Summary
Dynamin-related proteins (DRPs) are GTPases that reversibly assemble on cellular membranes[1]. Individual DRPs (for simplicity, we use “DRP” to include authentic dynamins) function in fission or tubulation of the plasma membrane, trans-Golgi network, mitochondria, peroxisomes, chloroplasts, and endosomes [1], and in mitochondrial fusion[2]. Many of these functions are widespread, present in animals, plants, trypanosomes, Giardia, ciliates, alga, and slime molds[3–8]. Lineage-specific expansions of the gene family created specialized DRPs including MxB in animals, reported to regulate nuclear pore transport[9]. While many unicellular organisms possess a small number of DRPs, expansions occurred in some protist lineages. The 8 DRPs in the ciliate Tetrahymena thermophila may contribute to aspects of ciliate complexity. Each ciliate cell contains distinct germ line and somatic nuclei, which must require distinct machinery for their differentiation and maintenance[10][11]. Here we show that Drp6p, previously shown to be targeted to the nuclear envelope[3], is required for macronuclear (mac) development. Drp6p activity, which is distinct from that of the only other known nuclear DRP, is modulated by a combination of stage-specific subcellular targeting and assembly dynamics. This work demonstrates a novel activity for a DRP, and presents a system in which key aspects of regulation can be easily manipulated by environmental and developmental cues.
Results and Discussion
A lineage-specific and recently-evolved clade of DRPs in Tetrahymena
The eight T. thermophila DRPs each possesses three canonical domains (GTPase, middle, and GTPase effector). Similarity between Drp6p and classical dynamin, including residues involved in GTP binding and hydrolysis, is shown in Fig S1A,B. DRP6 is related (~59% amino acid identity) to DRP3, 4 and 5. The four loci are adjacent on macronuclear chromosome CH670412 (http://www.ciliate.org/). The cluster is phylogenetically isolated from the other four T. thermophila DRPs (Fig S1C). In addition, no other sequenced protist genome encodes a DRP that falls into the DRP3–6 clade, including that of the ciliate Paramecium tetraurelia (fig S1D). We also queried a large EST library from Ichthyophthirius multifiliis, the ciliate most closely related to T. thermophila for which genome-scale data is currently available. While Ichthyophthirius DRPs related to T. thermophila DRP1, 2, 7, and 8 were easily identified (unpublished data), none related to DRP3–6 was found. Therefore, this set of DRPs in T. thermophila represents a family of relatively new genes. The unusual localization of Drp6p to the nuclear envelope hinted at convergent evolution, since this localization is shared with MxB in animals. We therefore asked whether Drp6p and MxB have similar functions.
Drp6p is not a protist MxB
Each Tetrahymena cell contains a polyploid transcriptionally-active mac and a diploid, silent germline micronucleus (mic). Two different tagged variants of Drp6p (GFP-drp6-4 and HA-drp6-1) localized at or near the envelopes of both nuclei in growing cells (fig 1A,B). Both tagged forms also appeared as cytoplasmic puncta that also bore features of the endoplasmic reticulum (fig S2). The transcript of the drp6-4 transgene, integrated at the MTT1 locus, was ~14-fold more abundant than the low level of the endogenous DRP6 transcript (not shown). While such overexpression carries the risk of spurious localization, it was necessary because we could not detect the Drp6p expressed at the low endogenous level. Reassuringly, individual cells showed a wide range of GFP-drp6-4p expression, and both mac, mic and cytoplasmic signals were seen even in cells with the lowest visible expression levels. We also caution that neither HA-Drp6p nor GFP-Drp6p, like tagged DRPs in most systems, appears to be a fully-active protein (see details below). To ask whether Drp6p is enriched at nuclear pore complexes (NPC), we co-expressed HA-drp6-1p together with a GFP-tagged NPC component, Nup3p (from D. Chalker, Wash. U.). GFP-nup3p was diffuse throughout the mac envelope in both living (not shown) and fixed cells (fig 1C, fig S3B). HA-drp6-1p on the same nuclei was patchily distributed and showed little co-localization with Nup3-GFP. GFP-drp6-4p in living cells showed a similar patchy distribution, distinct from Nup3p (fig S3A,B). We asked whether targeting of Nup3p depended on the presence of Drp6p, by generating ΔDRP6 cell lines lacking all mac alleles of DRP6 (fig S3C,D). ΔDRP6 cells were viable, and showed a wildtype distribution of GFP-nup3p (fig S3E). We saw no differences by electron microscopy between wildtype and ΔDRP6 in nuclear envelope ultrastructure of cryofixed cells.
