Abstract
Yeast Las17 protein is homologous to the Wiskott–Aldrich Syndrome protein, which is implicated in severe immunodeficiency. Las17p/Bee1p has been shown to be important for actin patch assembly and actin polymerization. Here we show that Las17p interacts with the Arp2/3 complex. LAS17 is an allele-specific multicopy suppressor of ARP2 and ARP3 mutations; overexpression restores both actin patch organization and endocytosis defects in ARP2 temperature-sensitive (ts) cells. Six of seven ARP2 ts mutants and at least one ARP3 ts mutant are synthetically lethal with las17Δ ts confirming functional interaction with the Arp2/3 complex. Further characterization of las17Δ cells showed that receptor-mediated internalization of α factor by the Ste2 receptor is severely defective. The polarity of normal bipolar bud site selection is lost. Las17-gfp remains localized in cortical patches in vivo independently of polymerized actin and is required for the polarized localization of Arp2/3 as well as actin. Coimmunoprecipitation of Arp2p with Las17p indicates that Las17p interacts directly with the complex. Two hybrid results also suggest that Las17p interacts with actin, verprolin, Rvs167p and several other proteins including Src homology 3 (SH3) domain proteins, suggesting that Las17p may integrate signals from different regulatory cascades destined for the Arp2/3p complex and the actin cytoskeleton.
INTRODUCTION
Arp2p and Arp3p are essential ubiquitous proteins of the actin-related family localized in actin-rich cortical structures (McCollum et al., 1996; Moreau et al., 1996; Mullins et al., 1997; Welch et al., 1997b; Winter et al., 1997). Analysis of yeast mutations shed light on their roles in the cortical cytoskeleton. Delocalization of actin patches and a slight accumulation of membranous vesicles resulted from a temperature-sensitive (ts) mutation in S. cerevisiae Arp2p (Moreau et al., 1996). This mutant was defective for dye uptake and for internalization of a membrane transporter, uracil permease (Moreau et al., 1997). A fission yeast cold-sensitive ARP3 mutation caused disorganization of actin patches and defects in polarized growth (McCollum et al., 1996). A ts mutation in Saccharomyces cerevisiae Arp3p impaired actin patch motility and caused accumulation of actin aggregates in the bud (Winter et al., 1997). Arp2p and Arp3p were first isolated by purification of a profilin-binding complex from Acanthamoeba, which contained five additional peptides, p40, p35, p19, p18, and p14 (Machesky et al., 1994). A homologous complex, purified from human platelets, localized at the leading edge of moving lamellipodia (Welch et al., 1997b). Purification of the Arp2/3 complex from S. cerevisiae (Winter et al., 1997) showed that it was homologous to the Acanthamoeba and human complexes with the exception that one subunit, p40 or Sop2p (Balasubramanian et al., 1996), did not copurify (Winter et al., 1997).
Structural modeling of Arp2p and Arp3p led to the hypothesis that Arp2p and Arp3p formed a heterodimer capable of nucleating actin polymerization at the pointed end of actin filaments (Kelleher et al., 1995), whereas electronic microscopy of purified Acanthamoeba Arp2/3 complex showed the complex bound to the sides of actin filaments in vitro (Mullins et al., 1997). Arp2/3 complex promotes actin polymerization at the surface of the intracellular pathogen Listeria monocytogenes by recruiting host proteins through a surface protein, ActA (Welch et al., 1997). The complex then nucleates actin filaments, generating propulsive force. However, Arp2/3 complex is also localized all along actin tails of moving bacteria and not just at actin polymerization sites on the Listeria cell surface (Welch et al., 1998). Recent evidence indicated that the Arp2/3 complex alone was a weak nucleator, whereas ActA alone was not. Their combined action had a dramatic synergistic effect on nucleation of actin filaments (Welch et al., 1998). Localization, nucleation, and cross-linking studies in Acanthamoeba have led to the proposition that the Arp2/3 complex promotes polymerization at the leading edge of membranes by attaching newly formed filaments to preexisting structures. Thus, as actin filaments grow at their membrane-proximal ends to drive motility, the bound Arp2/3 complex would be translocated away from the membrane (Mullins et al., 1998b). More recently, in addition to nucleating actin at the barbed end of filaments, the Arp2/3 complex was shown to have high-affinity pointed end capping activity to induce branched networks of filaments in vitro (Mullins et al., 1998a).
A novel yeast protein, Las17p/Bee1p, was shown to be a component of actin patches. Yeast cells deleted for the Wiskott–Aldrich Syndrome protein (WASP) homologue concentrated aberrant actin aggregates in the bud and were defective in actin patch assembly, budding, and cytokinesis (Li, 1997). Moreover, biochemical studies using permeabilized cells and rhodamine-labeled actin suggested that Las17p/Bee1p was required for actin polymerization and assembly in vitro (Lechler and Li, 1997). Las17p is proline-rich and shows homology to human WASP, mutations in which cause severe immunodeficiency (Amman and Hong, 1989). WAS patients show cytoskeletal abnormalities of lymphocytes and platelets (Molina et al., 1992), failure of B cells to respond to polysaccharide antigens, and defective chemotaxis in neutrophils (Ochs et al., 1980), suggesting that WASP is important in cytoskeletal reorganization necessary for the immunological response. WASP colocalized with polymerized actin in human cells and bound specifically to activated CDC42Hs (Apenström et al., 1996; Kolluri et al., 1996; Symons et al., 1996). A more ubiquitously expressed homologue, N-WASP, was shown to bind actin, calmodulin, and Ash/Grb2p (abundant Src homology/growth factor receptor-bound protein 2), an Src homology 2/Src homology 3 (SH3) domain protein implicated in signal transduction from membrane receptors to effectors of the cytoskeleton (Miki et al., 1996). Overexpression of either WASP or N-WASP in mammalian cells induced formation of actin clusters that colocalized with the overexpressed protein, indicating a role in actin polymerization (Miki et al., 1996; Symons et al., 1996). The vertebrate WASP family member Scar1/WAVE, like yeast Las17p, does not contain a recognizable GTPase binding site, but overexpression induced clustered actin filaments, demonstrating its involvement in actin reorganization (Miki et al., 1998). WASP and Scar1 have now been shown to interact with actin via their WASP homology 2 (WH2) domains and with the p21 subunit of the Arp2/3 complex via their C-terminal acidic domains. Overexpression of these C-terminal acidic fragments caused delocalization of the Arp2/3 complex with concomitant loss of actin spots and lamellipodia (Machesky and Insall, 1998).
We previously showed that in yeast, increased LAS17 expression suppressed arp2-1 growth thermosensitivity (Madania et al., 1997). Here we present genetic, cellular, and biochemical evidence that Las17p functions with the Arp2/3 complex and raise the possibility that Las17p may be part of signaling pathways directed to the Arp2/3 complex and actin.
MATERIALS AND METHODS
Plasmids, Strains, Gene Banks, and Genetic Manipulations
Plasmids and yeast strains used in this study are listed in Tables 1 and 2. Strains are derived from YPH and FY strains, which are S288C derivatives. Genomic DNA banks used were constructed in 2μ plasmids: Sau3A-derived fragments in pFL44 vector (from F. Lacroute), pGAD vector (from P. Bartel) (Fields and Song, 1989), multiple enzyme-derived fragments in pGAD424 (from P. James) (James et al., 1996), or mechanically derived fragments cloned in pACTII (from P. Legrain) (Fromont-Racine et al., 1997). The cDNA library used in two-hybrid screening was in pGAD-GH (from M. White). DNA manipulations were carried out by standard techniques (Sambrook et al., 1989). Yeast cell cultures and genetic manipulations were carried out essentially as described in by Guthrie and Fink (1991). Yeast cells were transformed by the LiAc method using single-stranded carrier DNA and DMSO (Schiestl and Gietz, 1989; Hill et al., 1991).
Table 1.
Plasmids used in this study
| Plasmid | Description |
|---|---|
| pIMW18-U | pUC18 containing ARP2 tagged with URA3 (this study) |
| pIMW300 | pUC18 containing ARP3 as an 1.85-kb BamH1 insert (this study) |
| pYCW204 | pUN60, CENIV, URA3, SUP11, ARP2 (Moreau et al., 1996) |
| pCMW306 | pUN60, CENIV, URA3, SUP11, ARP3, constructed by ligating the SalI–BamHI fragment of pIMW300 into the same sites of pUN60 (this study) |
| pCMW304 | pRS314 (Sikorski and Hieter, 1989), ARP3∶LEU2 (this study) |
| pYCW207 | pUN90, CENIV, HIS3, ARP2 (Moreau et al., 1996) |
| pYEW170 | pFL44, 2μD, URA3, LAS17 and flanking regions as a 4-kb BamHl insert (this study) |
| pAS2 | 2μD, TRP1, ADHpro-NLS-GAL4BD, HA, CYH2, bait vector (Durfee et al., 1993) |
| pAMW172 | pAS2, GAL4BD fused to full-length LAS17 (this study) |
| pDAB7 | pAS2 derived, CEN, GAL4BD fused to full-length ACT1 (Amberg et al., 1995) |
| pACT2 | 2μD, LEU2, ADHpro-NLS-GAL4AD, HA, prey vector (Durfee et al., 1993) |
| pAMW253 | pACT2, GAL4AD fused to intronless full length ARP2 (this study) |
| pAMW315 | pACT2, GAL4AD fused to full length ARP3 (this study) |
| pAMW173 | pACT2, GAL4AD fused to full length LAS17 (this study) |
| PGAD.GH | 2μD, LEU2, ADHpro-NLS-GAL4AD, prey vector (Chien et al., 1991) |
| pAMW200 | pGAD.GH, ARP2 (this study) |
| pAMW171 | PGAD.GH, LAS17 (2μ overexpression, this study) |
| pFA6a-kanMX4 | pFA6a, cassette for resistance to G418 (Wach et al., 1994) |
| pFA6a-HIS3MX6 | pFA6a, his3 complementing heterologous HIS5 ORF from S. pombe (Wach et al., 1997) |
| pFA6a GFPS65T-kanMX6 | Same as pFA6a-kanMX4, containing GFP (S65T) upstream of the kanMX6 module (Wach et al., 1997) |
Table 2.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| YPH499 | MATa, ade2-101, his3-Δ200, leu2-Δ1, lys2-801, trp1-Δ63, ura3-52 | P. Hieter |
| YPH500 | MATa , ade2-101, his3-Δ200, leu2-Δ1, lys2-801, trp1-Δ63, ura3-52 | P. Hieter |
| YPH501 | MATa, ade2-101, his3-Δ200, leu2-Δ1, lys2-801, trp1-Δ63, ura3-52 | P. Hieter |
| MATa, ade2-101, his3-Δ200, leu2-Δ1, lys2-801, trp1-Δ63, ura3-52 | ||
| YMW11 | MATa, arp2∷LEU 2, ade2-101, his3Δ200, leu2-Δ1, lys2-801, trp1-Δ63, ura3-52 (pYCW204) | Moreau et al. (1996) |
