Abstract
Repair of interstrand DNA cross-links (ICLs) in Escherichia coli can occur through a combination of nucleotide excision repair (NER) and homologous recombination. However, an alternative mechanism has been proposed in which repair is initiated by NER followed by translesion DNA synthesis (TLS) and completed through another round of NER. Using site-specifically modified oligodeoxynucleotides that serve as a model for potential repair intermediates following incision by E. coli NER proteins, the ability of E. coli DNA polymerases (pol) II and IV to catalyze TLS past N2-N2-guanine ICLs was determined. No biochemical evidence was found suggesting that pol II could bypass these lesions. In contrast, pol IV could catalyze TLS when the nucleotides that are 5′ to the cross-link were removed. The efficiency of TLS was further increased when the nucleotides 3′ to the cross-linked site were also removed. The correct nucleotide, C, was preferentially incorporated opposite the lesion. When E. coli cells were transformed with a vector carrying a site-specific N2-N2-guanine ICL, the transformation efficiency of a pol II-deficient strain was indistinguishable from that of the wild type. However, the ability to replicate the modified vector DNA was nearly abolished in a pol IV-deficient strain. These data strongly suggest that pol IV is responsible for TLS past N2-N2-guanine ICLs.
Interstrand DNA cross-links (ICLs)2 represent a major challenge to cellular survival by virtue of their ability to block progression of DNA replication and transcription. In Escherichia coli, biochemical and genetic data support a mechanism by which ICLs can be repaired through the combined activities associated with the nucleotide excision repair (NER) pathway and the homologous recombination (HR) damage avoidance pathway (1). The overall pathway requires recognition and initiation of ICL repair by the action of UvrABC, creating dual incisions by hydrolyzing the ninth phosphodiester bond 5′ and the third phosphodiester bond 3′ to the cross-link (2, 3). It has been proposed that this “unhooked” DNA can be structurally manipulated such that HR-driven strand invasion occurs via pairing with the intact strand that is still physically linked to the incised strand. Subsequent to recombination, in which a non-damaged homologous strand is positioned opposite the cross-linked strand, a second round of NER can excise the remaining damage, followed by gap synthesis and ligation.
Although the above mechanism has the capacity to fully repair ICL-containing DNA, an alternative pathway not requiring HR has been proposed (4, 5). This pathway utilizes the activity of the polB gene product, DNA polymerase (pol) II. The data demonstrated that pol II, but not pol IV or V, was functioning in this HR-independent ICL repair pathway. It was hypothesized that pol II could catalyze replication bypass of the incised but still covalently linked DNA (4, 5). However, no biochemical data supporting such an activity of pol II have subsequently appeared.
Using a series of synthetic oligodeoxynucleotide substrates that mimic various intermediates in the processing of NER-incised ICLs, our laboratory has recently explored the biochemical basis of translesion DNA synthesis (TLS) past N2-N2-guanine ICL intermediates using human DNA polymerases. These investigations revealed that pol κ was able to catalyze bypass of ICL lesions in which the nucleotides 3′ to the lesion had been removed (6).
Given a role for E. coli pol II in HR-independent ICL repair and that E. coli pol IV is an ortholog of human pol κ (7, 8), this study was designed to investigate the ability of pol II and pol IV to catalyze TLS past N2-N2-guanine ICLs.
EXPERIMENTAL PROCEDURES
Generation of Oligodeoxynucleotides Containing Site-specific ICLs—Modified oligodeoxynucleotides (ICL1, ICL2, ICL3, and ICL4) (see Fig. 1A) containing various model acrolein-mediated N2-N2-guanine cross-links (see Fig. 1B) were synthesized and purified as previously described (6) and were kind gifts of Drs. Carmelo J. Rizzo and Ivan D. Kozekov (Department of Chemistry, Vanderbilt University). A control non-damaged DNA substrate was obtained from the Molecular Microbiology and Immunology Research Core Facility of the Oregon Health & Science University.
FIGURE 1.