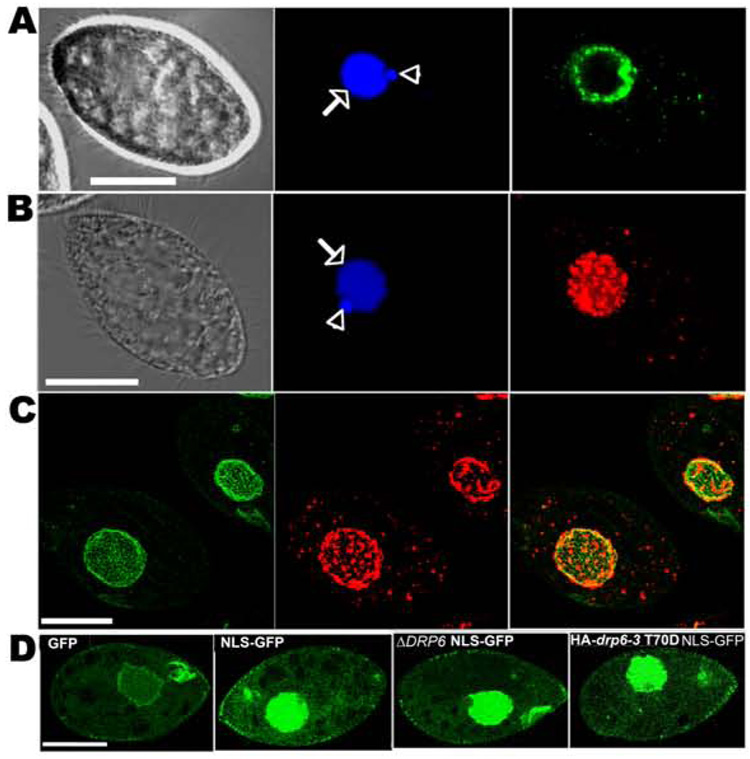
Figure 1. Drp6p localizes to nuclear envelopes but is not essential for nuclear import.
A. Left: phase-contrast image of fixed growing cell. Center: DAPI-stained nuclei: mac (arrow) and mic (arrowhead). Right: GFP-drp6-4p is at the periphery of both nuclei, and in cytoplasmic puncta. Bar=20µM. B. Left: phase-contrast image of fixed growing cell. Center: DAPI-stained nuclei. Right: HA-drp6-1p visualized by indirect immunofluorescence, shown as a confocal stack. Bar = 20 µM. C. Confocal stacks of cells co-expressing GFP-nup3p and HA-drp6-1p. GFP-nup3p is diffuse throughout the mac envelope (left) while HA-drp6-1p is in discrete patches (middle). The merged image is at the right. GFP-nup3p is also seen at the oral apparatus and at the cortex. Bar=20µM. D. Wildtype or ΔDRP6 cells, or cells expressing HA-drp6-3 T70D were transformed to express either GFP or GFP with a C-terminal nuclear localization signal (GFP-NLS). Confocal micrographs of fixed cells are shown. GFP-NLS accumulated strongly in nuclei of wildtype, ΔDRP6 and HA-drp6-3 T70D cells.
Unlike MxB, DRP6 does not appear to be involved in transport of nuclear localization sequence (NLS)-targeted proteins. GFP expressed in wildtype Tetrahymena distributed throughout the cytosol and mac, as expected since GFP can diffuse passively through NPCs[12](fig 1D). However, when an NLS was added to GFP, the protein was concentrated within the mac. Importantly, we obtained the same result when NLS-GFP was localized in ΔDRP6 cells. This result does not appear due to functional redundancy of Drp6p with Drp3p, 4p or 5p since none of these related DRPs, expressed as GFP-tagged constructs, localized to nuclei (not shown). Further evidence that Drp6p is not involved in nuclear import came from cells expressing HA-drp6-3 T70D, representing a substitution of aspartic acid for threonine 70, a conserved residue critical for efficient GTP hydrolysis in other DRPs (fig S1B). In particular, the corresponding mutation in MxB inhibited nuclear import[9]. While the inducible expression of HA-drp6-3 T70D in Tetrahymena did generate other phenotypes (below), it did not affect transport of NLS-GFP (fig 1D). These data indicate that Drp6p, though targeted to the nuclear envelope like MxB, plays a distinct role not previously documented for a DRP.