| YMW81 | MATa, arp2-1(H330L), ade2-101, his3-Δ200, leu2-Δ1, lys2-801, trp1-Δ63, ura3-52 | Moreau et al. (1996) |
| YMW201U | MATa, ARP2∶URA3, ade2-101, his3-Δ200, leu2-1, lys2-801, trp1-Δ63, ura3-52 | This study |
| YMW211U | MATa, arp2-1(H330L)∶URA3, ade2-101, his3-Δ200, leu2-Δ1, lys2-80, trp1-Δ63, ura3-52 | This study |
| YMW221U | MATa, arp2-2(G19D)∶URA3, ade2-101, his3-Δ200, leu2-Δ1, lys2-801, trp1-Δ63, ura3-52 | This study |
| YMW231U | MATa, arp2-3(E256K)∶URA3, ade2-101, his3-Δ200, leu2-Δ1, lys2-801, trp1-Δ63, ura3-52 | This study |
| YMW241U | MATa, arp2-4(L280P)∶URA3, ade2-101, his3-Δ200, leu2-Δ1, lys2-801, trp1-Δ63, ura3-52 | This study |
| YMW251U | MATa, arp2-5(L316S)∶URA3, ade2-101, his3-Δ200, leu2-Δ1, lys2-801, trp1-Δ63, ura3-52 | This study |
| YMW261U | MATa, arp2-6(G35S Y146N)∶URA3, ade2-101, his3-Δ200, leu2-Δ1, lys2-801, trp1-Δ63, ura3-52 | This study |
| YMW271U | MATa, arp2-7(F127S F203Y)∶URA3, ade2-101, his3-Δ200, leu2-Δ1, lys2-801, trp1-Δ63, ura3-52 | This study |
| YMW300T | MATa, arp3Δ∷TRP1, ade2-101, his3-Δ 200, leu2-Δ1, lys2-801, trp1-Δ63 ura3-52 | This study |
| MATa, ARP3, ade2-10, his3-Δ200, leu2-Δ1 lys2-801 trp1-Δ63 ura3-52 | ||
| YMW301T | MATa, arp3Δ∷TRP1 ade2-101 his3-Δ200, leu2-Δ1, lys2-801, trp1-Δ63, ura3-52 (+pCMW 306) | This study |
| YMW311L | MATa, ARP3∶LEU2, ade2-101, his3-Δ200, leu2-Δ1, lys2-801, trp1-Δ63, ura3-52 | This study |
| YMW321L | MATa, arp3-14 (P33L aa 48-108Δ)∶LEU2, ade2-101, his3-Δ200, leu2-Δ1, lys2-801, trp1-Δ63, ura3-52 | This study |
| YMW171K | MATa, las17Δ∷kanMX4, ade2-101, his3-Δ200, leu2-Δ1, lys2-801, trp1-Δ63, ura3-52 | This study |
| YMW172K | MATa, las17Δ∷kanMX4, ade2-101, his3-Δ200, leu2-Δ1, lys2-801, trp1-Δ63, ura3-52 | This study |
| YMW173K | MATa, las17Δ∷kanMX4, ade2-101, his3-Δ200, leu2-Δ1, lys2-801, trp1-Δ63, ura3-52 | This study |
| MATa, las17Δ∷kanMX4, ade2-101, his3-Δ200, leu2-Δ1, lys2-801, trp1-Δ63, ura3-52 | ||
| YMW175K | MATa, ura3-52, his3-Δ200, leu2-Δ1, arp2Δ∷LEU2, trp1-Δ63 LAS17-PROTA∶kanMX6 (pYEW248) | This study |
| RH4207 | MATa, las17Δ∷kanMX4, bar1∷LYS2, ade2-101, his3-Δ200, leu2-Δ1, trp1-Δ63, ura3-52 | This study |
| FY1679 | MATa, his3-Δ200, leu2-Δ1, trp1-Δ63, ura3-52, GAL2 | B. Dujon/F. Winston |
| MATa, HIS3, LEU2, TRP1, ura3-52, GAL2 | (EUROFAN strain) | |
| FDW23GK | MATa, ARP2-S65TGFP∶kanMX6, his3-Δ200, leu2-Δ1, trp1-Δ63, ura3-52, GAL2 | This study |
| MATa, ARP2-S65TGFP∶kanMX6, his3-Δ200, LEU2, TRP1, ura3-52, GAL2 | ||
| FDW24GK | MATa, ARP2, his3-Δ200, leu2-Δ1, trp1-Δ63, ura3-52 GAL2 | This study |
| MATa, ARP2-EGFP∶kanMX6, HIS3, LEU2, TRP1, ura3-52 GAL2 | ||
| FDW34GK | MATa, ARP3, his3-Δ200, leu2-Δ, 1 trp1-Δ63, ura3-52, GAL2 | This study |
| MATa, ARP3-EGFP∶kanMX6, HIS3, LEU2, TRP1, ura3-52, GAL2 | ||
| FMW17GK | MATa, LAS17-GFP∶kanMX4, his3-Δ200, leu2-Δ1, trp1-Δ63, ura3-52, GAL2 | This study |
| MATa, LAS17-GFP∶kanMX4, HIS3, LEU2, trp1-Δ63, ura3-52, GAL2 | ||
| FDW174H | MATa, las17Δ∷HISMX6, his3-Δ200, LEU2, TRP1, ura3-52, GAL2 | This study |
| MATa, las17Δ∷HISMX6, his3-Δ200, LEU2, TRP1, ura3-52, GAL2 | ||
| FDW180HK | MATa, las17Δ∷HISMX6, ARP2, his3-Δ200, LEU2, TRP1, ura3-52, GAL2 | This study |
| MATa, las17Δ∷HISMX6, ARP2-EGFP∶kanMX6, his3-Δ200, LEU2, TRP1, ura3-52, GAL2 | ||
| FDW190HK | MATa, las17Δ∷HISMX6, ARP3, his3-Δ200, LEU2, TRP1, ura3-52, GAL2 | This study |
| MATa, las17Δ∷HISMX6, ARP3-EGFP∶kanMX6, his3-Δ200, LEU2, TRP1, ura3-52, GAL2 | ||
| W303 | MATa, his3-Δ1, leu2-3,112, trp1-289, ura3-52 | P. Philippsen |
| MATa, his3-Δ1, leu2-3,112, trp1-289, ura3-52 | EUROFAN strain | |
| Y187 | MATa, gal4Δ, gal80Δ, his3, trp1-901, ade2-101, leu2-3,112, ura3-52, URA3∷GAL1(UAS)-lacZ, | Durfee et al. (1993) |
| transformed by the FRYL yeast DNA genomic library | James et al. (1996) | |
| Y190 | MATa, gal4Δ gal80Δ his3 trp-901 ade2-101 leu2-3,112 ura3-52 URA3∷GAL1(UAS)-lacZ | James et al. (1996) |
| LYS2∷GAL1(UAS)-HIS3 cyhr | ||
| CG1945 | MATa, gal4-542, gal80-538, his3, trp-901, ade2-101, leu2-3,112, ura3-52, URA3∷GAL4(3x17mers)CYC1(TATA)-lacZ,LYS2∷GAL1(UAS)-GAL1(TATA)-HIS3, cyhr | P. Legrain Froment-Racine et al. (1997) |
YMW strains and the RH strain are isogenic derivatives of YPH strains. Strain names beginning with F are isogenic FY1679 derivatives. Both of these series of strains are derivatives of S288C.
LAS17 Gene Deletion
Using PCR targeting with short flanking homology (Wach et al., 1994) the complete LAS17 coding region was deleted in three different diploid yeast strains: YPH501, FY1679, and W303 (Table 2). Oligonucleotides used to amplify the kanMX4 fragment on pFA6a-kanMX4 were 5′-AATTACAGTTCGTTACTTTAAGTGTTGATAG-GCGTGATTTAATGCTGCAGGTCGACGGATC-3′ and 5′-ACATA-TTTTCTATAACAGTAGTTTCATCTTTGTTTGCATTCCATCGAT-GAATTCGAGCTC-3′. Recombinants bearing the kanMX4 marker were selected on YPD plates containing G418 at 200 μg/ml. Correct integration of the kanMX4 cassette was verified by PCR analysis of genomic DNA. Haploid las17Δ strains (YMW17K1 and YMW17K2) were obtained by sporulation and dissection of heterozygous diploid disruptants; they appeared fully rescued at 37°C by plasmid-borne wild-type LAS17. The same methodology and oligonucleotides allowed us to create las17Δ::HIS3 strains using plasmid pFA6a-HIS3MX6 as template. FDW174H is the resulting las17Δ::HIS3 diploid strain.
Chromosomal Integration of Tagged ARP2 and ARP3 ts Alleles
Seven ARP2 (arp2-1 to arp2-7) ts mutations, initially obtained by PCR mutagenesis on plasmid pYCW207 (Moreau et al., 1996), were integrated at the ARP2 locus. The ARP2 gene was tagged with URA3 by cloning an XbaI–PvuII fragment containing the URA3 gene from pJJ244 (Jones and Prakash, 1990) blunt into the SspI site 130 bp downstream of the ARP2 stop codon in plasmid pBES18 (Moreau et al., 1996). This gave rise to pIMW18-U. Mutant alleles (arp2-1 to arp2-7) were exised as 1.38-kb SacI–SspI fragments from the mutant bank in pYCW207, purified, and ligated into the SacI–SalI (blunted) sites of pBMW18-U. Wild-type and resulting mutant plasmids were digested with SacI–KpnI, and the 3.4-kb fragments containing the ARP2 ts alleles with the downstream URA3 marker were purified, transformed into YPH499, and selected on SC − Ura. Ura+ clones were tested for thermosensitive growth then for correct integration of the URA3 cassette at the ARP2 locus by PCR analysis of genomic DNA (our unpublished results). Haploid-tagged wild-type and mutants were mated with YPH500, and diploids were sporulated. In all 20 tetrads dissected for each cross, two ts URA3 and two non-ts ura3 spores were obtained (Table 2, YMW201U and YMW211U–YMW271U). In a similar manner, wild-type ARP3 and the arp3-14 mutant allele were tagged with the LEU2 gene (Madania, 1998).
Construction of GFP Fusions with LAS17, ARP2, and ARP3
Stop codons of the LAS17, ARP2, and ARP3 genes were replaced with the coding sequence of green fluorescent protein (GFP) by short flanking homology PCR targeting using the kanMX6 marker (Wach et al., 1994). For LAS17:GFP, pFA6a-GFPS65TkanMX6 was used as template to amplify a 2.5-kb PCR fragment with oligonucleotides 5′-TAAAGTGGGAGCTCATGACGATATGGACAATGG-TGATGATTGGGCAGCGAGTAAAGGAGAAGAACTTT-3′ and 5′-ACATATTTTCTATAACAGTAGTTTCATCTTTGTTTGCATTCCA -TCGATGAATTCGAGCTC-3′ as primers. This fragment was transformed into diploid FY1679, and G418-resistant recombinants were verified for correct integration by PCR across both borders. Sporulation and dissection of a heterozygous G418-resistant diploid revealed a 2:2 segregation of the kanMX6 marker, which coincided with GFP fluorescence. Strain FMW173GK (homozygous for LAS17:GFP) was constructed by mating a and α spores.
For GFP fusion with ARP2, the pFA6a-GFPS65TkanMX6 plasmid was modified to add a 5× glycine-alanine linker following the ORF and then used as template to amplify a 2.5-kb PCR fragment with oligonucleotides 5′-TGGCAAGAAAGCGGGCCATCTGCAATGA-CTAAATTTGGTCCAAGAGGATCCGGAGCAGGTGCT-3′ and 5′-TCCATTTCCATTTTTCATTCGTAGATTTAAACTTTTTTATTAAT-TCTTTTGTCGATGAATTCGAGCTCGTTT-3′ as primers. A ARP2:GFP haploid strain was constructed and verified as above, and then α spores were mated to make strain FDW23GK. Growth of these strains was slow and temperature sensitive. To obtain ARP2 strains with these fusion proteins that grew better, the GFP of modified pFA6a-GFPS65TkanMX6 plasmid was converted to yEGFP (Cormack et al., 1997) and integrated into one allele of the diploid las17Δ strain, FDW174H, to create strain FDW180HK.
Similarly, to make a functional GFP fusion with ARP3, the stop codon of one allele of the ARP3 gene was replaced by yEGFP using oligonucleotides 5-TATGGTCCGGAAATTGTTAGAAATTTCAG-CCTTTTCAACATGGTTGGATCCGGAGCAGGTGCT-3′ (ARP3 in frame with GAGAGAGAGA:EGFP) and 5′-AGGGGCGTTTCAGTTATTTGCATTTGTTTTTGTACCTTTTTCCTTATAAGATCGATG -AATTCGAGCTGCTTT-3′ as primers, and the resulting fragment was transformed into strain FY1679 to create strain FDW34GK and into FDW174H to create strain FDW190HK.
Cell Fixation, Phalloidin and Antibody Staining, and GFP Observation
Early log phase cells were fixed and stained as previously described (Pringle et al., 1991), except that incubation with phalloidin was for 1 h with shaking at 4°C in the dark using 1 μM TRITC-labeled phalloidin (Sigma, St. Louis, MO) or 1 μM Alexa-594 phalloidin (Molecular Probes, Eugene, OR). Fixed cells were sometimes kept at 4°C for a maximum of 3 d before staining. For GFP observation, fixation time was reduced to 15 min. For in vivo observations of GFP fusion proteins, cells were grown in liquid YPD at 25°C to early log phase and then harvested, resuspended in 1 M sorbitol, and immediately observated using a GFP bandpass filter (excitation, 460–500; dichroic mirror, 505; emission, 510–560). Antibody staining of spheroplasts was as previously described (Moreau et al., 1996) with C4 monoclonal mouse immunoglobuliln G1 (IgG1) anti-actin antibody (Boehringer Mannheim, Indianapolis, IN ) and polyclonal rabbit anti-GFP antibody (a gift from K. Sawin, London, United Kingdom). Preparations were analyzed and photographed using an Optiphot-2 microscope (Nikon, Melville, NY) equipped with fluorescent optics. Photographs were taken using 400ASA film and then scanned (300 dots per inch final resolution) or direct acquisitions were made with a Photonics (Milpitas, CA) Science charge-coupled device Coolview camera equipped with the Gel Grab 2.02 software program.
