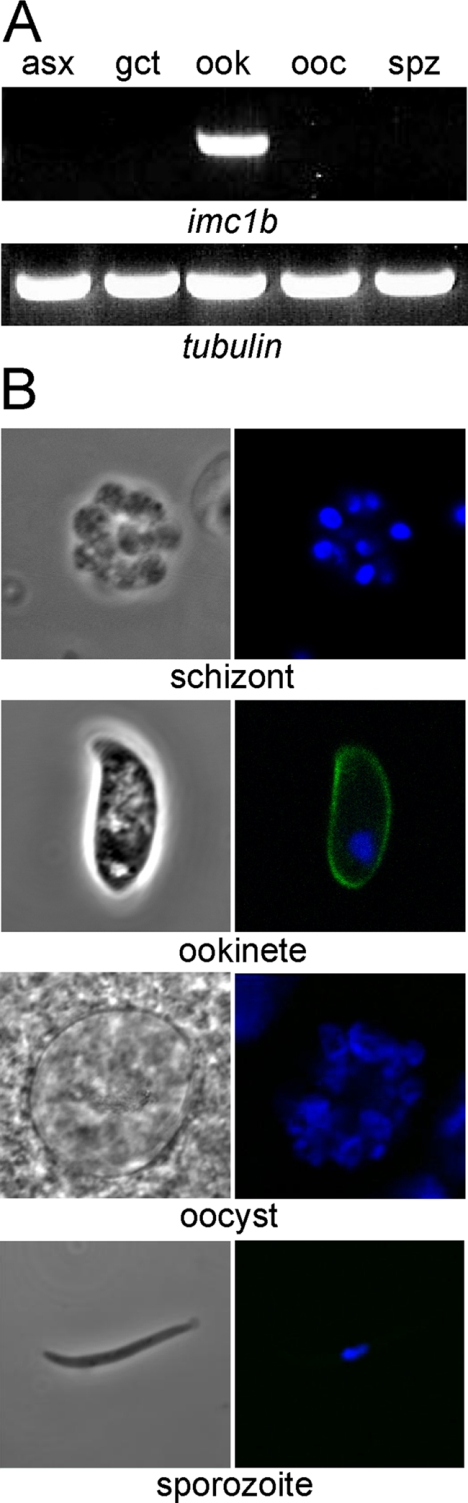
Life Stage Expression of IMC1b—We assessed transcription of the imc1b gene by RT-PCR of total RNA extracted from samples enriched for asexual blood stages, gametocytes, ookinetes, oocysts, and sporozoites, respectively. This assay revealed that transcription of the imc1b gene occurred predominantly in the enriched ookinete sample (Fig. 2A), pointing to an ookinete-specific expression of IMC1b. The expression of IMC1b protein was further studied using parasite line IMC1b/GFP. These parasites developed normally in mosquitoes and were readily transmitted by infected mosquito bites, demonstrating that the addition of the GFP tag to the carboxyl terminus of the IMC1b protein did not adversely affect parasite development. In support of the ookinete-specific IMC1b expression predicted from the imc1b transcription data (Fig. 2A), examination of IMC1b/GFP parasites by UV microscopy revealed strong GFP-based fluorescence in ookinetes, but not in asexual stages, oocysts, or sporozoites (Fig. 2B). GFP-based fluorescence in ookinetes was localized to the periphery of the cells (Fig. 2B), supporting a pellicular localization.
FIGURE 2.
Differential expression of IMC1b. A, RT-PCR analysis of purified parasite samples enriched for asexual blood stages (asx), gametocytes (gct), ookinetes (ook), day 10 oocysts (ooc), and sporozoites (spz) for the presence of imc1b mRNA. Tubulin 1 was used as a reference gene. B, confocal microscope bright field and fluorescence images of different developmental stages. GFP (green) and DAPI (blue) images are combined.