A role for DRP6 in nuclear differentiation
Starved Tetrahymena cells become competent for conjugation (fig 2A)[13]. Following pair formation between cells of complementary mating types, the mic in each cell undergoes elongation (“crescent formation”). Micronuclear meiosis produces haploid gametes that are exchanged between paired cells before fusing to form new pronuclei. Pronuclei divide and subsequently differentiate into two new mics and two developing macs (anlagen) in each cell. During this period, each cell also retains the old mac, which begins to degenerate 12 hours after pair formation. These nuclear events occur synchronously in mated wildtype cells and have been extensively studied with respect to chromosome rearrangements, but nothing is known about mechanisms underlying the implied restructuring of nuclear membranes.
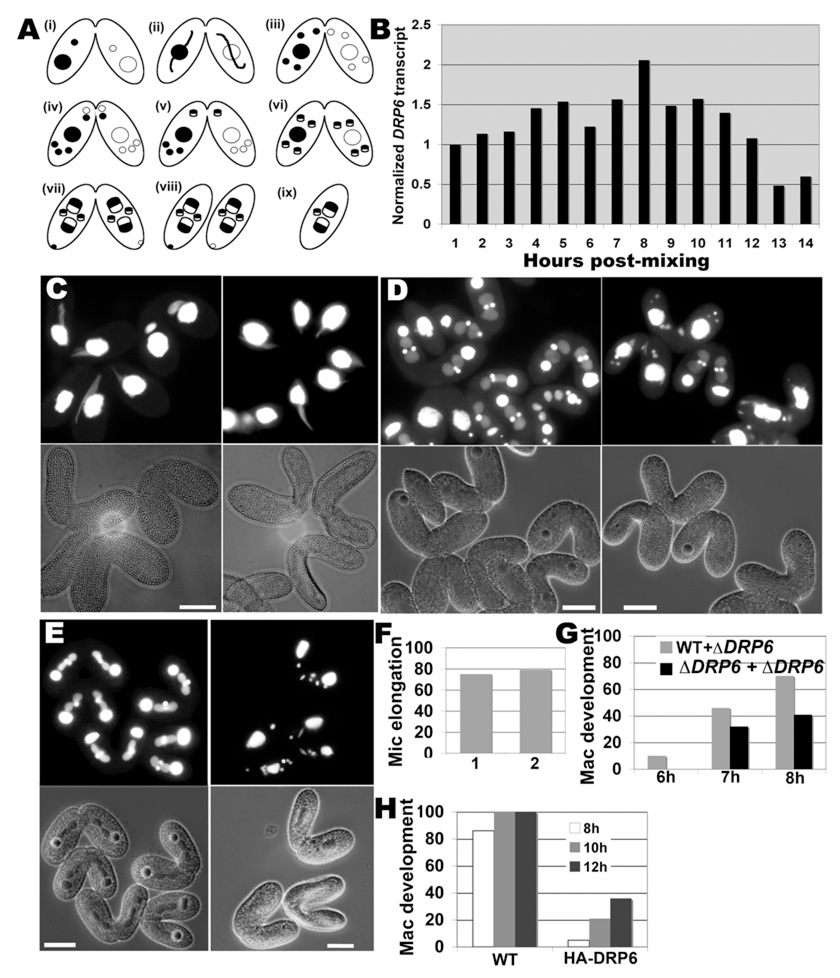
Figure 2. DRP6 is required for macronuclear development.