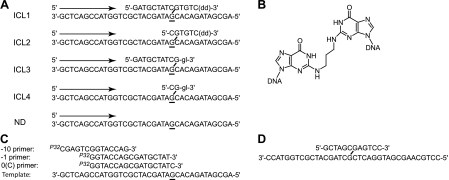
DNA substrates. A, schematic representation of DNA substrates: ICL1, ICL2, ICL3, and ICL4 containing an interstrand cross-link and a non-damaged substrate (ND). Arrows indicate the direction of DNA synthesis. The site of the N2-N2-guanine cross-link is underlined. dd, dideoxynucleotide; gl, glycerol. B, structure of N2-N2-guanine cross-link. C, primer sequences used in the in vitro reactions. D, schematic representation of an N2-N2-guanine cross-link-containing 36-mer oligodeoxynucleotide that was used for generating the pMS2-ICL vector.
Proteins—E. coli pol II and pol IV were purified as described previously (9, 10).
DNA Polymerase Bypass Assays—The primers (see Fig. 1C) were 32P-end-labeled and annealed with the DNA templates as described previously (11). The primer extension assays were conducted in a 10-μl reaction volume containing pol II or pol IV (as indicated in the figure legends), 5 nm primer-template, 5 mm MgCl2, 100 μm dNTPs, 25 mm Tris-HCl (pH 7.5), 25 mm dithiothreitol, 0.5 mg/ml bovine serum, and 10% glycerol. Reactions were carried out at 37 °C for 30 min, followed by termination of the reaction with a solution containing 95% (v/v) formamide, 10 mm EDTA, 0.03% (w/v) xylene cyanol, and 0.03% (w/v) bromphenol blue. DNA replication products were separated through a 15% (w/v) denaturing polyacrylamide gel at 2000 V and later analyzed on a PhosphorImager (GE Healthcare).
Steady-state kinetic assays were performed according to a standard procedure (12, 13). Briefly, reactions were conducted at 22 °C in the same buffer as the primer extension assays with the addition of NaCl (50 mm). The concentration of the primer-template DNA substrates was 10 nm. The concentrations of pol IV and incubation times were adjusted for each particular primer-template combination such that the formation of the product would not exceed 25%. Nucleotide concentrations (dCTP and dGTP) varied. Quantitative analyses were performed using ImageQuant 5.2 software (GE Healthcare). The rates of nucleotide incorporation were plotted as a function of nucleotide concentration, and the kcat and Km parameters were obtained from the best fit of the data to the Michaelis-Menten equation using KaleidaGraph 3.6 software (Synergy Software).
Bacterial Strains—The strains used in this study were derived from E. coli K-12 strain W3110 and are isogenic except for deletion of either dinB or polB (Table 1) (14). All strains were cultured in LB broth supplemented with spectinomycin (100 μg/ml) for the pol II-deficient strain and kanamycin (50 μg/ml) for the pol IV-deficient strain.
TABLE 1.
E. coli strains used in this study
Generation of a Vector Construct Carrying a Site-specific N2-N2-Guanine Cross-link—The characterization and preparation of a single-stranded pMS2 vector have been reported previously (15, 16). The single-stranded pMS2 DNA (15 pmol), which carries an EcoRV site in the hairpin region, was linearized by digestion with EcoRV (100 units) for 3 h at 37 °C and purified using Amicon 100K centrifugal filter devices according to the manufacturer's protocol. A 36-mer oligodeoxynucleotide (see Fig. 1D) carrying a site-specific N2-N2-guanine ICL was designed in such a way that the single-stranded regions were complementary to the peripheral regions of the linearized pMS2 vector. The 36-mer oligodeoxynucleotide (15 pmol) was phosphorylated using T4 polynucleotide kinase (50 units) for 1 h at 37 °C, added to the linearized pMS2 vector, annealed, and extended using the Klenow fragment of E. coli pol I (25 units). A double-stranded linear product was gel-purified and ligated overnight at 12 °C with T4 DNA ligase (4000 units). The ligated sample was designated pMS2-ICL and further used for transforming E. coli cells.