Latrunculin Treatment of GFP-labeled Cells
After growth to midlog phase at 25°C in liquid YPD, cells were concentrated to 108 cells/ml. Latrunculin-A (Molecular Probes) in DMSO (from a 10 mM stock to give 100 μM final concentration) or an equivalent volume of DMSO was added to a 100 μl aliquot for each condition tested. Cells were fixed by adding 10 vol of YPD containing 3.7% formaldehyde and incubating with gentle shaking for 15 min.
Lucifer Yellow and FM4-64 Staining and α-Factor Uptake
Lucifer yellow (LY)-carbohydrazine (Fluka, Buchs, Switzerland) uptake, an indication of fluid phase endocytosis, was done as described by Dulic et al. (1991). Staining with the lipophilic dye FM4-64 [N-(3-triethylammoniumpropyl)-4-(p-diethylaminophenylhexatrienyl) pyridinium dibromide] to follow uptake into membranes was as described by Vida and Emr (1995). 35S-Alpha factor uptake assays were performed as described (Dulic et al., 1991). All strains were preincubated at 37°C for 15 min before the addition of alpha factor. The samples were processed as described above. All uptake assays were performed twice.
Immunoprecipitation of Las17p and Arp2p
To tag Las17p with protein A, the stop codon of the LAS17 gene was replaced by the coding frame of the IgG binding domain of protein A using the kanMX6 selection marker as a PCR-synthesized fragment with short flanking homology on either side of the LAS17 stop codon. pYM7 was the template source of protein A (Knop et al., 1999), amplified with the same primers as for the LAS17:GFP fusion. The resulting 2.1-kb PCR fragment was transformed into the diploid strain FY1679, which was sporulated. G418-resistant progeny were verified for correct tag insertion by PCR. One of these haploids was crossed with a haploid arp2Δ rescued by hemagglutinin (HA)-tagged Arp2p (YMW15 + pYEW250) (Moreau et al., 1996), and a recombinant haploid derivative was selected (strain YMW175K).
Whole-cell extracts were made by the liquid nitrogen grinding in 50 mM HEPES, pH 7.5, 100 mM KCl, 3 mM MgCl2, 1 mM EGTA, 0.5% Triton X-100 buffer (Li, 1997) and resuspended at 10 mg/ml after clarification by 30 min of centrifugation at 20,000 × g. Eighty microliters of Las17-prot A extract and nontagged extract were incubated for 1 h at 4°C with 40 μl of IgG-Sepharose (6 Fast Flow; Pharmacia, Piscataway, NJ) suspension. Immunoprecipitation and washes were with starting buffer. Eluted proteins were precipitated with 10% trichloroacteic acid and resuspended in loading buffer. Samples were separated by 10% PAGE and blotted to nitrocellulose. Blots were incubated with appropriate primary and secondary antibodies and then developped with a Super Signal (Pierce, Rockford, IL) chemiluminescent kit for HRP.
IgG to reveal Las17-prot A was purified from nonimmunized rabbit serum by passage over a protein A-Sepharose column (Pharmacia CL-4B). Production and characterization of anti-Arp2p antibodies has been previously described (Moreau et al., 1996).
Construction of GAL4 Fusion Plasmids for Two-Hybrid Interactions
LAS17 was amplified with oligonucleotides 5′-GGCGTGATTTACCATGGGACTCC-3′ (with an NcoI site at the ATG of LAS17) and 5′-CATCTTCTCGAGCATTCCATTACCAA-3′ (with an XhoI site just after the stop codon) using pAMW171 as template. A 1.9-kb NcoI–XhoI PCR fragment was cloned into the NcoI–SalI sites of pAS2 in fusion with GAL4 binding domain (GAL4BD) to give pBMW172 and into the NcoI–XhoI sites of pACTII in fusion with GAL4 activation domain (GAL4AD) to give pAMW173. Both pBMW172 and pAMW173 were able to rescue YMW171K (las17Δ) at 25 and 37°C (our unpublished data). All GAL4 fusions constructed were systematically controlled by sequencing the fusion site. Two-hybrid GAL4AD constructions coding for wild-type Cdc42p and its activated form (Cdc42pG12V) were kindly provided by E. Leberer (Leberer et al., 1997) and by the Pringle laboratory (Cvrckova et al., 1995).
Two-Hybrid Screens with LAS17 as Bait
cDNA Screen.
Y190 strain (Durfee et al., 1993) containing pBMW172 (GAL4BD-LAS17) was transformed with a yeast cDNA library in pGAD-GH (kindly provided by Michael White, University of Texas Southwestern Medical School, Dallas, TX) and plated on SC − Trp, Leu, and His supplemented with 35 mM 3-aminotriazole (3-AT). After incubation for 1 wk at 30°C, 309 colonies among 200,000 initial transformants were patched onto SC − Trp, Leu, and His 35 mM 3-AT plates and allowed to grow at 30°C for 2 d. These clones were then tested in a 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) filter assay, and clones that turned blue were further analyzed. Plasmids were rescued and reintroduced into Y190 and Y190 containing pBMW172 and tested in the X-Gal colony filter assay. One hundred sixty plasmids giving a signal with pBMW172 were sequenced with oligonucleotides 5′-GGAATCACTACAGGGAT-G-3′ to control in-frame fusions and with 5′-GAAGTGAACTTGCGGGG-3′ to determine the size of the fused protein. After elimination of redundant fusions, 23 different positive clones representing 10 different genes were identified and tested in β-galactosidase liquid culture assays.
Genomic DNA Screen.
Yeast strain Y187 was transformed by standard procedures with DNA amplified from an aliquot of the FRYL library (Fromont-Racine et al., 1997). Ten million yeast transformants were collected and pooled, and aliquots were stored at −80°C. To screen by mating, a tube containing 107 transformants was equilibrated in fresh YPD then mixed with 2 × 107 log phase CG1945 cells previously transformed with plasmid pBMW172 (GAL4BD-LAS17) and incubated for 6 h. Cells representing 1.15 × 106 diploids were plated on SC − Trp, Leu, and His supplemented with 35 mM 3-AT and incubated at 30°C for 7 d. The clones thus obtained were assayed as described above then recycled, and plasmids were extracted and sequenced.
X-Gal Colony Filter Assay and β-Galactosidase Assay
β-Galactosidase activity of simple or double transformants of Y190 was assessed on nitrocellulose filters as described by Breeden and Nasmyth (1985). To quantitate β-galactosidase activity, freshly streaked Y190 double transformants were inoculated into 2.5 ml of liquid SC − Trp and Leu and allowed to grow overnight to OD600 nm of ∼1. β-Galactosidase activity was measured and expressed in Miller units (Kandels-Lewis and Seraphin, 1993).
RESULTS
LAS17 Is an Allele-specific Multicopy Suppressor of ARP2 ts Mutations
To identify proteins that interact with Arp2p, we screened yeast multicopy genomic libraries for clones whose expression was able to rescue the temperature sensitivity of the arp2-1 mutation in strain YMW81. In a preliminary screen using the Lacroute genomic bank we obtained several clones, the smallest of which contained a plasmid, pYEW170, with an insert of 4 kb covering several genes. Using the Fields and Bartel YL1/2/3 genomic bank, 24 plasmids were able to support growth at 37°C. Besides the ARP2 gene, seven groups of inserts were identified. Sequencing of insert borders revealed that all seven clones contained a full-length LAS17 coding sequence. Because the inserts of the two shortest clones did not overlap any adjacent coding regions, we concluded that LAS17 was responsible for suppression.
Suppression of previously isolated ts mutations arp2-1 to arp2-7 (Moreau et al., 1996) was tested to find out whether LAS17 suppression was allele specific. Sequence changes in the ARP2 gene were determined and are shown in Table 2. WT and mutant fragments tagged with the URA3 gene were integrated into strain YPH499 at the ARP2 locus (see MATERIALS AND METHODS), giving rise to strains YMW211U to YMW271U. We have previously shown that the arp2-1 ts mutation causes delocalization of polarized cortical actin patches at 37°C (Moreau et al., 1996). When strains YMW221U to YMW271U were stained with phalloidin, actin distribution appeared normal in most cells of arp2-2, arp2-4, arp2-5, and arp2-7 at 25°C, whereas arp2-3 and arp2-6 cells showed loss of actin organization at this temperature (Figure 1A). All mutants had lost the polarized distribution of actin patches after 2 h at the restrictive temperature. Actin clumps or aggregates in the bud or at the mother–daughter neck and an enlarged bud neck were observed in budding cells of each of the ts mutants. Surprisingly, none of these mutant actin phenotypes was qualitatively different from those observed for the original arp2-1 mutant. A similar ts phenotype also resulted from fusion of green fluorescent protein to the carboxyl terminus of the ARP2 coding sequence (see MATERIALS AND METHODS). Examination of the ARP2-GFP strain did not reveal any Arp2-gfp fluorescence in the many phalloidin-staining actin aggregates present in buds or in mother–bud necks, whereas both actin and Arp2p were present in the remaining nonpolarized patches (Figure 1B).
Figure 1.
Actin phenotypes of ARP2 ts mutants. (A) Phalloidin staining of wild-type YMW201U(a) and mutant YMW221U, YMW231U, YMW241U, YMW261U, and YMW271U (a) strains containing ARP2 point mutations. Cells were grown to early log phase in complete medium at 25°C. Part of the culture was shifted to 37°C for 2 h. (B) Strain FDW23GK (a/α) containing an ARP2:GFP C-terminal fusion. Cells were treated as in A and then fixed and stained with TRITC-phalloidin as described in MATERIALS AND METHODS. The left panels were photographed with a GFP bandpass filter, and the right panels were photographed with a rhodamine filter. Bar, 5 μm.
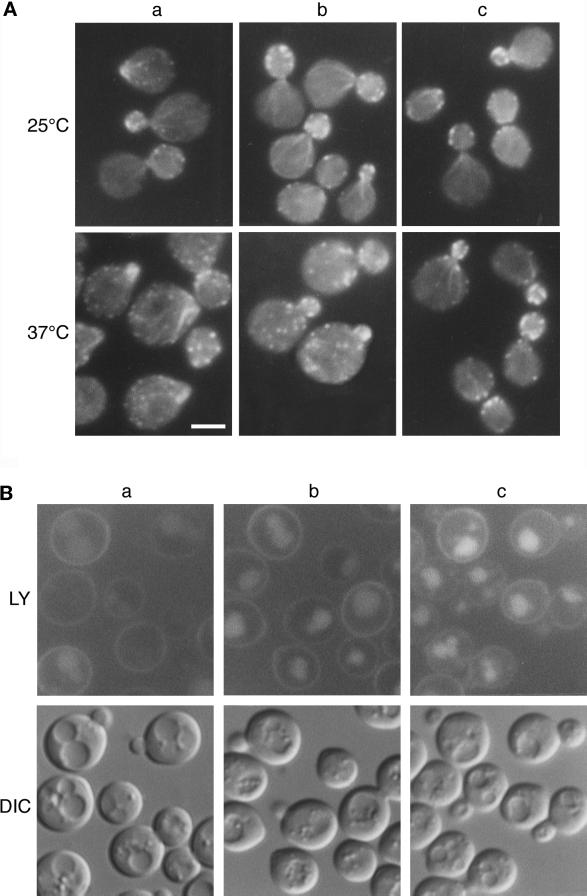
These ARP2 mutants were used to test allele specificity of LAS17 suppression. When ARP2 strains were transformed by a multicopy LAS17 plasmid, pAMW171, the arp2-2 mutant as well as the arp2-1 mutant initially used in the selection were rescued at 37°C. The thermosensitivity of the other mutants was not relieved (Figure 2). This clearly shows that suppression is specific to certain point mutations and suggests that a specific interaction could be involved. Major mutant phenotypes such as disorganization of the actin cytoskeleton and deficient endocytosis were examined in LAS17-suppressed strains YMW211U, YMW221U, and YMW241U. LAS17 overexpression suppressed actin cytoskeleton defects in the arp2-1 mutant and partially suppressed defects of the arp2-2 mutant at 37°C, in good correspondence with growth recovery. Figure 3A shows that even partial suppression in the arp2-2 mutant is clearly visible; the majority of arp2-2 ts cells containing overexpressed LAS17 no longer showed aberrant clumped actin in the bud at 37°C (Figure 3A, b, lower panel). Actin patches were less concentrated in the buds than in rescued cells (Figure 3A, c, lower panel). Suppression of endocytosis as judged by the ability of cells to take up and deliver the fluorescent neutral dye Lucifer yellow-carbohydrazine to the vacuole by fluid phase endocytosis (Riezman, 1985) followed the same pattern of allele specificity as growth and actin defects. Even at permissive growth temperature after extended times (60 min here vs. 10 min normally), uptake was inhibited in the arp2-2 mutant (Figure 3B, a, compared with the rescued mutant, c). The LAS17 overexpression allowed LY uptake and accumulation in the vacuole in the arp2-2 mutant (Figure 3B, b) although slightly less efficiently than for the arp2-1 allele (our unpublished data). Investigation of the arp2-4 allele revealed that the mutant was also endocytosis deficient but that LAS17 overexpression did not rescue this deficiency.