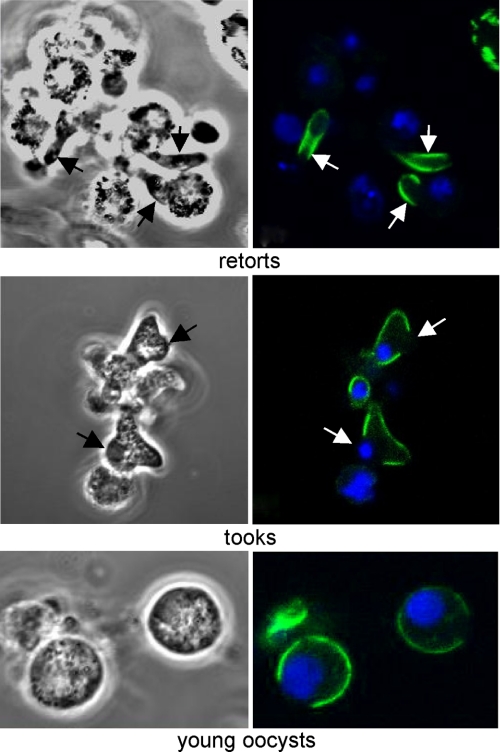
Subcellular Localization of IMC1b—To more closely follow the expression and subcellular localization of IMC1b, we cultured ookinetes and young oocyst stages in vitro. Very weak cytoplasmic fluorescence was observed in macrogametes and zygotes (data not shown). As the ookinete develops by forming a protrusion of the spherical zygote, a developmental stage called retort, strong fluorescence became localized to the periphery of the protrusion corresponding to the newly forming ookinete but not to the zygote part of the retort (Fig. 3). These observations argue against the targeting of IMC1b to the plasma membrane; in this scenario the protein would be localized to the periphery of entire retort, which is clearly not the case. The localized peripheral fluorescence did not extend all the way to the anterior end of the developing ookinetes. These observations are consistent with a localization of IMC1b to the ookinete pellicle structure, which support the prediction that IMC1b is a membrane skeleton protein.
FIGURE 3.
Subcellular localization of IMC1b during ookinete development and oocyst transition. Confocal microscope bright field and fluorescence images are shown of retorts (8 h into ookinete culture), tooks, and very young oocysts/fully rounded ookinetes. GFP (green) and DAPI (blue) images are combined.
We also studied the process of ookinete-to-oocyst transformation. This starts with the development of a round protrusion midway along the ookinete, a developmental stage called took. These protrusions were devoid of peripheral fluorescence (Fig. 3), indicating that IMC/SPN are absent from this part of the cell. As the rounding-up process of the ookinete completes, young spherical oocysts form; these displayed peripheral fluorescence only in part of the cell (Fig. 3). However, older multinucleate oocysts no longer displayed any fluorescence (data not shown) indicating loss of the entire IMC/SPN structures, consistent with documented oocyst architecture.
IMC1b Loss-of-Function Phenotypes—The function of IMC1b and its contribution to parasite development and infectivity were determined using the IMC1b-KO parasite line. IMC1b-KO parasites developed normally in mice and formed gametocytes. Gametogenesis occurred normally in vitro, and ookinetes developed in vitro and in vivo in numbers comparable with WT parasites (data not shown). RT-PCR analysis of IMC1b-KO ookinetes confirmed the absence of imc1b-specific messenger RNA in this parasite line, as predicted (supplemental Fig. S2C). Moreover, western blot analysis of ookinete samples using a commercially available anti-GFP antibody revealed bands with apparent sizes of ∼100 and 30 kDa in parasite lines IMC1b/GFP and IMC1b-KO, respectively (supplemental Fig. S2D). These bands correspond to the IMC1b/GFP fusion protein and to GFP fused to the amino-terminal 30 residues of IMC1b, respectively. This confirmed correct expression of the recombinant imc1b/gfp allele in parasite line IMC1b/GFP, as well as the absence of IMC1b expression in parasite line IMC1b-KO.