Transformation of E. coli Strains with pMS2-ICL and pBR322 Plasmids—Initial experiments were conducted using wild-type E. coli cells to determine the amount of pMS2-ICL that had comparable transformation efficiency with 0.5 ng of the reference plasmid, pBR322. For both pMS2-ICL and pBR322, selection of successful transformants was done using resistance to ampicillin. Next, a mixture of plasmids containing pMS2-ICL and pBR322 was prepared at quantities that would provide approximately equal transformation efficiencies, and this mixture was utilized to transform individual E. coli strains. Transformations were done by electroporation as described previously (16).
For further screening, the transformants were individually grown first in LB broth containing ampicillin (100 μg/ml) in 96-well plates at 37 °C for 4–6 h. A 20-μl aliquot from each 96 well was transferred to another 96-well plate containing LB broth with tetracycline (12.5 μg/ml) and grown overnight at 37 °C. Plasmids were isolated from tetracycline-sensitive colonies, thus positive for pMS2-ICL, and subjected to DNA sequencing using an 18-mer oligodeoxynucleotide (5′-AGCAACCATAGTCCCGCC-3′) as the primer.
RESULTS
Experimental Rationale and Substrate Design—Prior genetic evidence provides strong support for a role of E. coli pol II in an HR-independent ICL repair pathway (4, 5). Based on these data and previous models, it was hypothesized that pol II could be responsible for the replication bypass of a 12-mer DNA strand that was still covalently attached to the template strand. This structure would be representative of the product of a dual incision by E. coli UvrABC around an ICL site. Furthermore, replication bypass of N2-N2-guanine cross-links has been observed for the human ortholog of E. coli pol IV, pol κ (6). Thus, experiments were designed to test the ability of E. coli pol II and pol IV to catalyze TLS on DNA substrates containing a site-specific ICL (Fig. 1A). Specifically, DNA strands in each of the four ICL substrates (ICL1, ICL2, ICL3, and ICL4) are joined via N2-guanines in a CpG sequence context using a carbon bridge that models an acrolein-derived ICL (Fig. 1B). ICL1 models the product of incision by the UvrABC complex, whereas ICL2 and ICL3 represent potential repair intermediates in which nucleotides 5′ and 3′ to the ICL, respectively, have been removed. ICL4 contains a residual ICL in which nucleotides both 5′ and 3′ have been removed. The 3′-ends of ICL1 and ICL2 are terminated with a dideoxynucleotide ((dd)-3′) to prevent any synthesis from the cross-linked strand. Similarly, ICL3 and ICL4 are 3′-capped with a glycerol (gl-3′) to prevent replication from that site. 32P-Labeled primers (Fig. 1C) were designed to initiate synthesis 1 or 10 nucleotides 5′ to the cross-linked site in the template strand. A 0 primer was used to initiate the replication from C opposite the cross-linked G.
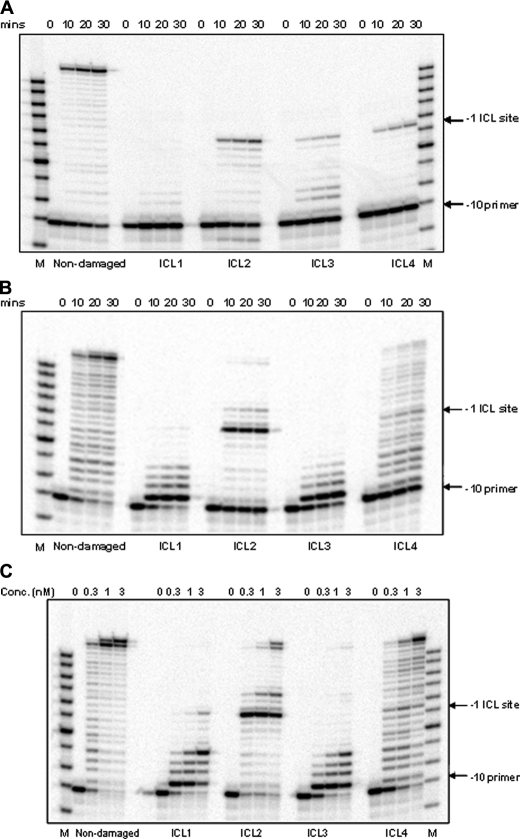
In Vitro Replication Bypass of ICL-containing DNA Substrates—Control non-damaged and ICL-containing DNA substrates were used to analyze replication from a –10 primer by pol II (Fig. 2A) and pol IV (Fig. 2, B and C). Using the non-damaged primer-template substrates, pol II catalyzed a highly processive polymerization to yield full-length primer extension products. However, under identical conditions, pol II was unable to carry out effective strand displacement synthesis on ICL1 and ICL3, whereas on ICL2 and ICL4, it could replicate up to one nucleotide prior to the cross-linked guanine, but no TLS past the lesion was observed (Fig. 2A). ICL bypass by pol II was not observed even when reactions were conducted with increased enzyme concentrations (up to a 1000-fold excess relative to the DNA substrate) (data not shown).
