Abstract
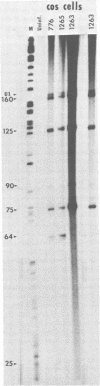
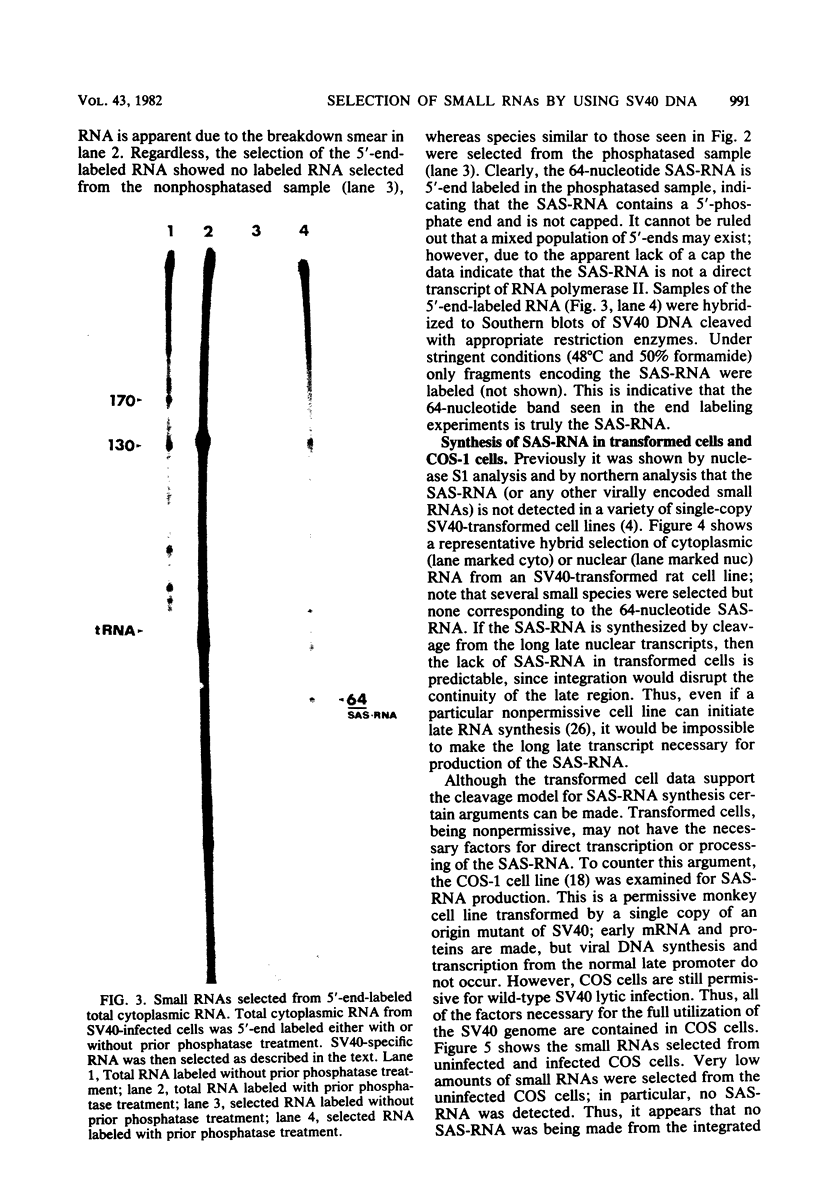
The simian virus 40 (SV40)-associated small RNA (SAS-RNA), approximately 64 nucleotides, is virally encoded within a region of the viral late (+) DNA strand which encodes no known protein. The SAS-RNA arises in abundance late in SV40 lytic infection. Previous data indicate that the synthesis of the SAS-RNA may be under the control of the normal late viral promoter; i.e., inhibition of transcription from the late promoter results in cessation of SAS-RNA synthesis. The synthesis of SAS-RNA was examined to determine whether the SAS-RNA is the product of cleavage from noncoding regions of nuclear late transcripts or an independent transcription product like 5S RNA, or the adenovirus VA-RNAs. The data described below suggest that SAS-RNA is cleaved from large late transcripts. In vitro transcription of DNA fragments containing the SAS-RNA coding region yielded no SAS-RNA synthesis; this result was supported by DNA sequence analysis, which indicated no promoter-like regions either within or flanking the SAS-RNA coding region. In support of a cleavage mechanism, the SAS-RNA has a 3′-phosphate end, an occurrence which is indicative of nuclease cleavage. In addition, 5′-end labeling of the SAS-RNA was possible only after calf alkaline phosphatase treatment; this indicates that the SAS-RNA is not capped. Hybrid selection analysis was used to demonstrate that separation of the SAS-RNA coding region from the normal late promoter resulted in elimination of SAS-RNA synthesis. This was demonstrated in SV40-transformed cells in which integration of a single copy of SV40 breaks the continuity of the late coding region, so that the SAS-RNA coding region is physically separated from the normal late promoter. The lack of SAS-RNA synthesis indicates that the SAS-RNA coding region cannot function as a primary transcription unit. The same result and conclusion were obtained by using a permissive cell line transformed by SV40 (COS-1 cells); here it was found that the integrated SAS-RNA coding region was not expressed even during a viable lytic infection in which the SAS-RNA could be expressed from the infecting viral genomes. The simplest conclusion drawn from the data is that the SAS-RNA is cleaved from larger late transcripts which initiate at the normal late promoter. This conclusion suggests that many of the small RNAs found in normal eucaryotic cells may be synthesized by specific cleavage rather than by primary transcription. In the course of these studies several small cellular RNAs were detected, due to their specific hybrid selection, by using SV40 DNA. Primary mapping and characterization data of these RNAs are also presented.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alwine J. C., Dhar R., Khoury G. A small RNA induced late in simian virus 40 infection can associate with early viral mRNAs. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1379–1383. doi: 10.1073/pnas.77.3.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alwine J. C., Kemp D. J., Stark G. R. Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5350–5354. doi: 10.1073/pnas.74.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alwine J. C., Khoury G. Simian virus 40-associated small RNA: mapping on the simian virus 40 genome and characterization of its synthesis. J Virol. 1980 Dec;36(3):701–708. doi: 10.1128/jvi.36.3.701-708.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J. M., Davidson N. Methylmercury as a reversible denaturing agent for agarose gel electrophoresis. Anal Biochem. 1976 Jan;70(1):75–85. doi: 10.1016/s0003-2697(76)80049-8. [DOI] [PubMed] [Google Scholar]