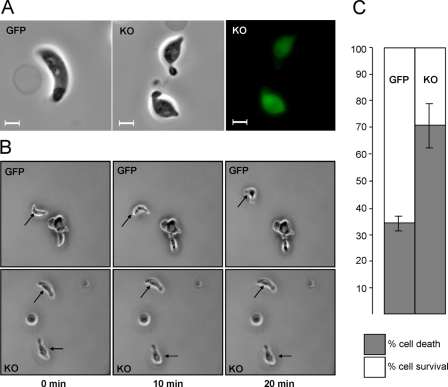
Upon closer examination, the morphology of IMC1b-KO ookinetes appeared abnormal (Fig. 4A). Compared with WT ookinetes, IMC1b-KO ookinetes were typically shorter (mean length 10.76 ± 0.15 μm for WT; 9.27 ± 0.10 μm for IMC1b-KO; n = 100), and wider (mean width 1.98 ± 0.03 μm for WT; 2.67 ± 0.05 μm for IMC1b-KO; n = 100). In particular, IMC1b-KO ookinetes possessed a bulging area typically in the central part of the cell (Fig. 4A). As expected, GFP was expressed in these ookinetes but was no longer targeted to the IMC/SPN resulting in cytoplasmic green fluorescence (Fig. 4A). To assess infectivity of IMC1b-KO parasites, mosquitoes were infected and analyzed for oocyst development. Oocyst numbers in IMC1b-KO parasite-infected mosquitoes were 8–10-fold lower than in WT parasite-infected mosquitoes (Table 1). These oocysts did, however, appear morphologically normal and produced sporozoites that successfully invaded the salivary glands. These sporozoites were infectious to mice following mosquito bite (data not shown). This is consistent with the observation that IMC1b is not expressed downstream of the ookinete stage (Fig. 2). Thus, parasites are capable of completing the Plasmodium life cycle without functional IMC1b, albeit at reduced efficiency. IMC1b-KO parasites that had been passaged through mosquitoes retained their loss-of-function phenotype both in terms of cell shape (data not shown) and infectivity (Table 1).
FIGURE 4.
IMC1b loss-of-function phenotypes. A, abnormal cell shape and cytoplasmic GFP fluorescence of IMC1b-deficient (KO) ookinetes. Scale bars represent 5 μm. B, ookinete gliding motility in parasite lines IMC1b/GFP (GFP) and IMC1b-KO (KO), by time-lapse microscopy. Frames are taken at 10-min intervals. Arrows point to moving ookinetes. C, percentage ookinete survival/death after hypo-osmotic shock (5 min exposure to 0.5× normal osmotic strength, values normalized to 100% viability in untreated cells). Error bars indicate standard deviations from two independent experiments. At least 100 ookinetes were scored for each sample.
TABLE 1.
Effects of IMC1b knock-out on P. berghei oocyst development in Anopheles stephensi mosquitoes
| Experiment | Parasite | Mean no. of oocysts/mosquito (range) | No. of mosquitoes dissected | % Infected |
|---|---|---|---|---|
| I | imc1b+ | 73 (0–250) | 19 | 95 |
| imc1b– | 9 (0–39) | 20 | 75 | |
| II | imc1b+ | 130 (2–401) | 20 | 100 |
| imc1b–a | 13 (0–89) | 20 | 75 |
Parasites were passaged once through mosquitoes
Gliding Motility and Mechanical Stability of IMC1b-KO Parasites—Gliding locomotion is a demonstrated prerequisite for cell invasion both in sporozoites (16, 17) and in ookinetes (8, 18, 19). To assess whether the reduced number of oocysts obtained in IMC1b-KO parasite-infected mosquitoes could be caused by an adverse effect of the absence of IMC1b on ookinete motility, gliding of KO ookinetes was examined in vitro. Indeed, gliding of IMC1b-KO ookinetes was markedly reduced compared with ookinetes expressing functional IMC1b (Fig. 4B). Over a period of 20 min, IMC1b/GFP ookinetes moved a mean distance of 22.06 ± 4.09 μm, whereas IMC1b-KO ookinetes traveled a mean distance of 5.75 ± 1.76 μm (n = 20). Accordingly, gliding motility of the IMC1b-KO ookinetes appears to be reduced ∼4-fold. Overall, these results show that the imc1b gene is not essential for ookinete gliding motility in vitro; however, its disruption does adversely affect, directly or indirectly, the ability of the ookinete to glide normally. Complete loss of gliding has been linked to an abolishment of oocyst development (8, 18, 19). Thus, the fact that reduced numbers of oocysts form in IMC1b-KO parasite-infected mosquitoes agrees with these ookinetes having reduced, rather than abolished, locomotion.
To experimentally assess whether IMC1b knock-out had affected the mechanical stability of the parasite, we subjected ookinetes to hypo-osmotic shock. Hypo-osmotic conditions cause cells to draw in water and swell, and the degree of hypoosmotic stress a cell can tolerate is a measure of its mechanical strength (20). Reproducibly, exposure to hypo-osmotic conditions (0.5 × normal osmotic strength) for 5 min caused twice as much cell death in IMC1b-KO ookinetes (70.5%) as it did in ookinetes expressing functional IMC1b (34.5%) (Fig. 4C). These results demonstrate that IMC1b is involved in the mechanical stability of ookinetes, which is consistent with it being an ookinete membrane skeleton component.