FIGURE 2.
A and B, primer extension by pol II and pol IV, respectively. Time course primer extension experiments were conducted using non-damaged and ICL DNA substrates (ICL1, ICL2, ICL3, and ICL4). A –10 primer (10 bases upstream of the adduct site) was annealed to the DNA template and incubated with pol II or pol IV (0.3 nm) for the indicated times. C, primer extension activity by pol IV is dose-dependent. The experiment was conducted using non-damaged and ICL1–4 templates primed with the –10 primer with increasing concentrations of pol IV at 37 °C for 30 min. M indicates the oligodeoxynucleotide size marker (Fisher).
Examination of the activities of pol IV on the same substrates revealed a less processive synthesis on the non-damaged DNA template and a very poor ability to catalyze strand displacement synthesis on ICL1 and ICL3 following the incorporation of the first nucleotide (Fig. 2B). Using the ICL2 primer-template, in which no strand displacement synthesis is necessary, pol IV was able to synthesize up to one nucleotide prior to the ICL, but was able to catalyze only minimal incorporation opposite the lesion; further synthesis was blocked two nucleotides beyond the cross-linked site. In contrast, pol IV synthesis on the ICL4 primer-template revealed that although multiple pause sites occurred prior to reaching the ICL, there was only modest blockage at the lesion (Fig. 2B). Following incorporation opposite the cross-linked nucleotide, synthesis continued with reduced processivity with full-length DNA products accumulating over time.
Given these data, 30-min reactions were conducted using increasing concentrations of pol IV (Fig. 2C). Again, very poor strand displacement synthesis was observed on ICL1 and ICL3, whereas replication bypass was readily measured on ICL4 and to a lesser extent on ICL2. These data suggest that pol IV can catalyze TLS past N2-guanine ICLs; however, 5′-resection leading up to the lesion and 3′-exonucleolytic processing increase the TLS efficiency.
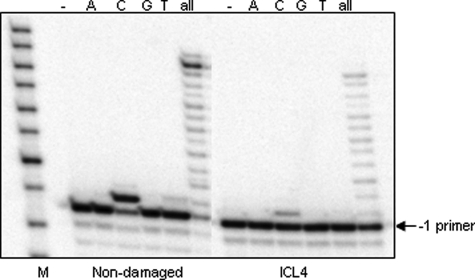
To determine the identity of nucleotide(s) inserted by pol IV opposite the cross-linked guanine, qualitative single nucleotide incorporation assays were conducted using a –1 primer annealed to ICL4. These data revealed that pol IV faithfully incorporated a dCTP opposite the lesion site (Fig. 3). Steady-state kinetic analyses showed that the catalytic efficiency (kcat/Km) of dCTP incorporation opposite the cross-linked G relative to the control G was reduced by ∼50-fold, whereas the efficiency of extension from a C opposite the cross-linked G was reduced by ∼2-fold (Table 2).
FIGURE 3.
Accuracy of pol IV-catalyzed nucleotide incorporation opposite the N2-N2-guanine cross-link. A single nucleotide incorporation assay was conducted in the presence of pol IV (0.3 nm) and individual nucleotides (25 μm) using the –1 primer annealed to non-damaged or ICL4 substrate (5 nm). These reactions were carried out for 30 min at 37 °C. M indicates the oligodeoxynucleotide size marker (Fisher).