A. Conjugation stages in Tetrahymena. (i) pair formation, (ii) crescent stage (3h post mixing), (iii) meiosis (4.5h post mixing), (iv) pronuclear exchange (5h), (v) pronuclear fusion (5.5h), (vi) 2nd postzygotic mitosis (6.5h), (vii) new mac development (8h), (viii) pair separation (12h) and (ix) mac resorption and mic degradation (16h). B. Starved cultures of two complementary mating types were mixed and RNA samples were isolated subsequently. DRP6 mRNA in each sample, determined by reverse transcription PCR, was normalized with respect to alpha tubulin mRNA. Maximum expression is at 8h post-mixing, corresponding to mac development. C. ΔDRP6 cells were crossed with wildtype (left) or with a complementary ΔDRP6 strain (right). Pairs fixed at 3h were visualized by phase-contrast (lower panel) or DAPI-fluorescence (upper panel). Mic elongation, equivalent in both crosses, is quantified in (F) (1=wt × ΔDRP6; 2=ΔDRP6 × ΔDRP6; n=100 to 150). D. Same as (C) but fixed at 8h post mixing. Most conjugants in the ΔDRP6 × WT crosses showed two developing macs in each partner (left), while developing macs are only present in a minority of the ΔDRP6 × ΔDRP6 pairs (right). Results are quantified in (G) for pairs fixed at 6, 7 and 8h (n=150 to 200). E. A WT strain expressing endogenous DRP6 was crossed either with a second WT strain, or with cells transformed to also over-express HA-drp6-1. Pairs were fixed at 8h and visualized as in (C). Most WT × WT pairs show two developing macs in each partner (left), while developing macs are seen in a minority of pairs in which one partner expresses HA-drp6-1 (right). Bar=20µM. Results are quantified in (H) for pairs fixed at 8, 10 and 12h (n=150 to 200).
The DRP6 transcript could be detected in vegetative cells and becomes somewhat less abundant in starvation (not shown). Peak expression of DRP6 occurred in conjugating pairs at 8 hours post-mixing, suggesting a role at this stage (fig 2B). We therefore asked whether DRP6 were specifically required for any stage in conjugation. Note that DRP6 in conjugants can be transcribed both from the parental macs and, starting at ~8h, from the newly developing macs. Eliminating all DRP6 transcription in conjugants therefore requires disruption of all mic and mac alleles, but we were unable to disrupt DRP6 in the mic. We therefore took two complementary approaches to analyze DRP6 function during conjugation. First, we generated two ΔDRP6 (mac) strains with complementary mating types and set up pairings of ΔDRP6 × ΔDRP6, ΔDRP6 × wt, and wt × wt. The latter two were indistinguishable, but ΔDRP6 × ΔDRP6 conjugants had clear stage-specific nuclear defects. Mic elongation (fig 2C,F) and meiosis (fig S4) were unaffected, but the appearance of developing macs at 8h was ~2-fold inhibited (fig 2D,G). The lack of complete inhibition could be accounted for by transcription from the intact DRP6 alleles in the new macs developing at that stage. Consistent with this, over-expression from the inducible MTT1 promoter in conjugants of tagged DRP6 alleles, or of GTP binding or hydrolysis mutants, all resulted in dominant-negative inhibition of mac development starting at 8h (fig 2E,H; fig S5)[14]. Since overexpression of the epitope-tagged wildtype allele or the GTPase mutants gave similar results, we do not know whether the latter were acting in a classical dominant-negative fashion. The inhibition by any of these constructs persisted so that few cells developed visible new macs even in conjugation endpoints (fig S5,6). The defect was gene-specific since it was not recapitulated by comparable over-expression of tagged DRP3, 4 or 5 (fig S7). Conjugants expressing DRP6 transgenes were also defective in a subsequent step, the active degrading of parental macs at ~12h (fig S6). Transcription from the new mac is required for this step, so persistence of the parental macs in pairs expressing DRP6 transgenes can be considered an indirect result of the inability to build new macs[15].
Localization studies provided important support for a direct role of DRP6 in mac formation. At early stages in conjugation, GFP-drp6-4p (or HA-drp6-1p) did not decorate nuclei but instead localized to cytoplasmic puncta (fig 3A). However, beginning at ~8h, Drp6p associated strongly with the nuclear envelopes (fig 3B.) For the minority of mating pairs expressing GFP-drp6-4p that proceeded to stage viii shown in fig 2A, Drp6p was still associated with the developing macs as well as the mics (fig 3C). Thus the targeting of Drp6p to nuclear membranes is stage-specific during conjugation. It is also nucleus-specific, since a consistent feature was that HA-drp6-4p clearly failed to re-associate with the parental mac, normally destined for degradation (fig 3B). Taken together, the results indicate that Drp6p plays a specific role in conjugation that is likely to be exerted most directly at the stage of mac development.