DISCUSSION
Although membrane skeletons have important roles in cell shape, structural integrity, and development (21), not much is known about the proteins that compose, or are otherwise associated with, membrane skeletons in malaria parasites. In this study we characterize a putative membrane skeleton protein, IMC1b, in the rodent malaria parasite P. berghei. Our findings show that IMC1b expression is restricted to the ookinete, where it localizes to the pellicle, and we demonstrate that IMC1b has central roles in ookinete cell shape, motility, mechanical stability, and infectivity. These roles are consistent with IMC1b being a component of the ookinete membrane skeleton, and provide experimental evidence to support the view that membrane skeletons form an integral structure of apicomplexan zoites and function to provide rigidity to the pellicular membrane complex. This rigidity could help provide directionality to the motive force of the glideosome and thereby improves the efficiency of the invasion process.
For this study we have developed a genetic tool to facilitate the swift generation of DNA constructs that allow the stable replacement of Plasmodium genes with GFP-tagged versions. We have tested this system successfully with the putative membrane skeleton protein IMC1b, demonstrating that this is a fast, effective, and arguably more reliable alternative to generating specific antibodies and immunofluorescence. In addition, we show that removing the IMC1b coding sequence from the same DNA construct facilitates the placement of GFP under control of the imc1b promoter, resulting in a gene knock-out parasite with a GFP reporter. Potential interference of the GFP epitope tag with normal IMC1b protein function can be assessed by comparing parasites expressing GFP-tagged IMC1b with WT parasites on the one hand and with IMC1b knock-out parasites on the other hand. In this study, IMC1b/GFP parasites behaved like WT rather than IMC1b-KO parasites. From this we infer that the carboxyl-terminal GFP tagging of IMC1b did not adversely affect IMC1b localization or function.
The genetically modified parasites generated have allowed us to assess the expression and localization of IMC1b throughout the Plasmodium life cycle, as well as to study formation of the ookinete and its transformation into oocyst. Peripheral localization of IMC1b is first observed in retort stages, concomitant with the developing ookinete protruding from the zygote body (Fig. 3), a process reminiscent of sporozoites budding from the sporoblast. This fluorescence pattern is consistent with IMC1b localizing to the pellicle structures, which are laid down as the developing ookinete protrudes from the zygote body (3, 22, 23). The weak cytoplasmic fluorescence already detected in macrogametes/zygotes indicates the start of IMC1b synthesis, in preparation for its supramolecular arrangement into SPN at the retort stage. Notably, IMC1b localization does not extend all the way to the apical end of ookinetes, indicating that the SPN does not extend all the way to the anterior end of the cell. This is plausible, because the cytoskeletal organization at the anterior end of ookinetes is markedly distinct from the rest of the pellicle, consisting of a complex, multilayered structure known as the collar (23). As ookinetes start to transform into oocysts, a bulging area is formed that is devoid of IMC1b labeling and is likely to constitute plasma membrane devoid of IMC/SPN. We postulate that this is newly synthesized plasma membrane required to relax the ookinete structure, allowing it to round up. Consistent with this hypothesis, the next phase of oocyst transition is a spherical, mononucleate cell, which still appears to possess its original IMC/SPN structure as documented (22). In older oocysts, however, IMC1b protein is absent, consistent with the documented absence of IMC/SPN at this point of development (22).
We show that IMC1b-KO ookinetes possess reduced locomotion. Movement of ookinetes occurs by gliding motility, a substrate-dependent form of eukaryotic locomotion that neither requires cilia or flagella nor any major changes in cell morphology (such as pseudopod formation in amoeba). Gliding motility is essential for three important activities of apicomplexan parasites: migration, host-cell invasion, and egress (24). In apicomplexans, gliding motility is actin- and myosin-dependent, and the actin-myosin motor that drives parasite motility is located between the plasma membrane and the IMC (3, 25–28). Given this association between the pellicle and the actin-myosin motor, it is conceivable that the absence of IMC1b in the membrane skeleton could adversely affect ookinete motility. Another cause of the reduced IMC1b-KO oocyst numbers observed could be the diminished mechanical strength of the ookinetes. Ookinetes undergo considerable constrictions as they migrate through the peritrophic matrix and through midgut epithelial cells (29, 30), and structurally weakened IMC1b-KO ookinetes might be less able to successfully invade or be more prone to damage during such events.