TABLE 2.
Steady-state kinetic parameters for ICL bypass by E. coli pol IV
ND, non-damaged DNA substrate.
| DNA substrate | Primer | dNTP | kcat | Km | kcat/Km | Relative efficiency |
|---|---|---|---|---|---|---|
| min-1 | mm | μm-1 min-1 | ||||
| ND | -1 | dCTP | 0.90 ± 0.08 | 168 ± 30 | 5.4 × 10-3 | 1 |
| ICL4 | -1 | dCTP | 0.020 ±0.001 | 183 ± 18 | 0.11 × 10-3 | 0.02 |
| ND | 0 | dGTP | 3.9 ± 0.2 | 85 ± 8 | 46 × 10-3 | 1 |
| ICL4 | 0 | dGTP | 0.75 ± 0.02 | 36 ± 3 | 21 × 10-3 | 0.46 |
Replication of ICL-containing Plasmid DNAs in E. coli—To explore a cellular role for E. coli polymerases in the processing of N2-N2-guanine ICLs, we generated a double-stranded plasmid vector carrying a site-specific N2-N2-guanine cross-link (Fig. 1D) and utilized this modified DNA to transform wild-type and pol II and pol IV deletion E. coli strains. The efficiency of transformation was evaluated relative to a reference plasmid, unmodified pBR322. The pBR322 plasmid encodes resistance to both ampicillin and tetracycline, whereas pMS2-ICL can be selected only when cells are challenged with ampicillin. This feature allowed us to distinguish between the cells transformed with pBR322 versus the ones transformed with pMS2-ICL. The bacterial cells were electroporated with a mixture of pBR322 and pMS2-ICL and grown on LB agar plates containing ampicillin. For further screening, 192 transformed colonies were selected per strain, and a ratio of transformants carrying pBR322 versus those transformed with pMS2-ICL was determined by growing them first in LB broth with ampicillin, followed by transferring an aliquot of culture to LB broth containing tetracycline.
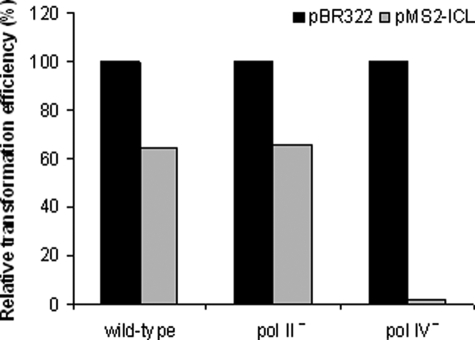
For the wild-type and pol II-deficient strains, of 192 transformants tested, 76 and 75 transformants, respectively, were found to be tetracycline-negative and thus positive for carrying plasmids that originated from pMS2-ICL. Plasmids were isolated from a subset of pMS2-ICL transformants (10 for the pol II-deficient strain and 52 for the wild-type strain), and the region of modification was analyzed by DNA sequencing. In all these plasmids, the insert sequences were present. Therefore, the relative transformation efficiency measured for the wild-type and pol II-deficient strains (Fig. 4) indicated that pol II has no effect on the intracellular replication of N2-N2-guanine ICL-containing DNA.
FIGURE 4.
Relative colony-forming ability of E. coli strains following transformation with an ICL-containing plasmid. For each strain, the percentage of pMS2-ICL transformants was calculated relative to pBR322 transformants. The apparent transformation efficiency with the reference plasmid, pBR322, was comparable for all the strains tested.
Using the pol IV-deficient strain, the relative efficiency of transformation with pMS2-ICL was, in contrast, extremely low; of 192 transformants, only five were tetracycline-negative. When plasmids isolated from these transformants were subjected to DNA sequencing, three of five contained the insert sequences, whereas two others were homologous to religated pMS2 vectors without inserts. Thus, relative to the wild type, the efficiency of transformation with plasmids containing N2-N2-guanine ICL was reduced by ∼40-fold in the pol IV-deficient strain (Fig. 4). These data strongly support our hypothesis that pol IV is essential for cellular processing of N2-N2-guanine ICLs. It is important to emphasize that the yield of non-adducted pBR322-transformed progenies remained comparable for the wild-type and pol II- and pol IV-deficient strains.