- Benecke B. J., Penman S. A new class of small nuclear RNA molecules synthesized by a type I RNA polymerase in HeLa cells. Cell. 1977 Dec;12(4):939–946. doi: 10.1016/0092-8674(77)90158-1. [DOI] [PubMed] [Google Scholar]
- Benoist C., Chambon P. Deletions covering the putative promoter region of early mRNAs of simian virus 40 do not abolish T-antigen expression. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3865–3869. doi: 10.1073/pnas.77.7.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenhagen D. F., Sakonju S., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: II. The 3' border of the region. Cell. 1980 Jan;19(1):27–35. doi: 10.1016/0092-8674(80)90385-2. [DOI] [PubMed] [Google Scholar]
- Campos R., Jovanovich S., Villarreal L. P. A small RNA complementary to an intervening sequence is produced late in SV40 infection. Nature. 1981 May 28;291(5813):344–346. doi: 10.1038/291344a0. [DOI] [PubMed] [Google Scholar]
- Cole C. N., Crawford L. V., Berg P. Simian virus 40 mutants with deletions at the 3' end of the early region are defective in adenovirus helper function. J Virol. 1979 Jun;30(3):683–691. doi: 10.1128/jvi.30.3.683-691.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFranco D., Schmidt O., Söll D. Two control regions for eukaryotic tRNA gene transcription. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3365–3368. doi: 10.1073/pnas.77.6.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande A. K., Jakowlew S. B., Arnold H. H., Crawford P. A., Siddiqui M. A. A novel RNA affecting embryonic gene functions in early chick glastoderm. J Biol Chem. 1977 Sep 25;252(18):6521–6527. [PubMed] [Google Scholar]
- Dwyer N., Blobel G. A modified procedure for the isolation of a pore complex-lamina fraction from rat liver nuclei. J Cell Biol. 1976 Sep;70(3):581–591. doi: 10.1083/jcb.70.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England T. E., Bruce A. G., Uhlenbeck O. C. Specific labeling of 3' termini of RNA with T4 RNA ligase. Methods Enzymol. 1980;65(1):65–74. doi: 10.1016/s0076-6879(80)65011-3. [DOI] [PubMed] [Google Scholar]
- Ford P. J. Polymerase III control region defined. Nature. 1980 Sep 11;287(5778):109–110. doi: 10.1038/287109a0. [DOI] [PubMed] [Google Scholar]
- Fowlkes D. M., Shenk T. Transcriptional control regions of the adenovirus VAI RNA gene. Cell. 1980 Nov;22(2 Pt 2):405–413. doi: 10.1016/0092-8674(80)90351-7. [DOI] [PubMed] [Google Scholar]
- Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981 Jan;23(1):175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- Grosschedl R., Birnstiel M. L. Spacer DNA sequences upstream of the T-A-T-A-A-A-T-A sequence are essential for promotion of H2A histone gene transcription in vivo. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7102–7106. doi: 10.1073/pnas.77.12.7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellung-Larsen P., Frederiksen S. Occurrence and properties of low molecular weight RNA components from cells at different taxonomic levels. Comp Biochem Physiol B. 1977;58(3):273–281. doi: 10.1016/0305-0491(77)90202-4. [DOI] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Jelinek W. R., Toomey T. P., Leinwand L., Duncan C. H., Biro P. A., Choudary P. V., Weissman S. M., Rubin C. M., Houck C. M., Deininger P. L. Ubiquitous, interspersed repeated sequences in mammalian genomes. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1398–1402. doi: 10.1073/pnas.77.3.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinek W., Leinwand L. Low molecular weight RNAs hydrogen-bonded to nuclear and cytoplasmic poly(A)-terminated RNA from cultured Chinese hamster ovary cells. Cell. 1978 Sep;15(1):205–214. doi: 10.1016/0092-8674(78)90095-8. [DOI] [PubMed] [Google Scholar]