Figure 2.
LAS17 overexpression suppresses certain ARP2 ts mutants. Seven ARP2 ts strains (arp2-1 to 2-7/YMW211U–YMW271U) and wild-type (YMW201U) described in Table 2 were transformed with pGAD (vector) (A), pAMW171 (LAS17 in pGAD) (B), and pAMW200 (ARP2 in pGAD) (C). Transformants were streaked on SC − Ura and Leu plates and incubated at 37°C for 4 d. Note that the arp2-5 ts mutant grows at 37°C on minimal medium but is more ts on complete medium.
Figure 3.
Suppression of arp2-2 actin and endocytosis phenotypes by LAS17. Strain YMW221U (bearing the arp2-2:URA3 ts allele) was transformed with vector (a), with pAMW171 (LAS17 2μ) (b), and with pAMW200 (ARP2 in pGAD) (c). (A) Transformants were grown at 25°C in liquid YPD to log phase and shifted to 37°C for 2 h. Cells were fixed, stained with TRITC-phalloidin, and photographed as described in MATERIALS AND METHODS. Bar, 5 μm. (B) Transformants were grown in liquid YPD at 25°C to log phase and incubated for 1 h with LY as described in MATERIALS AND METHODS. Cells were photographed using an FITC filter (LY) or Nomarski optics (differential interference contrast [DIC]).
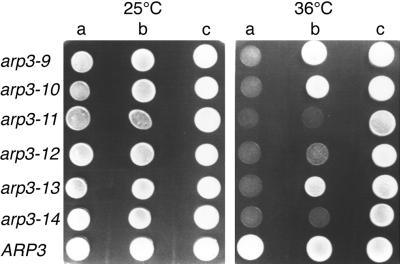
LAS17 Overexpression Can Also Rescue Conditional Mutations in the ARP3 Gene
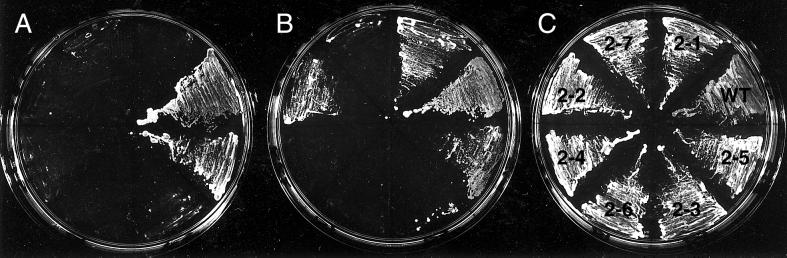
Because Arp2p and Arp3p are part of the same macromolecular complex (Machesky et al., 1994; Welch et al., 1997b; Winter et al., 1997), we questioned whether LAS17 overexpression would also suppress the phenotype of ARP3 conditional mutations. We have isolated recessive ARP3 ts alleles on plasmids in an arp3Δ strain (Madania, 1998). The capacity of LAS17 overexpression to rescue growth at nonpermissive temperatures for six different mutants, arp3-9 to arp3-14, was tested. Only one ts mutant, arp3-9, was well suppressed at 37°C. At 36°C LAS17 overexpression allowed good growth of arp3-9, arp3-10, and arp3-13 mutants and some growth of arp3-12 mutant, whereas growth of arp3-11 and arp3-14 mutants was not rescued (Figure 4). Thus, LAS17 is also an allele-specific suppressor of ARP3 mutants indicating that Las17p interaction with the Arp2/3 complex is important for at least one essential function.
Figure 4.
LAS17 allele-specific suppression of ARP3 ts mutants. The arp3Δ (YMW301T) strain rescued by five different ARP3 ts alleles (arp3-9 to arp3-13 and ARP3) and strain YMW321L (bearing the arp3-14: LEU2 allele) were transformed by vector (a), pYEW170 (LAS17, 2μ) (b), or pCMW304 rescue plasmid (ARP3:LEU2) (c). Freshly cultured transformants were spotted onto YNB − Leu and Ura plates, which were incubated at 25°C for 5 d or at 36°C for 3 d before being photographed.
Synthetic Enhancement of LAS17 Deletion with ARP2 and ARP3 Mutants
To gain further information on Las17p interactions with Arp2p and Arp3p, we looked for synthetic enhancement between las17Δ and ARP2 or ARP3 mutants. The las17Δ strain YMW172K was crossed with seven URA3-tagged ARP2 and one LEU2-tagged ARP3 integrated mutants (see MATERIALS AND METHODS). Viable haploid spores after meiosis were tested for G418 resistance (kanMX4 marker for las17Δ) and for uracil and leucine prototrophy (Table 3). As demonstrated by the absence of viable double mutants, six of seven different ARP2 ts mutants, arp2-1, arp2-2, arp2-3, arp2-4, arp2-5, and arp2-6, are lethal in the absence of LAS17. Analysis of a cross between strain YMW321L bearing arp3-14:LEU2 and strain YMW171K bearing las17Δ revealed no viable kanR Leu+ spores (Table 3). These synthetic lethality results between las17Δ and ARP2 and ARP3 ts mutants confirmed the importance of Las17p in the vital functions of Arp2p and Arp3p, prompting us to further investigate the cellular functions and interactions of Las17p.
Table 3.
Sythetic lethality of las17Δ with ARP2 and ARP3 mutant alleles
| Cross | No. tetrads | No. spores Ura3+kanR | No. spores Leu2+kanR | No. viable spores | No. dead spores |
|---|---|---|---|---|---|
| las17Δ × arp2-1 | 35 | 28 | 0 | 28 | |
| las17Δ × arp2-2 | 18 | 15 | 0 | 15 | |
| las17Δ × arp2-3 | 17 | 19 | 0 | 19 | |
| las17Δ × arp2-4 | 17 | 16 | 0 | 16 | |
| las17Δ × arp2-5 | 17 | 15 | 0 | 15 | |
| las17Δ × arp2-6 | 15 | 12 | 0 | 12 | |
| las17Δ × arp2-7 | 15 | 14 | 14 | 0 | |
| las17Δ × ARP2 | 17 | 13 | 12 | 1 | |
| las17Δ × arp3-14 | 44 | 37 | 0 | 37 | |
| las17Δ × ARP3 | 45 | 53 | 48 | 5 |
Strain YMW172K (MATαlas17 ) was crossed with strains YMW201U (ARP2∶URA3) and YMW211U to YMW271U (arp2ts∶URA3). Strain YMW172K (MATα las17 Δ∷kanMX4) was crossed with strains YMW311L (MATa ARP3∶LEU2) and YMW321L (MATa arp3-14∶LEU2). Diploids were sporulated, tetrads were dissected, and viable spores were tested for G418 resistance and for growth on SC − Ura at 25°C. The genotype of dead spores was inferred from that of viable spores in each tetrad tested. For each cross the number of tetrads was analyzed, and the number of double mutants and the number of viable and dead spores among these double mutants is indicated.
Phenotypes of las17Δ Mutant Cells
Deletion of LAS17 in the S288C strain background has been reported to cause temperature-sensitive growth, budding, and cytokinesis defects with aberrant actin bundles and lack of patches in the buds (Li, 1997). las17Δ cells showed misshapen unpinched bud necks and actin cortical patch defects as described (Li, 1997), except that actin-containing patches were visible in both the bud and the mother cell, as has been since reported (Karpova et al., 1998). We found that deletion of LAS17 results in lethality or temperature sensitivity, depending on the strain background. The phenotype was more severe in the W303 background than in the YPH or FY1679 background. Testing of las17Δ ts cells in the presence of osmoregulators showed that they were acutely osmosensitive. Growth was completely inhibited at 25°C with 0.30 M NaCl or 1.8 M sorbitol added to complete medium.
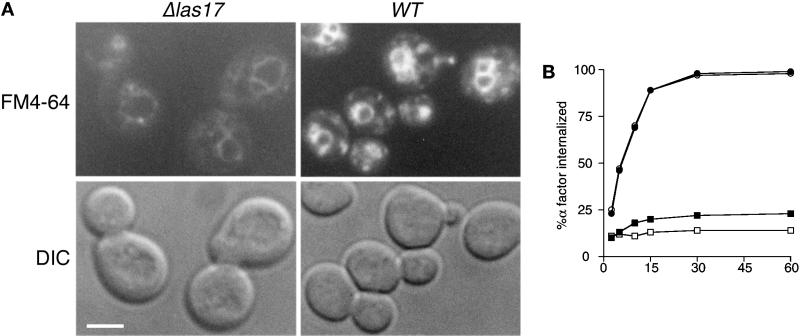
Because LAS17 overexpression suppressed defective endocytosis in certain ARP2 and ARP3 mutants, we also investigated endocytosis in las17Δ cells using a variety of methods. Three different reporter systems were used to reveal different aspects of endocytosis; LY for fluid phase endocytosis, the lipophilic vital stain FM4-64 to follow membrane internalization (Vida and Emr, 1995), and α-factor pheromone uptake to follow ligand-dependent receptor-mediated (Ste2p) endocytosis (Singer and Riezman, 1990; Riezman, 1993). Naqvi et al. (1998) have recently reported deficient LY endocytosis by las17Δ cells. Our independent results showed severe deficiency at the permisive growth temperature with no visible accumulation in the vacuole. We also examined strain YMW171K after incubation with FM4-64 to follow the intracellular fate of the dye. Very little dye was taken up by mutant relative to wild-type cells (Figure 5A). Nevertheless, trafficking through membrane compartments without accumulation in intermediary (endosomal) structures in addition to accumulation of weak fluorescence in membranes around the vacuole suggests that the dye progressed to its normal destination. Thus, the pathway from the plasma membrane to the vacuole appears to function normally.
Figure 5.
Endocytosis defects in las17Δ cells. (A) FM4-64 uptake by las17Δ cells. YMW171K (las17Δ) and YPH499 (WT) cells were grown to log phase in liquid YPD at 25°C and then concentrated and incubated for 15 min in 20 μM FM4-64 followed by a 45-min chase in fresh YPD. Cells were photographed using an FITC filter (top panel) or DIC optics (bottom panel). Bar, 5 μm. (B) Alpha factor internalization by las17Δ cells. las17Δ strain RH 4207 and its equivalent wild type were preincubated at 37°C for 15 min before the addition of 35S-alpha factor. The samples were processed as described by Dulic et al. (1991). Results are from one of two independent experiments, which gave nearly identical results. ●, WT, 24°C; ▪, las17Δ, 24°C; ○, WT, 37°C; □, las17Δ, 37°C.
Pheromone recognition and receptor internalization are the first steps in receptor-mediated endocytosis. The ability of cells to bind and internalize α-factor is constant throughout the cell cycle, allowing measurement of ligand-dependent internalization of the Ste2p receptor in growing cells (Zanolari and Riezman, 1991). To increase sensitivity to α-factor, strain YMW171K and its parent were first disrupted for the BAR1 gene (MacKay et al., 1988). Radioactive uptake of biosynthetically labeled α-factor across the plasma membrane was then quantified. Figure 5B shows that in cells grown at 24°C, tested before and after shift up to 37°C, α-factor binds to las17Δ cells. However, receptor-mediated endocytosis is severely deficient at 24°C, and uptake is indistinguishable from background at 37°C. Taken together these results allow us to conclude that Las17p is essential for the internalization step of endocytosis.