The expression, localization, and loss-of-function phenotypes of IMC1b reported here are reminiscent of those of the sporozoite-expressed membrane skeleton protein IMC1a (5). IMC1a localizes to the pellicle structure of sporozoites. In addition, IMC1a-deficient sporozoites were smaller in size compared with WT sporozoites and possessed a single, enlarged area associated with the position of the nucleus. These sporozoites displayed reduced gliding motility and mechanical stability (5). It thus appears that IMC1a and IMC1b are functional homologues in different invasive life stages. This is not unprecedented. For example, the extracellular proteins CTRP and TRAP are both structurally and functionally homologous, involved in gliding motility of ookinetes and sporozoites, respectively, via interaction with host substrates (8, 16–19). The functional homology between IMC1a and IMC1b supports the view that membrane skeletons form an integral part of the pellicle in invasive stages of apicomplexan parasites. Moreover, the similarities in loss-of-function phenotype between IMC1a and IMC1b indicate that the function of the membrane skeleton of ookinetes and sporozoites is conserved. The fact that ookinetes and sporozoites do not, and probably cannot, use the same IMC1 protein suggests that their respective membrane skeletons possess some distinct properties as well. Indeed, ookinetes and sporozoites invade different structures and tissues. Accordingly, different strengths and/or flexibilities may be required of these cells.
At least eight conserved and structurally related IMC1 proteins have been identified in Plasmodium species, named IMC1a–h (5). Recently, high throughput mass spectrometry-based protein analysis of different life stages of P. berghei has provided some information on the expression of these family members (31). This analysis highlights the ookinete as one of the most interesting life stages regarding the expression of IMC1 proteins, as at least five of them (IMC1b, -c, -e, -f, and -h) appear to be expressed in this life stage. By contrast, only two members (IMC1a and -h) were shown to be expressed in sporozoites (31). The different number of IMC1 proteins expressed by ookinetes and sporozoites further supports the notion that their respective membrane skeletons possess distinct properties. We are currently investigating the function of the other ookinete-specific IMC1 family members and their potential interactions with IMC1b, as well as with other parasite proteins.
Supplementary Material
Acknowledgments
Sequence data of P. berghei, P. knowlesi, and P. gallinaceum were produced by the Plasmodium Sequencing Groups at the Sanger Institute and can be obtained online. We thank Elizabeth Brooks and Rachel Gregory for assistance with the confocal microscopy.
This work was supported by the Wellcome Trust. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.

Author's Choice—Final version full access.
Footnotes
The abbreviations used are: IMC, inner membrane complex; SPN, subpellicular network; RT, reverse transcription; DAPI, 4′,6-diamidino-2-phenylindole; GFP, green fluorescent protein; EGFP, enhanced GFP; PBS, phosphate-buffered saline; UTR, untranslated region; WT, wild-type; BisTris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
References
- 1.Sachs, J., and Malaney, P. (2002) Nature 415 680-685 [DOI] [PubMed] [Google Scholar]
- 2.Raibaud, A., Lupetti, P., Paul, R. E., Mercati, D., Brey, P. T., Sinden, R. E., Heuser, J. E., and Dallai, R. (2001) J. Struct. Biol. 135 47-57 [DOI] [PubMed] [Google Scholar]
- 3.Morrissette, N. S., and Sibley, L. D. (2002) Microbiol. Mol. Biol. Rev. 66 21-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mann, T., and Beckers, C. (2001) Mol. Biochem. Parasitol. 115 257-268 [DOI] [PubMed] [Google Scholar]
- 5.Khater, E. I., Sinden, R. E., and Dessens, J. T. (2004) J. Cell Biol. 167 425-432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arai, M., Billker, O., Morris, H. R., Panico, M., Delcroix, M., Dixon, D., Ley, S. V., and Sinden, R. E. (2001) Mol. Biochem. Parasitol. 116 17-24 [DOI] [PubMed] [Google Scholar]