- Lai C. J., Dhar R., Khoury G. Mapping the spliced and unspliced late lytic SV40 RNAs. Cell. 1978 Aug;14(4):971–982. doi: 10.1016/0092-8674(78)90351-3. [DOI] [PubMed] [Google Scholar]
- Lange M., May E., May P. Ability of nonpermissive mouse cells to express a simian virus 40 late function(s). J Virol. 1981 Jun;38(3):940–951. doi: 10.1128/jvi.38.3.940-951.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner M. R., Boyle J. A., Mount S. M., Wolin S. L., Steitz J. A. Are snRNPs involved in splicing? Nature. 1980 Jan 10;283(5743):220–224. doi: 10.1038/283220a0. [DOI] [PubMed] [Google Scholar]
- Lerner M. R., Steitz J. A. Snurps and scyrps. Cell. 1981 Aug;25(2):298–300. doi: 10.1016/0092-8674(81)90047-7. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Ohe K., Weissman S. M., Cooke N. R. Studies on the origin of a low molecular weight ribonucleic acid from human cells infected with adenoviruses. J Biol Chem. 1969 Oct 10;244(19):5320–5332. [PubMed] [Google Scholar]
- Pettersson U., Philipson L. Location of sequences on the adenovirus genome coding for the 5.5S RNA. Cell. 1975 Sep;6(1):1–4. doi: 10.1016/0092-8674(75)90066-5. [DOI] [PubMed] [Google Scholar]
- Ringuette M., Liu W. C., Jay E., Yu K. K., Krause M. O. Stimulation of transcription of chromatin by specific small nuclear RNAs. Gene. 1980 Jan;8(2):211–224. doi: 10.1016/0378-1119(80)90038-4. [DOI] [PubMed] [Google Scholar]
- Rosa M. D., Gottlieb E., Lerner M. R., Steitz J. A. Striking similarities are exhibited by two small Epstein-Barr virus-encoded ribonucleic acids and the adenovirus-associated ribonucleic acids VAI and VAII. Mol Cell Biol. 1981 Sep;1(9):785–796. doi: 10.1128/mcb.1.9.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakonju S., Bogenhagen D. F., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: I. The 5' border of the region. Cell. 1980 Jan;19(1):13–25. doi: 10.1016/0092-8674(80)90384-0. [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Kédinger C., Corden J., Brison O., Chambon P. Specific in vitro initiation of transcription on conalbumin and ovalbumin genes and comparison with adenovirus-2 early and late genes. Nature. 1980 Jun 5;285(5764):367–373. doi: 10.1038/285367a0. [DOI] [PubMed] [Google Scholar]
- Weinberg R. A., Penman S. Small molecular weight monodisperse nuclear RNA. J Mol Biol. 1968 Dec;38(3):289–304. doi: 10.1016/0022-2836(68)90387-2. [DOI] [PubMed] [Google Scholar]
- Weinmann R., Brendler T. G., Raskas H. J., Roeder R. G. Low molecular weight viral RNAs transcribed by RNA polymerase III during adenovirus 2 infection. Cell. 1976 Apr;7(4):557–566. doi: 10.1016/0092-8674(76)90206-3. [DOI] [PubMed] [Google Scholar]
- Weinmann R., Roeder R. G. Role of DNA-dependent RNA polymerase 3 in the transcription of the tRNA and 5S RNA genes. Proc Natl Acad Sci U S A. 1974 May;71(5):1790–1794. doi: 10.1073/pnas.71.5.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zain S., Gingeras T. R., Bullock P., Wong G., Gelinas R. E. Determination and analysis of adenovirus-2 DNA sequences which may include signals for late messenger RNA processing. J Mol Biol. 1979 Dec 5;135(2):413–433. doi: 10.1016/0022-2836(79)90444-3. [DOI] [PubMed] [Google Scholar]
- Zieve G. W. Two groups of small stable RNAs. Cell. 1981 Aug;25(2):296–297. doi: 10.1016/0092-8674(81)90046-5. [DOI] [PubMed] [Google Scholar]
- Zieve G., Penman S. Small RNA species of the HeLa cell: metabolism and subcellular localization. Cell. 1976 May;8(1):19–31. doi: 10.1016/0092-8674(76)90181-1. [DOI] [PubMed] [Google Scholar]