Las17p Is Also a Determinant for Polarity Development
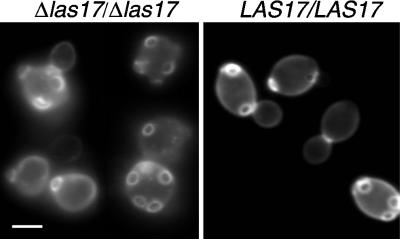
The actin cytoskeleton is required for the diploid-specific bipolar budding pattern normally seen in wild-type cells, and mutations in a number of cortical patch components affect this pattern (Rodriguez and Paterson, 1990; Sivadon et al., 1995; Vaduva et al., 1997; Yang et al., 1997). When the bipolar budding pattern was examined, most las17Δ diploid cells had randomly located bud scars, whereas wild-type cells showed a normal bipolar budding pattern (Figure 6). Thus, the choice of the bud site, an initial step in the determination of a growth polarity, appears to be the primary defect rather than bud emergence or growth.
Figure 6.
Bipolar budding defects in las17Δ/las17Δ cells. Strain FMW173K was grown overnight in YPD at 25°C. Cells were fixed and stained with calcofluor, washed, and observed in the microscope with a UV filter. Bar, 5 μm.
Las17-gfp Localizes to Cortical Patches Independently of Polymerized Actin
Las17p has been shown to coprecipitate with G-actin and is important for in vitro reconstitution of cortical actin assembly sites (Lechler and Li, 1997; Li, 1997). To visualize Las17p in living cells, an in situ fusion with GFP was constructed to replace LAS17 (see MATERIALS AND METHODS). LAS17:GFP spores grew well at all temperatures, confirming functionality. GFP fluorescence in strain FMW17GK localized in cortical patch structures concentrated at sites of bud emergence, in small buds and at the bud neck before cytokinesis. When LAS17:GFP cells were fixed and stained with phalloidin, actin and Las17-gfp patches colocalized (control cells; Figure 7). This in vivo colocalization confirms that established using an overexpressed Myc-tagged Las17/Bee1p (Li, 1997). We then examined the consequences of disrupting the (polymerized) actin cytoskeleton with latrunculin-A on Las17p localization. The drug latrunculin-A binds G-actin (Couéet al., 1987) and has been shown to rapidly disrupt the yeast actin cytoskeleton by inhibiting actin filament assembly (Ayscough et al., 1997). FMW17GK cells expressing Las17-gfp were treated for 5, 15, or 30 min with 100 μM latrunculin-A and then briefly fixed, washed, and stained with phalloidin. Actin filaments were no longer detectable by the 5 min time point, whereas Las17-gfp remained localized in cortical patch-like structures at presumtive bud sites, in small buds, and at the mother–daughter bud neck of dividing cells. This localization remained up to 30 min after latrunculin-A treatment (Figure 7A), although cells were slightly larger and more rounded than at the 5-min time point, indicating anisotropic cell growth. Las17p localization is thus maintained in the absence of actin cytoskeletal structures needed for polarized growth. Because Las17p might bind actin monomers or dimers not revealed by phalloidin staining, we also examined latrunculin-A-treated cells using anti-actin antibodies. After 30 min of latrunculin-A treatment, control cells (Figure 7B, upper panels) showed normal polarized distribution of both actin and Las17-gfp. However, no concentration of actin was detected in patch structures, whereas anti-GFP antibodies clearly revealed Las17-gfp concentrated in polarized patches (Figure 7B, lower panels) in the majority of cells.
Figure 7.
Functional Las17-gfp remains localized independently of actin. (A) Localization of Las17p at 25°C after treatment of cells with latrunculin-A. Strain YMW173G homozygous for LAS17:GFP was grown in liquid YPD to log phase at 25°C and incubated with latrunculin-A (100 μM final concentration) in DMSO or an equivalent volume of DMSO. Images show cells fixed after 30 min exposure to latrunculin-A, which were briefly fixed and then stained with Alexa-phalloidin. Bar, 5 μm. (B) Cells were grown as in A, fixed for 1 h, and then digested with zymolyase to make spheroplasts. Spheroplasts were labeled with rabbit anti-GFP IgG (left panels) and mouse monoclonal anti-actin IgG (right panels), which were revealed by goat anti-rabbit FITC and goat anti-mouse Cyanine 3 secondary antibodies using appropriate filters. No overlap between filter channels was detected in control exposures.
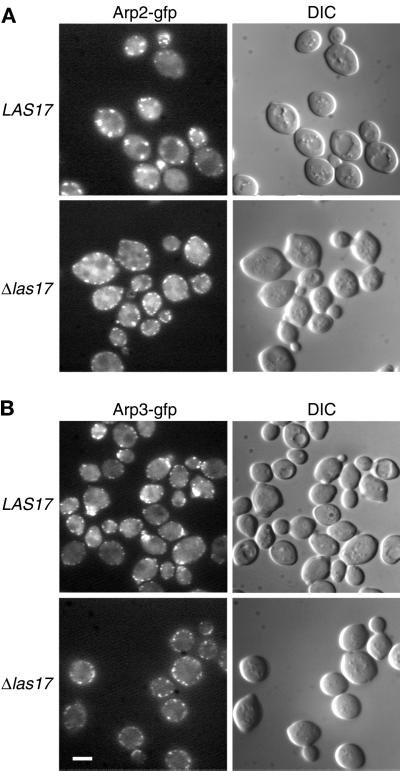
Las17p Is Necessary for the Polarized Localization of Arp2p and Arp3p
We then addressed the question of whether Las17p was necessary for polarized localization of Arp2p and Arp3p. Because Arp2-gfp and Arp3-gfp fusion proteins as unique source of Arp caused ts growth in haploids (our unpublished observations), we tagged one allele in a diploid strain to render these fusion proteins functional. ARP2:EGFP and ARP:EGFP were integrated on one allele of a las17Δ/las17Δ strain (FDW174H) giving rise to strains FDW180HK and FDW190HK, respectively. Diploid strains were no longer ts, and both Arp2-egfp (Figure 8A, strain FDW24GK) and Arp3-egfp (Figure 8B, strain FDW34GK) were localized in a polarized manner. In the absence of Las17p, Arp2-egfp and Arp3-egfp patches were clearly visible at the cortex (Figure 8, A and B, lower panels) but were not organized in a polarized manner. This depolarization of Arp2p patches was also observed (using Arp2p antibody) after latrunculin-A treatment (Ayscough et al., 1997). Loss of patch organization indicates that Las17p is a necessary polarity determinant for the localization of the Arp2/3 complex as well as for actin patches as shown above.
Figure 8.
Loss of Arp2p and Arp3p polarity in las17Δ strains. (A) Localization of Arp2-gfp in wild-type and las17Δ/las17Δ cells. One allele of ARP2 was replaced by an ARP2:EGFP (Cormack et al., 1997) allele in FY1679 (WT) and in strain YMW173K (las17Δ/las17Δ) as described in MATERIALS AND METHODS. Cells were grown at 25°C in YPD, harvested, resuspended in 1 M sorbitol, and immediately put on slides for microscopic examination. Images were recorded using a GFP filter. (B) Localization of Arp3-gfp in wild-type and las17Δ/las17Δ cells. Experimental conditions were as in A. Bar, 5 μm.
Does Las17p Interact Physically with the Arp2/3 Complex ?
Because LAS17 showed genetic and functional interactions with ARP2 and ARP3, we asked whether Las17p could interact directly with Arp2p and/or Arp3p using the two-hybrid system. Constructions are described in MATERIALS AND METHODS. Las17p bait showed activation with actin as expected, but significant Las17p interaction was not detected with Arp2p or Arp3p fusions (Table 4). In light of specific genetic interactions between Las17p and Arp2p and Arp3p, we pursued the possibility of direct interaction between Las17p and the Arp2/3 complex by immunoprecipitation of Las17p from crude extracts. The LAS17 gene was tagged in situ with a protein A-coding DNA fragment as described in MATERIALS AND METHODS. This strain was then crossed to an arp2Δ strain complemented by 3HAC′ARP2 on pYEW248 (Moreau et al., 1996) and recombinant doubly tagged spores were selected, giving rise to strain YMW175K.
Table 4.
Interactions of two-hybrid fusion proteins with Las17p and actin baits
| GAL4-AD (activation domain) | GAL4-BD (DNA binding domain) β-galactosidase activity (Miller units)
|
|
|---|---|---|
| Las17p1–643 (pBMW172) | Act1p1–375 (pDAB7) | |
| Vector (pACTII) | 00.1 ± 0.02 | 0.2 ± 0.06 |
| Act1p−24–375 (pAIP70) | 3.5 ± 0.5 | 1.4 ± 0.07 |
| Arp2p1–391 (pAMW253) | ≤0.1 | ≤0.2 |
| Arp3p1–448 (pAMW315) | ≤0.1 | ≤0.2 |
| Las17p1–643 (pAMW173) | 1.0 ± 0.1 | 1.2 ± 0.3 |
| Act1p−28–375 (2×) | 2.0 ± 0.1 | 1.1 ± 0.1 |
| Rvs167p69–482 (2×) | 50 ± 1 | 1.0 ± 0.1 |
| Rvs167p191–482 (pAIP38) | 54 ± 5 | 0.7 ± 0.2 |
| Rvs167p239–482 (2×) | 122 ± 3 | ≤0.2 |
| Rvs167p400–482 (2×) | 16 ± 2 | ≤0.2 |
| Vrp1p492–818 (2×)* | 62 ± 9 | ≤0.2 |
| Vrp1p783–818 (1×)* | 29 ± 7 | ≤0.2 |
| Lsb1p8–241 (37×) | 58 ± 5 | ≤0.2 |
| Lsb2p3–215 (9×) | 96 ± 11 | ≤0.2 |
| Lsb3p186–451 (8×) | 15 ± 4 | ≤0.2 |
| Ysc84p/Lsb4p270–468 (10×) | 6 ± 0.2 | ≤0.2 |
| Lsb5p40–213 (11×) | 34 ± 1 | ≤0.2 |
| Lsb6p67–607 (5×) | 8 ± 1 | ≤0.2 |
Vectors encoding fusions with the GAL4-(AD) were transformed into Y190 cells expressing GAL4-DB fusions with Las17p (pBMW172) or Act1p (pDAB7) and plated on SC − Trp and Leu plates. At least three colonies from each transformation were cultivated overnight in liquid selective medium (SC − Trp and Leu). β-Galactosidase activities were measured as described in MATERIALS AND METHODS. Values shown ± SD. Most double transformants that did not show blue color in the filter test were considered as approximately equal (≤) to background and were not assayed in liquid cultures. Plasmids marked with an asterisk were found in the FYRL genomic library; nonmarked entries are from the White cDNA library. Redundancy of the AD plasmids found is shown in parentheses. Not all candidates found are shown; interactions of the strongest and shortest fusions of each candidate are shown; longest fusions for Rvs167p and Vrp1p are also shown.
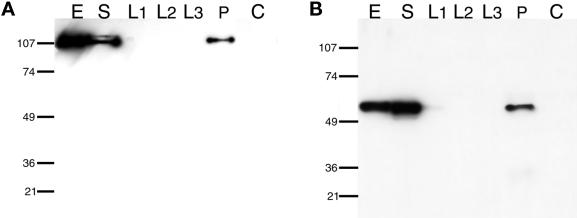
To immunoprecipitate Las17p, protein A-Sepharose beads were incubated with IgG and then with whole cell extract from strain YMW175K as described in MATERIALS AND METHODS. Las17-prot A precipitated from the supernatant was visualized after SDS gel electrophoresis on blots with IgG (Figure 9A). Arp2p clearly precipitated with Las17-prot A after multiple washes (Figure 9B). Neither Las17-prot A nor Arp2p precipitated in a parallel control experiment from a nontagged wild-type extract (Figure 9, A and B, lane C). Analysis of the same fractions with specific anti-Arc35p/End9p antibodies (Brodbeck-Schärer, unpublished results) confirmed the presence of this Arp2/3 complex subunit in the immunoprecipitate with Arp2p. This result confirms that the genetic data presented above represents specific physical interaction between Las17p and the Arp2/3 complex.
Figure 9.
Coimmunoprecipitation of Las17p and Arp2p (Arp2/3 complex). Las17-prot A was immunoprecipitated from crude extracts of strain YMW175K, which contained a chromosomally integrated allele of protein A-tagged Las17p and plasmid-borne HA tagged Arp2p (described in MATERIALS AND METHODS). After precipitation with TCA, cell extract and each step of the immunoprecipitation were subjected to 10% SDS-PAGE and blotted to nitrocellulose. E, extract; S, supernatant; L1, wash 1; L2, wash 2; L3, wash 3; P, immunoprecipitate; C, control immunoprecipitate, mock IP from crude extract of the YMW175K parental strain without protein A tag. (A) Las17-protA detected with purified rabbit IgG. (B) HA-tagged Arp2p detected with mouse monoclonal anti-HA IgG.