Interestingly, for all the strains, sequence analysis of the screened progenies originating from pMS2-ICL did not reveal any deletions or base substitutions at the adducted site or the neighboring bases. Thus, recombination-independent repair of N2-N2-guanine ICLs in E. coli is essentially non-mutagenic. Given an accurate bypass of these ICLs by pol IV in vitro and on the basis of our results from plasmid-based assays, we speculate that pol IV would be primarily responsible for the non-mutagenic TLS past N2-N2-guanine cross-links in vivo.
DISCUSSION
For over a decade, genetic evidence in E. coli has demonstrated that repair of ICLs can proceed via an HR-independent, pol II-dependent pathway. A model to account for these data hypothesized that following an NER-mediated unhooking of one of the strands associated with the ICL by the activity of the UvrABC complex, pol II could catalyze TLS past the intact 12-mer still attached to the complementary DNA strand. In this investigation, we sought to obtain biochemical data to support a role for pol II in catalyzing this reaction. However, our data revealed that at least in the case of N2-N2-guanine ICLs, pol IV (but not pol II) could catalyze TLS on ICL-containing substrate that had been designed to model the product of exonucleolytic processing of the eight nucleotides 5′ to the ICL site. Additionally, the efficiency of TLS could be enhanced by removal of nucleotides 3′ to the ICL, implying that an exonuclease activity on both ends of the incised strand may be required to stimulate the bypass activity of pol IV at an ICL site.
The biochemical observations suggesting a role for pol IV in TLS past N2-N2-guanine ICLs were further substantiated by plasmid reactivation assays, indicating that the pol IV-deficient strain is significantly impaired in its ability to replicate DNA containing a unique N2-N2-guanine ICL. In contrast, both biochemical and plasmid-based approaches did not generate any evidence for a possible involvement of pol II in bypass of such ICLs.
However, the cross-linking agents used in the previous studies by Loechler and co-workers (4, 5) produced different chemical linkages than those described in this report. In these studies, the largest differential cytotoxicity arising from the deficiency in pol II was observed with nitrogen mustard, an agent that induces ICLs via N-7 of guanines (a major groove adduct). It should be noted that the bypass activity of pol IV was not assessed in the above studies. In contrast, the acrolein-mediated ICLs used in this study are between the exocyclic amino groups of guanine residues in a CpG sequence context (17–19). This linkage is within the minor groove of DNA, and this difference may strongly dictate which polymerase might be capable of catalyzing TLS past specific ICLs. Thus, these data suggest that the repair of cross-linked DNAs via a mechanism not requiring HR may rely on different polymerases or combinations of polymerases to carry out TLS and that the conformational flexibility of both the unhooked or incised strand and the active site of the polymerase will dictate the efficiency and fidelity of the bypass reaction. In this regard, we have previously demonstrated that mutagenic bypass of intrastrand DNA cross-links was absolutely dependent on the activity of pol II (20). In that study, the linkages were two, three, or four carbon tethers between adjacent adenines via N6 atoms.
In summary, we propose a model in which error-free TLS-assisted repair of ICL-containing DNA (with an N2-N2-guanine linkage) is mediated by pol IV. Our model is based on the following observations: 1) an efficient and accurate bypass of N2-N2-guanine ICLs by pol IV in vitro and 2) an indispensable role of pol IV in replication of N2-N2-guanine ICL-containing plasmids.
Acknowledgments
We thank Drs. Carmelo J. Rizzo and Ivan D. Kozekov for synthesis and purification of the ICL-containing oligodeoxynucleotides. We also thank Dr. Masaaki Moriya for providing the pMS2 vector and Lauriel F. Earley for purification of the single-stranded pMS2 vector. We gratefully acknowledge Drs. Amanda K. McCullough and Jodi L. Johnson for suggestions and comments.