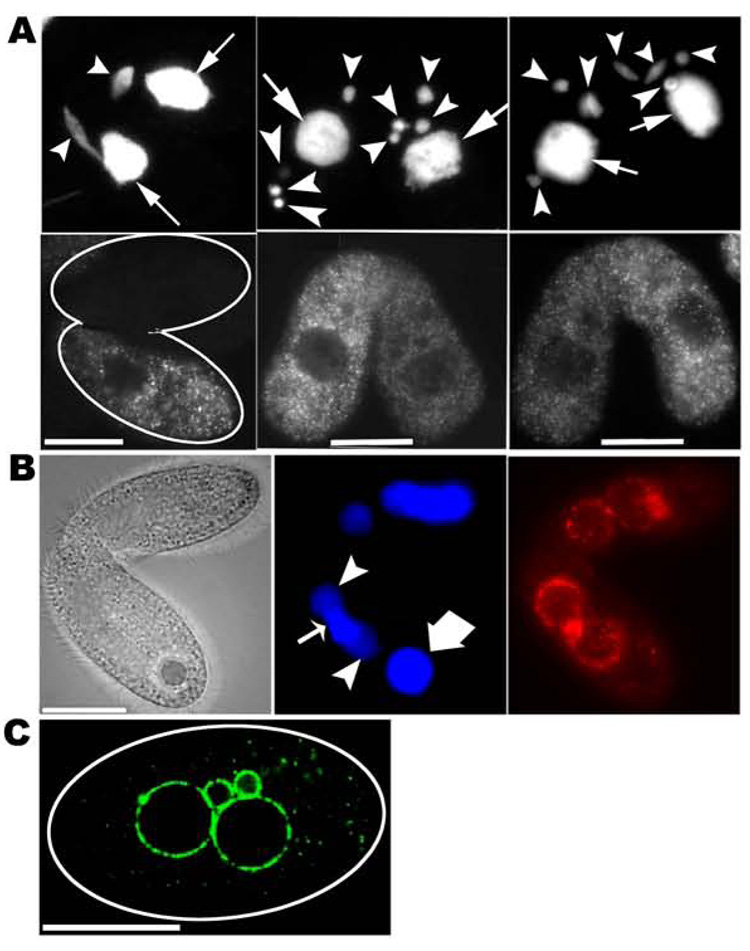
Figure 3. Drp6p localization in conjugating cells.
A. (left) Pair at 3h post mixing in which one partner is expressing GFP-drp6-4p. (top) DAPI-stained nuclei. (bottom) GFP-drp6-4p is largely restricted to the expressing cell, and is dispersed in cytoplasmic puncta. (middle and right panels) Pairs, with one cell expressing GFP-drp6-4p, at pronuclear selection stage (figure 5A, iii). (top) DAPI-stained mics (arrowheads) and macs (arrows). (bottom) GFP-drp6-4p is dispersed. B. Pair in which one partner expresses HA-drp6-1p, fixed at 9h post mixing. (left) phase-contrast. (middle) DAPI-stained old parental mac (thick arrow); developing macs (solid arrowheads); mic (thin arrow)(note: a second mic is not visible in this plane). The developing macs at this stage have a lower ploidy than the old mac and thus stain less brightly. (right) HA-drp6-1p, visualized by indirect immunostaining, is targeted to developing macs and the mic, but not to the old mac. C. A cell expressing GFP-drp6-4p, derived from a mating pair at 16h post mixing. GFP-drp6-4p is localized to both mics and newly developed macs, and in cytoplasmic puncta. Bar=20µM.
DRP6 activity during vegetative growth
DRP6 may be required for rapid expansion of the nuclear envelope that occurs during mac development in conjugants. Nuclear envelope expansion also occurs during the vegetative cell cycle, and DRP6 transcript levels in elutriation-synchronized vegetative cultures (RNA generously provided by D. Romero, U. Minnesota) peaked shortly after cell division, a period when cells enter macronuclear S-phase (fig S8). ΔDRP6 cells grew slowly (fig S9A) and accumulated excess DNA (fig S9B). These defects were also observed in transformants expressing DRP6 transgenes including GTP binding and hydrolysis mutants (fig S9A,C). The defects were specific, since strains bearing the NEO3 cassette at other loci (for example, ΔGRL6) or bearing the same construct to over-express other proteins (for example, GFP-rab36) showed no comparable growth inhibition (fig S9A). Thus nuclei in growing cells, like developing macs in conjugating cells, depend upon DRP6 function, albeit less critically.