The human homologue of Las17p, WASP, regulates actin polymerization and cytoskeleton rearrangements and has been shown to interact directly with activated CDC42Hs (Apenström et al., 1996; Miki et al., 1996; Symons et al., 1996). We were intrigued to know whether Las17p interacted directly with Cdc42p in S. cerevisiae because the Cdc42p consensus binding sequence is absent. To test these interactions in the two-hybrid system, a Las17p bait was tested for interaction with wild-type and activated Cdc42p (Cvrckova et al., 1995; Leberer et al., 1997), in addition to profilin and actin (Amberg et al., 1995) as known interactants. No significant interactions with wild-type or activated Cdc42p were detected with these constructions.
To learn more about the interactions of Las17p, we used the two-hybrid system to screen a yeast cDNA library and a genomic DNA library for potential Las17p interacting proteins (see MATERIALS AND METHODS). In these screens (neither of which represent saturation) we found clones representing three known cytoskeletal proteins actin, verprolin, and Rvs167p (reduced viability upon starvation), and six proteins of unknown function, named LSB for Las seventeen binding proteins: Lsb1p (YGR136W), Lsb2p (YPR154w), Lsb3p (YFR024c-A), Lsb4p/Ysc84p (YHR016c), Lsb5p (YCL034w), and Lsb6p (YJL100w). Plasmids expressing these fusions were purified and transformed into strain Y190 expressing either Gal4DB-Las17p (pBMW172) or Gal4DB-Act1p (pDAB7). Results of β-galactosidase assays for the most informative in-phase clones are summarized in Table 4.
Of the known proteins found, a fusion containing actin with 27 additional aa at the amino terminus was found, similar to an extended actin (pAIP70) found in a screen against actin itself (Amberg et al., 1995). Las17p showed strong two-hybrid interaction with multiple derivatives of the SH3 domain protein Rvs167p. Nine different truncations (from the cDNA library) gave strong interaction with the Las17p bait. Rvs167p, originally identified along with Rvs161p as being important for recovery from starvation, affects cytoskeletal organization (Bauer et al., 1993; Sivadon et al., 1995). The smallest interacting fusion (83 aa) contains only the SH3 domain plus 21 aa of the GPA (for rich in Gly, Pro, and Ala) domain, whereas the strongest interacting fusion (239–482) contains the SH3 domain plus the GPA domain. In addition, four more N-terminally extended clones, beginning at aa 220, 191, 183, and 69, showed interaction with actin, as was reported by Amberg et al. (pAIP38 aa 189–549), whereas shorter clones did not. Strikingly, for verprolin (Vrp1p/End5p), eight different genomic fragments (and one cDNA fragment) all contained the C-terminal extremity of the protein. The smallest fusion contained only the C-terminal 36 aa, and the strongest interacting fusion had 327 aa. These results “map” a major Las17p interaction with Vrp1p to the Vrp1p C-terminus.
Among the unknown candidates, sequence comparisons revealed that Lsb1p, Lsb2p, Lsb3p, and Lsb4/Ysc84 contain SH3 domains with high similarity to the SH3 domain of Rvs167p and to that of human Grb2p. Lsb1p/V25 (241 aa) and Lsb2p (215 aa) contain an N-terminal SH3 domain followed by a proline-rich acidic region. With 64% identity to each other they form a strongly interacting minimal family. Except for the SH3 domain, no significant homologies for Lsb1p/Lsb2p were detected with other proteins in databases. Lsb1p has been characterized in a comparative mRNA study as being highly expressed in high-density cultures and upon diauxic shift, but deletion of the gene showed no particular phenotype (Wang et al., 1997). Lsb3p (451 aa) and Ysc84p/Lsb4p (468 aa) are 67% identical to each other, forming another minimal family with a C-terminal SH3 domain. BLAST searches and sequence alignment revealed homology between these two proteins and an SH3-domain containing protein of unknown function from mouse (340 aa, cDNA clone ge:D85926). YSC84/LSB4 was studied as an example of a divergently transcribed gene whose expression increased 5- to 10-fold during sporulation. However, its deletion caused no apparent phenotype for vegetative growth, progression into meiosis, or sporulation (Rocco et al., 1993). These SH3 domain proteins are currently under investigation. Candidates Lsb5p and Lsb6p are unknown, but both have possible homologues in other organisms. Lsb5p (265 aa) is similar to a hypothetical Schizosaccharomyces pombe protein. Lsb6p (607 aa) is similar to a hypothetical Caenorhabditis elegans protein C56A3.8. Although the existence of possible homologues in other species suggests that these ORFs code for functional proteins, the biological relevance to Las17p remains to be established.
DISCUSSION
LAS17 Is Important for the Function of the Arp2/3 Complex
The phylogenetic conservation of the Arp2/3 complex and its localization to actin-rich cortical regions of cells suggest that it fulfills a common essential function in the cortical actin cytoskeleton. The Arp2/3 complex functions to bind and cross-link actin filaments (Mullins et al., 1997, 1998b) and to nucleate actin polymerization required to produce movement at the cell surface (Mullins et al., 1998b; Welch et al., 1997a, 1998). Las17p/Bee1p has been shown to be involved in the actin cytoskeleton (Li, 1997) and in actin assembly in vitro (Lechler and Li, 1997). We addressed here the question of how Las17p produces its effects on actin organization. Our genetic evidence shows that Las17p plays an important role in the functioning of Arp2 and Arp3 proteins, thus linking it to the function(s) of the Arp2/3 complex. First, LAS17 is an allele-specific suppressor of the temperature-sensitive growth of ARP2 and ARP3 mutants. Defects in polarity of actin distribution and in fluid phase endocytosis were also suppressed. That is, no separation of function between actin defects and deficient endocytosis was apparent. Second, synthetic lethality of las17Δ with six of seven ARP2 mutants and with an ARP3 mutant indicates that these proteins affect the same cellular processes. Taken together, these results would tell us that Las17p plays an important role in the functions of the Arp2/3 complex but do not differentiate between stabilization of Arp2/3 complex interactions from regulation of actin-polymerizing function and/or signaling mechanisms mediated by Las17p.
LAS17Δ and ts Mutations in ARP2 and ARP3 Cause Similar Phenotypes
Deletion of LAS17 perturbs the organization of the actin cytoskeleton at permissive growth temperature (Li, 1997; this study) and is lethal at 37°C or is essential at all temperatures depending on the strain context. We have found a striking similarity between the las17Δ phenotype and the phenotypes of ARP2 ts mutants. In addition to depolarization of actin patches, many las17Δ cells show actin aggregates in the bud and at the bud neck of large budded cells as is seen in ARP mutant cells. An independent study showed that aberrant aggregates in las17Δ cells, as do cables, contain Sac6p but lack normal patch components cofilin and Cap2p (Li, 1997). Nor do aggregates appear to contain Arp2p or Arp3p. Interestingly, Arp2p-gfp still colocalized with residual actin patches (Figure 1B; Dumoulin, unpublished observations). Recent microscopic studies have confirmed that cables and patches depend on each other for assembly and function (Karpova et al., 1998). The fact that overexpression of LAS17 suppresses the formation of such structures in arp2-1 and arp2-2 mutants implicates Las17p and the Arp2/3 complex together in filament organization. Abnormally enlarged necks between mother and daughter cells are a further common defect of las17Δ and certain ARP2 mutant cells, suggesting that Las17p and Arp2p may both be important for cytokinesis and/or cell division. This is consistent with the previously shown genetic interactions between arp2-1 and the septin cdc10-1 mutations (Moreau et al., 1996). In further characterization of las17Δ cells, deficient α factor uptake lets us conclude that Las17p is essential for ligand-dependent receptor-mediated internalization in addition to fluid phase endocytosis, whereas later steps of the endocytosis pathway are apparently functional. Internalization was severely deficient at 24°C, at which cells continued to divide, as we have previously described for arp2-1 cells (Munn et al., 1995; Moreau et al., 1997). This step of endocytosis has also been shown to require actin and fimbrin and many other cytoskeletal proteins, including End3p, Sla2p/End4p, Vrp1p/End5 Rvs161/End6p, and Rvs167p (Raths et al., 1993; Munn and Riezman, 1994; Munn et al., 1995).
We also investigated the in vivo relationship of Las17p to polarity and actin organization. Loss of the bipolar budding pattern in growing diploid cells points to a possible role for Las17p in establishment of cell growth polarity. Furthermore, in the presence of latrunculin-A, functional Las17-gfp, probably with other proteins, remains in cortical patches in a polarized distribution in the absence of any detectable concentration of filamentous or total actin. Las17p must therefore be upstream of actin with respect to the flow of polarity information. Interestingly, certain other proteins important for polarity establishment, such as Cdc42p and Bem1p (Ayscough et al., 1997) as well as verprolin (Vaduva et al., 1997), also show polarized localization independently of polymerized actin in virtually all cells, whereas others such as Bud6/Aip3p, Myo2p, and calmodulin appear to maintain only partial polarity. Still other proteins that are important for the actin cytoskeleton and secretion, such as Arp2p, Aip1p, Sec4p, Sec8p, and Smy1p are dependent on actin structures for their polarized localization (Ayscough et al., 1997). The fact that las17Δ strains show increased sensitivity to latrunculin-A in growth tests (our unpublished results), in addition to being necessary for actin, Arp2p, and Arp3p localization, suggests that Las17p normally plays a role in polarized localization and in stabilization of the actin-polymerizing complex at the cortex. When the actin filament cytoskeleton is destroyed by latrunculin-A treatment, despite nonpolarized growth of cells (in the absence of increase of cell numbers), proteins such as Las17p and verprolin remain polarized, suggesting that the flow of polarity information necessary for polarized growth may also require interaction with actin filaments for these proteins to function with the Arp2/3 complex. Although Las17p is not essential for growth in all conditions, it is clearly a major determinant in multiple cellular processes implicating the actin cytoskeleton.
Elucidating Las17p Functions through its Interactions
Genetic evidence for functional interaction between las17Δ and ARP2 and ARP3 mutations is extended biochemically to indicate physical interaction with the coimmunoprecipitation of Arp2p and Las17p from crude extracts. How does high-copy expression of LAS17 overcome specific ARP2 and ARP3 ts mutations? Las17p might interact directly with Arp2p or Arp3p despite the fact that we were unable to detect interactions between Las17p and Arp2p or Arp3p in two-hybrid tests. In this case, Arp2p or Arp3p might bind less tightly to Las17p, but higher expression of Las17p could compensate weak binding. Another interpretation of our results is that intermediary protein components are necessary to allow functional interaction of Las17p with the Arp2/3 complex. Las17p might interact with the Arp2/3 complex through contact with actin, with other subunits, or both. In vitro protein interaction between Las17p and actin was demonstrated by coimmunoprecipitation (Li, 1997), and the two proteins interact directly in the two-hybrid system. Direct interaction between bovine N-WASP and actin has also been shown in vitro. The WH2 domain or “verprolin homology” and the “cofilin homology” domains of N-WASP participated in actin binding (Miki et al., 1996). Similar direct physical interactions with actin were confirmed for both WASP and SCAR (Machesky and Insall, 1998). Because the WH2 domain is rather well conserved in the C-terminal region of the yeast homologue, it likely to be the region of Las17p that interacts with actin:GRGALLDQIRQ . . GIQLNKT (WASP_human Swiss prot) GRDALLDQIRQ . . GIQL . KS (N-WASP; DDBJ accession D88460) GRDALLASIRGAGGIGALRKV (La17_yeast; Swiss prot).
SCAR and WASP were recently found to regulate the actin cytoskeleton through the Arp2/3 complex. The C-terminal WA fragment (WH2 and acidic domains) of both the WASP and the SCAR homologues bind the p21-Arc subunit (equivalent to S. cerevisiae Arc18p) of the Arp2/3 complex (Machesky and Insall, 1998). The ARP2 allele-specific interactions we found with las17Δ are not inconsistent with a similar interaction in S. cerevisiae, because certain mutations could affect small subunit binding indirectly. However, we did not detect two-hybrid interactions between a full-length Las17p and Arp2p, Arp3p, Arc35p, Arc40p, or any of the small subunits Arc19p, Arc18p, or Arc15p tested as activation fusions (Table 4; our unpublished results).