This work was supported, in whole or in part, by National Institutes of Health Grant ES05355 (to R. S. L.) and Center Grant ES000267 and National Institutes of Health Grants R37 GM21244 and ES012259 (to M. F. G.). This work was also supported by National Science Foundation Career Award MCB-0237975 (to S. E. F.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: ICL, interstrand cross-link; NER, nucleotide excision repair; HR, homologous recombination; pol, DNA polymerase; TLS, translesion DNA synthesis.
References
- 1.Cole, R. S. (1973) Proc. Natl. Acad. Sci. U. S. A. 70 1064–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Houten, B., Gamper, H., Holbrook, S. R., Hearst, J. E., and Sancar, A. (1986) Proc. Natl. Acad. Sci. U. S. A. 83 8077–8081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Houten, B. (1990) Microbiol. Rev. 54 18–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berardini, M., Mackay, W., and Loechler, E. L. (1997) Biochemistry 36 3506–3513 [DOI] [PubMed] [Google Scholar]
- 5.Berardini, M., Foster, P. L., and Loechler, E. L. (1999) J. Bacteriol. 181 2878–2882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Minko, I. G., Harbut, M. B., Kozekov, I. D., Kozekova, A., Jakobs, P. M., Olson, S. B., Moses, R. E., Harris, T. M., Rizzo, C. J., and Lloyd, R. S. (2008) J. Biol. Chem. 283 17075–17082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerlach, V. L., Feaver, W. J., Fischhaber, P. L., and Friedberg, E. C. (2001) J. Biol. Chem. 276 92–98 [DOI] [PubMed] [Google Scholar]
- 8.Lee, C. H., Chandani, S., and Loechler, E. L. (2006) J. Mol. Graph. Model. 25 87–102 [DOI] [PubMed] [Google Scholar]
- 9.Cai, H., Yu, H., McEntee, K., and Goodman, M. F. (1995) Methods Enzymol. 262 13–21 [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi, S., Valentine, M. R., Pham, P., O'Donnell, M., and Goodman, M. F. (2002) J. Biol. Chem. 277 34198–34207 [DOI] [PubMed] [Google Scholar]
- 11.Minko, I. G., Washington, M. T., Kanuri, M., Prakash, L., Prakash, S., and Lloyd, R. S. (2003) J. Biol. Chem. 278 784–790 [DOI] [PubMed] [Google Scholar]
- 12.Creighton, S., Bloom, L. B., and Goodman, M. F. (1995) Methods Enzymol. 262 232–256 [DOI] [PubMed] [Google Scholar]
- 13.Creighton, S., and Goodman, M. F. (1995) J. Biol. Chem. 270 4759–4774 [DOI] [PubMed] [Google Scholar]
- 14.Yeiser, B., Pepper, E. D., Goodman, M. F., and Finkel, S. E. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 8737–8741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moriya, M., Takeshita, M., Johnson, F., Peden, K., Will, S., and Grollman, A. P. (1988) Proc. Natl. Acad. Sci. U. S. A. 85 1586–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanuri, M., Minko, I. G., Nechev, L. V., Harris, T. M., Harris, C. M., and Lloyd, R. S. (2002) J. Biol. Chem. 277 18257–18265 [DOI] [PubMed] [Google Scholar]
- 17.Kozekov, I. D., Nechev, L. V., Moseley, M. S., Harris, C. M., Rizzo, C. J., Stone, M. P., and Harris, T. M. (2003) J. Am. Chem. Soc. 125 50–61 [DOI] [PubMed] [Google Scholar]
- 18.Kim, H. Y., Voehler, M., Harris, T. M., and Stone, M. P. (2002) J. Am. Chem. Soc. 124 9324–9325 [DOI] [PubMed] [Google Scholar]
- 19.Noll, D. M., Mason, T. M., and Miller, P. S. (2006) Chem. Rev. 106 277–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanuri, M., Nechev, L. V., Kiehna, S. E., Tamura, P. J., Harris, C. M., Harris, T. M., and Lloyd, R. S. (2005) DNA Repair 4 1374–1380 [DOI] [PubMed] [Google Scholar]
- 21.Zambrano, M. M., Siegele, D. A., Almirón, M., Tormo, A., and Kolter, R. (1993) Science 259 1757–1760 [DOI] [PubMed] [Google Scholar]