Drp6p is activated in starved and conjugating cells
Drp6p associates with nuclei in growing cells but dissociates during early conjugation and then re-associates with selected nuclei at later stages. Drp6p in starved, non-conjugating cells showed an intermediate localization, with clear nuclear targeting but also prominent cytoplasmic puncta (fig S10). The localization of DRPs, like that of many cytoplasmic proteins, is dynamic rather than static and depends on association and dissociation from target membranes driven by GTP-fueled rounds of assembly and disassembly. We used FRAP (fluorescence recovery after photobleaching) to confirm that GFP-Drp6p localization was dynamic. For comparison we also analyzed GFP-tagged Drp1p, a DRP involved in clathrin-mediated endocytosis[3]. GFP-drp1-1p in growing cells showed rapid exchange, with recovery of fluorescence in photobleached spots after ~2 minutes (fig 4A,B). In contrast, GFPdrp6-4p showed very slow recovery (fig 4A,B). Similarly slow rates were seen at either the mac or mic (not shown). Strikingly, the mobility of Drp6p increased dramatically upon starvation, and in fact became roughly equivalent to that of Drp1p (fig 4C,D). Drp6p mobility was equally high when measured by initial bleaching of either the mac or mic. Drp6p was also highly mobile in starved conjugating cells at the 8h stage when the protein is targeted to nuclear membranes. The changes in mobility are likely to reflect modulation of Drp6p assembly/disassembly at the nuclear envelope.
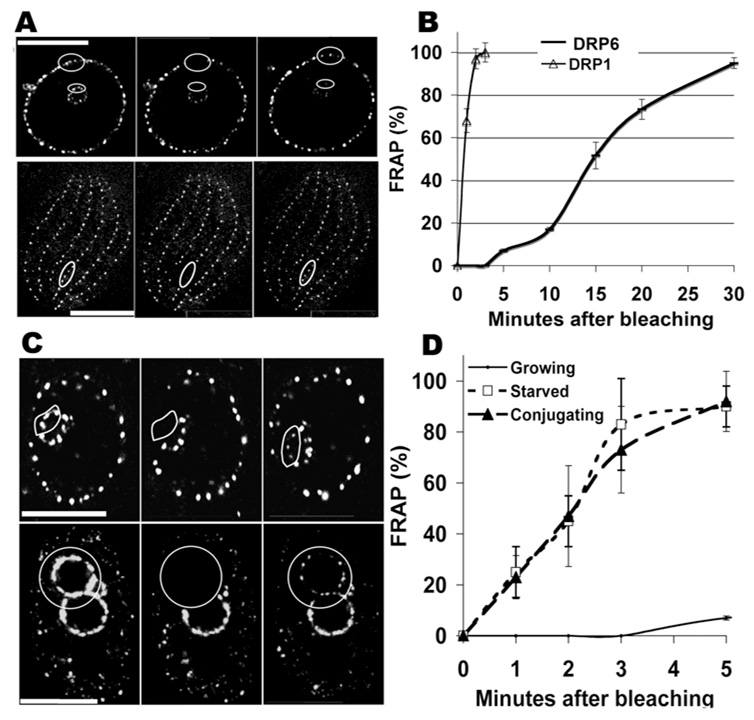
Figure 4. Drp6p is activated in starved and conjugating cells.
A. FRAP in growing cells of GFP-drp6-4p and GFP-drp1-1p. The upper panel focuses on the nuclei within a cell expressing GFP-drp6-4p; the oval is the area of the mac to be photobleached. The 2nd frame shows the cell immediately after photobleaching. The nuclei at 30’ post-bleach appear in the 3rd frame. Bar=10µM. FRAP of nuclear GFP-drp6-4p in growing cells is quantified (for n=3) in panels B and D. Lower panel: GFP-drp1-1p is present at regularly-spaced endocytic sites, whose distribution along cortical rows in a single cell is shown. The zone to be photobleached is shown by an oval. The cell immediately after bleaching appears in the 2nd frame, and at 2’ post-bleach in the 3rd frame. FRAP is quantified for GFP-drp1-1p in growing cells (n=3) in panel B. Bar=20µM. C. FRAP of GFP-drp6-4p in a starved cell (upper panel) and in an exconjugant (lower panel). The distributions of GFP-drp6-4p on the respective nuclei are shown in the left panels, outlining the areas to be photobleached. The 2nd frames show the nuclei immediately after photobleaching. The recovering signals at 2’ post-bleach are shown in the 3rd frame. The upper panel shows photobleaching of Drp6p in the mic; identical results were obtained for photobleaching of mac-localized Drp6p. FRAP results for starved and conjugating cells is quantified (for n=3) in panel D. Similar FRAP results were obtained in exconjugant cells.