- 7.Carter, V., Nacer, A. M., Underhill, A., Sinden, R. E., and Hurd, H. (2007) Int. J. Parasitol. 37 1221-1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dessens, J. T., Beetsma, A. L., Dimopoulos, G., Wengelnik, K., Crisanti, A., Kafatos, F. C., and Sinden, R. E. (1999) EMBO J. 18 6221-6227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dessens, J. T., Mendoza, J., Claudianos, C., Vinetz, J. M., Khater, E., Hassard, S., Ranawaka, G. R., and Sinden, R. E. (2001) Infect. Immun. 69 4041-4047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dessens, J. T., Siden-Kiamos, I., Mendoza, J., Mahairaki, V., Khater, E., Vlachou, D., Xu, X.-J., Kafatos, F. C., Louis, C., Dimopoulos, G., and Sinden, R. E. (2003) Mol. Microbiol. 49 319-329 [DOI] [PubMed] [Google Scholar]
- 11.Le Chat, L., Sinden, R. E., and Dessens, J. T. (2007) Mol. Biochem. Parasitol. 153 41-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Claudianos, C., Dessens, J. T., Trueman, H. E., Arai, M., Mendoza, J., Butcher, G. A., Crompton, T., and Sinden, R. E. (2002) Mol. Microbiol. 45 1473-1484 [DOI] [PubMed] [Google Scholar]
- 13.Carter, V., Shimizu, S., Arai, M., and Dessens, J. T. (2008) Mol. Microbiol. 68 1560-1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waters, A. P., Thomas, A. W., van Dijk, M. R., and Janse, C. J. (1997) Methods (San Diego) 13 134-147 [DOI] [PubMed] [Google Scholar]
- 15.Mann, T., Gaskins, E., and Beckers, C. (2002) J. Biol. Chem. 277 41240-41246 [DOI] [PubMed] [Google Scholar]
- 16.Menard, R., Sultan, A. A., Cortes, C., Altszuler, R., van Dijk, M. R., Janse, C. J., Waters, A. P., Nussenzweig, R. S., and Nussenzweig, V. (1997) Nature 385 336-340 [DOI] [PubMed] [Google Scholar]
- 17.Spaccapelo, R., Naitza, S., Robson, K. J., and Crisanti, A. (1997) Lancet 350 335. [DOI] [PubMed] [Google Scholar]
- 18.Yuda, M., Sakaida, H., and Chinzei, Y. (1999) J. Exp. Med. 190 1711-1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Templeton, T. J., Kaslow, D. C., and Fidock, D. A. (2000) Mol. Microbiol. 36 1-9 [DOI] [PubMed] [Google Scholar]
- 20.Menke, A., and Jockusch, H. (1991) Nature 349 69-71 [DOI] [PubMed] [Google Scholar]
- 21.Pumplin, D. W., and Bloch, R. J. (1993) Trends Cell Biol. 3 113-117 [DOI] [PubMed] [Google Scholar]
- 22.Garnham, P. C., Bird, R. G., Baker, J. R., Desser, S. S., and el-Nahal, H. M. (1969) Trans. R. Soc. Trop. Med. Hyg. 63 187-194 [DOI] [PubMed] [Google Scholar]
- 23.Sinden, R. E., Hartley, R. H., and Winger, L. (1985) Parasitology 91 227-244 [DOI] [PubMed] [Google Scholar]
- 24.Soldati, D., and Meissner, M. (2004) Curr. Opin. Cell Biol. 16 32-40 [DOI] [PubMed] [Google Scholar]
- 25.Heintzelman, M. B. (2003) Curr. Biol. 13 R57-R59 [DOI] [PubMed] [Google Scholar]
- 26.Dobrowolski, J. M., and Sibley, L. D. (1996) Cell 84 933-939 [DOI] [PubMed] [Google Scholar]
- 27.Dobrowolski, J. M., Carruthers, V. B., and Sibley, L. D. (1997) Mol. Microbiol. 26 163-173 [DOI] [PubMed] [Google Scholar]
- 28.Meissner, M., Schluter, D., and Soldati, D. (2002) Science 298 837-840 [DOI] [PubMed] [Google Scholar]
- 29.Han, Y. S., Thompson, J., Kafatos, F. C., and Barillas-Mury, C. (2000) EMBO J. 19 6030-6040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vernick, K. D., Fujioka, H., and Aikawa, M. (1999) Exp. Parasitol. 91 362-366 [DOI] [PubMed] [Google Scholar]
- 31.Hall, N., Karras, M., Raine, J. D., Carlton, J. M., Kooij, T. W., Berriman, M., Florens, L., Janssen, C. S., Pain, A., Christophides, G. K., James, K., Rutherford, K., Harris, B., Harris, D., Churcher, C., Quail, M. A., Ormond, D., Doggett, J., Trueman, H. E., Mendoza, J., Bidwell, S. L., Rajandream, M. A., Carucci, D. J., Yates, J. R., III, Kafatos, F. C., Janse, C. J., Barrell, B., Turner, C. M., Waters, A. P., and Sinden, R. E. (2005) Science 307 82-86 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.