During the revision of this manuscript several in vitro studies pertinent to our results have been published. Las17p (GST-Bee1p) and the Arp2/3 complex have been coimmunoprecipitated in yeast, an independent demonstration of the physical interaction we report here between Las17-protA and the Arp2/3 complex. This interaction required both Arc19p and Arc15p (Winter et al., 1999). Simultaneous requirement of two or more subunits could be an explanation for the lack of two-hybrid interactions. Further knowledge of the Arp2/3 complex topology will be necessary to understand whether the mammalian and yeast results represent the same overall interactions. In addition to the activation of the S. cerevisiae Arp2/3 complex by Las17p in vitro (Winter et al., 1999), the direct role of WASP (Yarar et al., 1999) and SCAR (Machesky et al., 1999) in the activation of the in vitro nucleation activity of the Arp2/3 complex to generate new filament barbed ends has been elucidated in mammalian cells.
It has been shown previously that Cdc42, activated with guanosine 5′-3-O-(thio)triphosphate, induces actin polymerization in neutrophils (Zigmond et al., 1998). The Arp2/3 complex is one of the elements required for Cdc42-induced actin polymerization in Acanthamoeba (Mullins and Pollard, 1999) and Xenopus extracts (Ma et al., 1998). Now, it has also been shown that the stimulation of actin nucleation by N-WASP binding to the Arp2/3 complex requires both the actin binding domain and the Arp2/3 complex binding domain of N-WASP, as is the case for SCAR activation of the Arp2/3 complex (Machesky et al., 1999). Actin polymerization in the Xenopus system is enhanced by both Cdc42 and phosphatidylinositol-(4,5)-biphosphate, linking signal transduction pathway molecules to the stimulation of actin polymerization (Rohatgi et al., 1999). Evidence that the combined action of WASP family members and the Arp2/3 complex are also directly dependent on Cdc42 in vivo (Castellano et al., 1999) comes from experiments which used a modifed plasma membrane receptor capable of inducible local recruitment of Cdc42 (or WASP) to show that activated Cdc42 (or WASP) triggered actin polymerization and subsequent formation of protrusions in whole cells.
Las17p Shows Two-Hybrid Interaction with SH3 Domain Proteins
Previous studies showed that proline-rich stretches of various proteins bind to SH3 domains (Ren et al., 1993). SH3 domains are small regions (∼60–80 aa) present in a large number of proteins showing similarity to a domain in c-src and v-src proto-oncogene proteins. The presence of several proline-rich stretches in Las17p suggests that it could interact directly with one or several SH3 domain proteins. In two-hybrid screens with Las17p bait, five of the nine proteins we identified as Las17p interactants contained an SH3 domain. Although the strength of possible in vivo interactions between complete proteins cannot be implied directly from the activation values of two-hybrid interaction, clones representing the SH3 domain with or without other parts of these proteins interacted strongly with full-length Las17p. One of these SH3 domain proteins was Rvs167p, which contains three domains, BAR/RVS, GPA, and SH3. The SH3 domain with a few additional amino acids of the neighboring GPA domain was sufficient to show strong interaction with Las17p. Thus, in addition to the SH3 domain, Las17p-Rvs167p interaction may require participation of the GPA domain. Rvs167p is known to function with Rvs161p/End6p (Navarro et al., 1997), which was also fished out in a screen for endocytosis-defective mutants as mentioned above (Munn et al., 1995). In this screen we did not pick up Rvs161p (which is homologous to Rvs167p except for lack of the C-terminal SH3 domain) but it does interact with Las17p when tested directly (Recordon-Navarro, 1998), further suggesting that SH3 domains may not be solely responsible for interaction with Las17p. Rvs167p was previously found as an actin partner (Amberg et al., 1995). Our results show that only Rvs167p fusions containing at least part of the N-terminal domain of Rvs167p interact with actin in two-hybrid tests (≥aa 220–482; Table 4; our unpublished observations).
We also found that Las17p interacts strongly with verprolin (Vrp1/End5p). Verprolin has been shown to be part of the actin cytoskeleton and, like Las17p, is important for endocytosis and is a polarity determinant that localizes to polarized cortical patches independently of polymerized actin (Munn et al., 1995; Vaduva et al., 1997). Results of an independent communication have been presented showing that the C-terminal half of Las17p binds verprolin (Naqvi et al., 1998). Our results show that interactions of several different fragments from a random genomic bank map the binding site of verprolin to the C terminus; a fragment of only 35 C-terminal amino acids is sufficient for strong binding to Las17p. This interaction is comparable with that of WASP (as bait) with the human homologue of verprolin, WIP (WASP-interacting protein), isolated in a two-hybrid screen. In two-hybrid tests a C-terminal 87-aa truncation of WIP no longer bound to WASP, in good agreement with our mapping of the verprolin-Las17 interaction. Overexpression of WIP affected cortical actin assembly, inducing the formation of actin-containing projections at the cell surface of human B cells (Ramesh et al., 1997).
Given that the las17Δ genetic interaction with ARP2 and ARP3 mutants is allele specific, and that Las17p and Arp2p interact physically, Las17p is shown to be an effector of the Arp2/3 complex. These results coincide with multiple demonstrations in many systems of WASP family members’ interaction with the Arp2/3 complex. We have also shown that Las17p interacts directly with actin, Rvs167p, and verprolin by two-hybrid analyses. Finally, results of our two-hybrid screen suggest that Las17p might interact with at least six other unknown proteins, four of which are SH3 domain proteins. The fact that Lsb1p has been localized in the nucleus by transposon tagging of genes with increased expression in stationary phase and that LSB4/YSC84 expression is increased 5–10 times during sporulation raises the question of how Las17p might integrate signals from different developmental processes. Possible implication of these new proteins in the actin cytoskeleton or in signaling mechanisms are currently under investigation.
Whereas Arp2p and Arp3p bind as a complex to actin filaments (Mullins et al., 1997, 1998b), Las17p may bind only actin monomers, by analogy to SCAR and WASP (Machesky and Insall, 1998) and verprolin (Vaduva et al., 1997), which bind actin monomers in vitro. The absence of a concentration of actin in Las17p patches after latrunculin-A treatment does not eliminate the possibility that Las17p recruits the first monomers to cortical sites, but further studies are needed to clarify this possibility. The suppression of mutations in Arp2/3 complex components by LAS17 overexpression could then be explained by increasing monomeric actin at nucleation sites. The polarized localization of Las17p reflects its anchorage in areas of cell surface growth, hence its potential for involvement in both plasma membrane and intracellular signaling. An attractive possibility is that Las17p, in association with verprolin and Rvs167p (and perhaps with other proteins such as the Lsbp reported here), forms the essential part of a complex that remains anchored at the plasma membrane, receives diverse signals, and transmits them to the actin-polymerizing complex where Las17p plays a direct role in amplifying actin nucleation by the Arp2/3 complex.
ACKNOWLEDGMENTS
We thank S. Elledge, S.P. Bartel, A. Wach, and B Cormack for plasmids and M. White, P. James, and M. Fromont-Racine for two-hybrid banks. This work was financed by French National Research (Centre National de la Recherche Scientifique) funding to the Unité Propre de Recherche 9005 laboratory and by a French Cancer Association (Association pour la Recherche sur le Cancer) grant to B.W. A.M. was supported by the Syrian Atomic Energy Commission, and H.K. was supported by the Japanese National Institute of Agrobiological Resources.
REFERENCES
- Amberg DC, Basart E, Botstein D. Defining protein interaction with yeast actin in vivo. Nat Struct Biol. 1995;2:28–35. doi: 10.1038/nsb0195-28. [DOI] [PubMed] [Google Scholar]
- Amman AJ, Hong R. Disorders of the T-cell system. In: Stiehm ER, editor. Immunological Disorders in Children. Philadelphia: W.B. Saunders; 1989. pp. 257–315. [Google Scholar]
- Apenström P, Lindberg U, Hall A. Two GTPases, Cdc42 and Rac, bind directly to a protein implicated in the immunodeficiency disorder Wiskott–Aldrich syndrome. Curr Biol. 1996;6:70–75. doi: 10.1016/s0960-9822(02)00423-2. [DOI] [PubMed] [Google Scholar]
- Ayscough K, Stryker J, Pokala N, Sanders M. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian MK, Feoktistova A, McCullum D, Gould KL. Fission yeast Sop2: a novel and evolutionarily conserved protein that interacts with Arp3p and modulates profilin function. EMBO J. 1996;15:6426–6437. [PMC free article] [PubMed] [Google Scholar]
- Bauer F, Urdaci M, Aigle M, Crouzet M. Alteration of a yeast SH3 protein leads to conditional viability with defects in cytoskeletal and budding patterns. Mol Cell Biol. 1993;13:5070–5084. doi: 10.1128/mcb.13.8.5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeden L, Nasmyth K. Regulation of the yeast HO gene. Cold Spring Harb Symp Quant Biol. 1985;50:643–650. doi: 10.1101/sqb.1985.050.01.078. [DOI] [PubMed] [Google Scholar]
- Castellano FMP, Guillemot JC, Gouin E, Machesky L, Cossart P, Chavrier P. Inducible recruitment of Cdc42 or WASP to a cell-surface receptor triggers actin polymerization and filopodium formation. Curr Biol. 1999;9:351–360. doi: 10.1016/s0960-9822(99)80161-4. [DOI] [PubMed] [Google Scholar]
- Chien CT, Bartel PL, Sternglanz R, Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack B, Bertram G, Egerton M, Gow N, Falkow S, Brown A. Yeast-enhanced green fluorescent protein (yEGFP), a reporter of gene expression in Candida albicans. Microbiology. 1997;143:303–311. doi: 10.1099/00221287-143-2-303. [DOI] [PubMed] [Google Scholar]
- Coué M, Brenner S, Spector I, Korn E. Inhibition of actin polymerization by latrunculin A. FEBS Lett. 1987;213:316–318. doi: 10.1016/0014-5793(87)81513-2. [DOI] [PubMed] [Google Scholar]
- Cvrckova F, De Virgilio C, Manser E, Pringle J, Nasmyth K. Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes & Dev. 1995;9:1817–1830. doi: 10.1101/gad.9.15.1817. [DOI] [PubMed] [Google Scholar]
- Dulic V, Egerton M, Elguindi I, Raths S, Singer B, Riezman H. Yeast endocytosis assays. Methods Enzymol. 1991;194:697–710. doi: 10.1016/0076-6879(91)94051-d. [DOI] [PubMed] [Google Scholar]
- Durfee T, Becherer K, Chen P, Yeh S, Yang Y, Kilburn AE, Lee W, Elledge S. The retinoblastoma protein associates with protein phosphatase type I catalytic subunit. Genes & Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Fromont-Racine M, Rain JC, Legrain P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Methods in Enzymology. San Diego: Academic Press; 1991. Guide to yeast genetics and molecular biology; pp. 1–933. [PubMed] [Google Scholar]
- Hill J, Ian KA, Donald G, Griffiths DE. DMSO-enhanced whole cell yeast transformation. Nucleic Acids Res. 1991;19:5791. doi: 10.1093/nar/19.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig E. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JS, Prakash L. Yeast Saccharomyces cerevisiae selectable markers in pUC18 polylinkers. Yeast. 1990;6:363–366. doi: 10.1002/yea.320060502. [DOI] [PubMed] [Google Scholar]
- Kandels-Lewis S, Seraphin B. Involvement of U6 snRNA in 5′ splice site selection. Science. 1993;262:2035–2039. doi: 10.1126/science.8266100. [DOI] [PubMed] [Google Scholar]