Discussion
As shown in this paper, a recently-evolved DRP in T. thermophila has acquired a novel function at the nuclear envelope. Drp6p is targeted primarily to both mac and mic envelopes, and some protein was also found in cytoplasmic puncta that appeared long-lived and mobile in continuous imaging of live cells. These may be comparable to vesicular forms of ER of unknown function in higher plants[16]. Further work will be required to investigate their potential functional significance, and to confirm that active Drp6p, expressed at endogenous levels, is targeted to these bodies.
The activity of Drp6p is clearly different from that of the sole previously-characterized nuclear envelope DRP, MxB in animals, which by some unknown mechanism regulates import at nuclear pores. In contrast, neither null nor variant alleles of DRP6 interfered with accumulation of NLS-GFP in growing cells. It remains possible that DRP6 shifts its function to nuclear transport in starved or conjugating cells. We consider this unlikely, but for technical reasons such cells could not be assayed for NLS-GFP import. Our results instead suggest that Drp6p facilitates specific nuclear membrane dynamics. The most dramatic DRP6-dependent phenotypes were seen during conjugation but the gene also plays a function in vegetative cultures. DRP6 transcript levels in growing cells show cell cycle periodicity, peaking during macronuclear S phase. DRP6 is essential for rapid vegetative growth, a striking result since the gene evolved only recently and may be restricted to the Tetrahymena lineage. The growth defect was seen in ΔDRP6 cells and even more acute in cells expressing a DRP6 variant that no longer binds to the nuclear envelope. Similar alleles of DRPs result in dominant inhibition in other systems[6, 17]. Whether the DRP6 variants act via a classical dominant-negative mechanism in Tetrahymena is not yet clear since we found that many DRP6 phenotypes could be recapitulated by simple overexpression of tagged DRP6 alleles. We could not study expression of these tagged alleles at the level of the endogenous gene because DRP6 is transcribed at too low a level.
Either the absence of DRP6 (ΔDRP6) or Drp6p overexpression resulted in dramatic nuclear defects during conjugation. While our data cannot yet define a precise site of action for Drp6p, there are strong hints. DRP6 mRNA peaks at 8h after conjugation begins. At this stage, but not in previous stages, Drp6p is targeted to nuclei, specifically the new mics and developing macs. Strikingly, a large fraction of such macs failed to develop in cells where parental Drp6p was absent, or when tagged or mutated alleles were over-expressed. These defects were specific for DRP6 since mac formation was not comparably inhibited by over-expression of related DRPs. One possibility is that Drp6p functions, like a subset of mitochondrial DRPs, to promote membrane fusion[2, 18]. At the mac envelope, fusion with compatible vesicles or tubules could fuel membrane expansion. It is intriguing that Drp6p is not required for crescent elongation, a step requiring expansion of the mic envelope[19]. The difference between crescent mics and developing macs in their dependence on DRP6 suggests that Tetrahymena use more than one mechanism of nuclear envelope expansion.
All GTPases are regulated by interaction with GTPase activating proteins (GAPs). For DRPs, the GAP activity resides on the same polypeptide as the GTPase domain. These domains interact in trans when the protein self-assembles, thereby linking oligomerization, localization, and GTP turnover[20]. The activity of DRPs can also be regulated by post-translational modification. This is well illustrated for the DRP involved in mitochondrial scission, Drp1[21, 22]. While specific residues and their post-translational modifications, as well as associated mitochondrial phenotypes, have been identified, it is less clear whether or how those modifications affect the Drp1 assembly/disassembly cycle, GTPase activity, or localization.