- Karpova TS, McNally JG, Moltz SL, Cooper JA. Assembly and function of the actin cytoskeleton of yeast: relationships between cables and patches. J Cell Biol. 1998;142:1501–1517. doi: 10.1083/jcb.142.6.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher JF, Atkinson SJ, Pollard TD. Sequences, structural models and cellular localization of the actin-related proteins Arp2 and Arp3 from Acanthamoeba. J Cell Biol. 1995;131:385–397. doi: 10.1083/jcb.131.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Kolluri R, Tolias KF, Carpenter CL, Rosen FS, Kirchhausen T. Direct interaction of the Wiskott–Aldrich syndrome protein with the GTPase Cdc 42. Proc Natl Acad Sci USA. 1996;93:5615–5618. doi: 10.1073/pnas.93.11.5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer E, Wu C, Leeuw T, Fourest-Lieuvin A, Segall JE, Thomas DY. Functional characterization of the Cdc42 binding domain of yeast Ste20p protein kinase. EMBO J. 1997;16:83–97. doi: 10.1093/emboj/16.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechler T, Li R. In vitro reconstitution of cortical actin assembly sites in budding yeast. J Cell Biol. 1997;138:95–103. doi: 10.1083/jcb.138.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R. Bee1, a yeast protein with homology to Wiscott–Aldrich syndrome protein, is critical for the assembly of cortical actin cytoskeleton. J Cell Biol. 1997;136:649–658. doi: 10.1083/jcb.136.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Rohatgi R, Kirschner MW. The Arp2/3 complex mediates actin polymerization induced by the small GTP-binding protein Cdc42. Proc Natl Acad Sci USA. 1998;95:15362–15367. doi: 10.1073/pnas.95.26.15362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky L, Mullins R, Higgs H, Kaiser D, Blanchoin L, May R, Hall M, Pollard T. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc Natl Acad Sci USA. 1999;96:3739–3744. doi: 10.1073/pnas.96.7.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Atkinson SJ, Ampe C, Vandekerckhove J, Pollard TD. Purification of a cortical complex containing two unconventional actins from Acanthamoaba by affinity chromatography on profilin-agarose. J Cell Biol. 1994;127:107–115. doi: 10.1083/jcb.127.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Insall RH. Scar1 and the related Wiscott–Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr Biol. 1998;8:1347–1356. doi: 10.1016/s0960-9822(98)00015-3. [DOI] [PubMed] [Google Scholar]
- MacKay VL, Welch SK, Insley MY, Manney TR, Holly J, Saari GC, Parker M. The Saccharomyces cerevisiae BAR1 gene encodes an exported protein with homology to pepsin. Proc Natl Acad Sci USA. 1988;85:55–59. doi: 10.1073/pnas.85.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madania A. Etude fonctionelle des proteines apparentées à l’actine, Arp2p et Arp3p; recherche de proteines interactant chez la levure Saccharomyces cerevisiae. Institut de Biologie Moleculaire et Cellulaire, Strasbourg, France: Universite Louis Pasteur; 1998. pp. 1–113. [Google Scholar]
- Madania A, Moreau V, Martin R, Winsor B. LAS17 is a multi-copy suppressor of S. cerevisiae arp2-1. Yeast. 1997;13:S144. [Google Scholar]
- McCollum D, Feoktistova A, Morphew M, Balasubramanian M, Gould KL. The Schizosaccromyces pombe actin-related protein, Arp3, is a component of the cortical actin cytoskeleton and interacts with profilin. EMBO J. 1996;15:6438–6446. [PMC free article] [PubMed] [Google Scholar]
- Miki H, Miura K, Takenawa T. N-WASP, a novel actin-depolymerizing protein, regulates the cortcal cytoskeleton rearrangement in a PIP2-dependent manner downstream of tyrosine kinases. EMBO J. 1996;15:5526–5335. [PMC free article] [PubMed] [Google Scholar]
- Miki H, Seutsugo S, Takenawa T. WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. EMBO J. 1998;17:6932–6941. doi: 10.1093/emboj/17.23.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina I, Kenny D, Rosen F, Remold-O’Donnell E. T cell lines characterize events in the pathogenesis of the Wiscott–Aldrich syndrome. J Exp Med. 1992;176:867–874. doi: 10.1084/jem.176.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau V, Galan J-M, Devilliers G, Haguenauer-Tsapis R, Winsor B. The yeast actin-related protein Arp2 is required for the internalization step of endocytosis. Mol Biol Cell. 1997;8:1361–1375. doi: 10.1091/mbc.8.7.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau V, Madania A, Martin RP, Winsor B. The Saccharomyces cerevisiae actin-related protein Arp2 is involved in the actin cytoskeleton. J Cell Biol. 1996;134:117–132. doi: 10.1083/jcb.134.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins R, Heuser J, Pollard T. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci USA. 1998a;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins R, Pollard T. Rho-family GTPases require the Arp2/3 complex to stimulate actin polymerizationin Acanthamoeba extracts. Curr Biol. 1999;9:405–415. doi: 10.1016/s0960-9822(99)80187-0. [DOI] [PubMed] [Google Scholar]
- Mullins RD, Kellerher JF, Xu J, Pollard TP. Arp2/3 complex from Acanthamoeba binds profilin and cross-links actin filaments. Mol Biol Cell. 1998b;9:841–852. doi: 10.1091/mbc.9.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins RD, Stafford WF, Pollard TP. Structure, subunit topology and actin-binding activity of the ARP2/3 complex from Acanthamoeba. J Cell Biol. 1997;136:331–343. doi: 10.1083/jcb.136.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn AL, Riezman H. Endocytosis is required for the growth of vacuolar H+-ATPase-defective yeast: identification of six new END genes. J Cell Biol. 1994;127:373–386. doi: 10.1083/jcb.127.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn AL, Stevenson BJ, Geli MI, Riezman H. end5, end6, end7: mutations that cause actin delocalization and block the internalization step of endocytosis in Saccharomyces cerevisiae. Mol Biol Cell. 1995;6:1721–1742. doi: 10.1091/mbc.6.12.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi S, Zahn R, Mitchell DA, Stevenson BJ, Munn AL. The WASp homologue Las17p functions with the WIP homologue End5/verprolin and is essential for endocytosis in yeast. Curr Biol. 1998;8:959–962. doi: 10.1016/s0960-9822(98)70396-3. [DOI] [PubMed] [Google Scholar]
- Navarro P, Durrens P, Aigle M. Protein-protein interaction between the RVS161 and RVS167 gene products of S. cerevisiae. Biochim Biophys Acta. 1997;1343:187–192. doi: 10.1016/s0167-4838(97)00108-8. [DOI] [PubMed] [Google Scholar]
- Ochs H, Slichter S, Harker L, Von B, Clark R, Wedgewood R. The Wiscott–Aldrich syndrome: studies of lymphocytes, granulocytes and platelets. Blood. 1980;55:243–252. [PubMed] [Google Scholar]
- Pringle JR, Adams AE, Drubin DG, Haarer BK. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- Ramesh N, Anton IM, Hartwig JH, Geha RS. WIP, a protein associated with Wiscott–Aldrich syndrome protein, induces actin polymerization and redistribution in lymphoid cells. Proc Natl Acad Sci USA. 1997;94:14671–14676. doi: 10.1073/pnas.94.26.14671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raths S, Rohrer J, Crausaz F, Riezman H. end3 and end4: two mutants defective in receptor-mediated and fluid-phase endocytosis in Saccharomyces cerevisiae. J Cell Biol. 1993;120:55–65. doi: 10.1083/jcb.120.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recordon-Navarro P. Interactions between Rvs161p and Rvs167p and Their Partners Related to the Actin Cytoskeleton in Saccharomyces cerevisiae. Bordeaux: Victor Segalen; 1998. [Google Scholar]
- Ren R, Mayer P, Cicchetti P, Baltimore D. Identification of a ten amino acid proline-rich SH3 binding site. Science. 1993;259:1157–1161. doi: 10.1126/science.8438166. [DOI] [PubMed] [Google Scholar]
- Riezman H. Endocytosis in yeast: several of the yeast secretory mutants are defective in endocytosis. Cell. 1985;40:1001–1009. doi: 10.1016/0092-8674(85)90360-5. [DOI] [PubMed] [Google Scholar]
- Riezman H. Yeast endocytosis. Trends Cell Biol. 1993;3:273–277. doi: 10.1016/0962-8924(93)90056-7. [DOI] [PubMed] [Google Scholar]
- Rocco V, Daly MJ, Matre V, Lichten M, Nicolas A. Identification of two divergently transcribed genes centromere-proximal to the ARG4 locus on chromosome VIII of Saccharomyces cerevisiae. Yeast. 1993;9:1111–1120. doi: 10.1002/yea.320091012. [DOI] [PubMed] [Google Scholar]
- Rodriguez JR, Paterson BM. Yeast myosin heavy chain mutant: maintenance of the cell type specific budding pattern and the deposition of chitin and cell wall components requires an intact myosin heavy chain gene. Cell Motil Cytoskeleton. 1990;17:301–308. doi: 10.1002/cm.970170405. [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, Kirschner M. The interaction between N-WASP, and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97:221–231. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleid acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in S. cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer B, Riezman H. Detection of an intermediate compartment involved in transport of alpha-factor from the plasma membrane to the vacuole in yeast. J Cell Biol. 1990;110:1911–1922. doi: 10.1083/jcb.110.6.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivadon P, Bauer F, Aigle M, Crouzet M. Actin cytoskeleton and budding pattern are altered in the yeast rvs161 mutant: the Rvs161 protein shares common domains with the brain protein amphiphysin. Mol Gen Genet. 1995;246:485–495. doi: 10.1007/BF00290452. [DOI] [PubMed] [Google Scholar]
- Symons M, Derry JMJ, Karlak B, Jiang S, Lemahieu V, McCormick F, Francke U, Abo A. Wiskott–Aldrich syndrome protein, a novel effector for the GTPase CDC42Hs, is implicated in actin polymerization. Cell. 1996;84:723–734. doi: 10.1016/s0092-8674(00)81050-8. [DOI] [PubMed] [Google Scholar]
- Vaduva G, Martin NC, Hopper A. Actin-binding verprolin is a polarity development protein required for the morphogenesis and function of the yeast actin cytoskeleton. J Cell Biol. 1997;139:1821–1833. doi: 10.1083/jcb.139.7.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida TA, Emr S. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A, Brachat A, Alberti-Segui C, Rebischung C, Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targetting in Saccharomyces cerevisiae. Yeast, 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pöhlmann R, Philippsen P. New heterologous modules for classical or PCR-based disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Wang WY, Nishikawa T, Isono K. Isolation and characterization of Saccharomyces cerevisiae genes: differential expression under different growth conditions. J Gen Appl Microbiol. 1997;43:217–224. doi: 10.2323/jgam.43.217. [DOI] [PubMed] [Google Scholar]
- Welch MD, Iwamatsu A, Mitchison T. Actin polymerization is induced by the Arp2/3 complex at the surface of Listeria monocytogenes. Nature. 1997a;385:265–269. doi: 10.1038/385265a0. [DOI] [PubMed] [Google Scholar]
- Welch MD, DePace AH, Verma S, Iwamatsu A, Mitchison T. The human Arp2/3 complex is composed of evolutionarily conserved subunits and is localized to cellular regions of dynamic actin filament assembly. J Cell Biol. 1997b;138:375–384. doi: 10.1083/jcb.138.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch MD, Rosenblatt J, Scoble J, Portnoy DA, Mitchison TJ. Interaction of human Arp2/3 complex and the Listeria monocytogenes ActA protein in actin filament nucleation. Science. 1998;281:105–108. doi: 10.1126/science.281.5373.105. [DOI] [PubMed] [Google Scholar]
- Winter D, Lechler T, Li R. Activation of the yeast Arp2/3 complex by bee1p, a WASP-family protein. Curr Biol. 1999;9:501–504. doi: 10.1016/s0960-9822(99)80218-8. [DOI] [PubMed] [Google Scholar]
- Winter D, Podteljikov AV, Mann M, Li R. The complex containing actin-related proteins Arp2 and Arp3 is required for the motility and integrity of yeast actin patches. Curr Biol. 1997;7:519–529. doi: 10.1016/s0960-9822(06)00223-5. [DOI] [PubMed] [Google Scholar]
- Yang S, Ayscough KR, Drubin DG. A role for the actin cytoskeleton of Saccharomyces cerevisiae in bipolar bud-site selection. J Cell Biol. 1997;136:111–123. doi: 10.1083/jcb.136.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarar D, To W, Abo A, Welch MD. The Wiskott–Aldrich syndrome protein directs actin-based motility by stimulating actin nucleation with the Arp2/3 complex. Curr Biol. 1999;20:555–558. doi: 10.1016/s0960-9822(99)80243-7. [DOI] [PubMed] [Google Scholar]
- Zanolari B, Riezman H. Quantitation of alpha-factor internalization and response during the Saccharomyces cerevisiae cell cycle. Mol Cell Biol. 1991;11:5251–5258. doi: 10.1128/mcb.11.10.5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond SH, Joyce M, Yang C, Brown K, Huang M, Pring M. Mechanism of Cdc42-induced actin polymerization in neutrophil extracts. J Cell Biol. 1998;142:1001–10012. doi: 10.1083/jcb.142.4.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]