Tetrahymena Drp6p represents a striking example of a novel role for DRPs and also offers a clear case of physiological regulation at multiple levels. First is regulation of transcript abundance during the cell cycle and particularly during conjugation. Secondly, Drp6p shows at least three stage-specific patterns of localization. The protein is targeted in growing cells to the nuclear envelope, but dissociates when cells are starved so that a large proportion of Drp6p shifts to cytoplasmic puncta. This shift is exacerbated when starved cells are mated, so that Drp6p is no longer visibly associated with nuclear envelopes at early stages in conjugation. Subsequently the protein re-associates strongly with the new mics and the developing macs, but not with the parental macs. The last aspect of Drp6p regulation is that recycling of the GFP-tagged protein at nuclear envelopes, as measured by FRAP, increases dramatically in starved or conjugating cells compared to vegetative cells. We hypothesize that post-translational Drp6p modifications, linked to changes in metabolism and during conjugation, are modulating the GTPase activity to control the assembly/disassembly cycle. Since starvation as well as conjugation can be easily synchronized in Tetrahymena, this offers a highly accessible system for probing mechanisms involved in DRP regulation.
Supplementary Material
Supplemental Experimental procedures and ten figures are available at:
Acknowledgements
For valuable reagents we thank D. Romero (U. Minnesota), D. Chalker (Washington U., St. Louis), and M. Gorovsky and J. Bowen (U. Rochester, NY); for other help we thank E. Cole (St. Olaf College, MN), C. Jahn (NWU Medical School, Chicago), J. Frankel (U. Iowa) and members of this laboratory: N. Khuong, L. Bright, N. Kambesis, and M. Zampa. We received EM help from Jotham Austin II (U. Chicago), and Tom Giddings, Alex Stemm-Wolf, and Mark Winey (U. Colorado Boulder). Light microscopy was done at the Integrated Light Microscopy Core Facility (U. Chicago); flow cytometry was done at the Cancer Research Center Facility (U. Chicago). NCE is an Ellison Medical Foundation Fellow of the Life Sciences Research Foundation. This work was supported by NSF-MCB-0422011 and NIH GM077607 to APT.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Praefcke GJ, McMahon HT. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- 2.Meeusen S, DeVay R, Block J, Cassidy-Stone A, Wayson S, McCaffery JM, Nunnari J. Mitochondrial inner-membrane fusion and crista maintenance requires the dynamin-related GTPase Mgm1. Cell. 2006;127:383–395. doi: 10.1016/j.cell.2006.09.021. [DOI] [PubMed] [Google Scholar]
- 3.Elde NC, Morgan G, Winey M, Sperling L, Turkewitz AP. Elucidation of Clathrin-Mediated Endocytosis in Tetrahymena Reveals an Evolutionarily Convergent Recruitment of Dynamin. PLoS Genet. 2005;1:e52. doi: 10.1371/journal.pgen.0010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Segui-Simarro JM, Austin JR, 2nd, White EA, Staehelin LA. Electron tomographic analysis of somatic cell plate formation in meristematic cells of Arabidopsis preserved by high-pressure freezing. Plant Cell. 2004;16:836–856. doi: 10.1105/tpc.017749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wienke DC, Knetsch ML, Neuhaus EM, Reedy MC, Manstein DJ. Disruption of a dynamin homologue affects endocytosis, organelle morphology, and cytokinesis in Dictyostelium discoideum. Mol Biol Cell. 1999;10:225–243. doi: 10.1091/mbc.10.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaechter V, Schraner E, Wild P, Hehl AB. The single dynamin family protein in the primitive protozoan giardia lamblia is essential for stage conversion and endocytic transport. Traffic. 2008;9:57–71. doi: 10.1111/j.1600-0854.2007.00657.x. [DOI] [PubMed] [Google Scholar]
- 7.Chanez AL, Hehl AB, Engstler M, Schneider A. Ablation of the single dynamin of T. brucei blocks mitochondrial fission and endocytosis and leads to a precise cytokinesis arrest. J Cell Sci. 2006;119:2968–2974. doi: 10.1242/jcs.03023. [DOI] [PubMed] [Google Scholar]
- 8.Nishida K, Takahara M, Miyagishima SY, Kuroiwa H, Matsuzaki M, Kuroiwa T. Dynamic recruitment of dynamin for final mitochondrial severance in a primitive red alga. Proc Natl Acad Sci U S A. 2003;100:2146–2151. doi: 10.1073/pnas.0436886100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.King MC, Raposo G, Lemmon MA. Inhibition of nuclear import and cell-cycle progression by mutated forms of the dynamin-like GTPase MxB. Proc Natl Acad Sci U S A. 2004;101:8957–8962. doi: 10.1073/pnas.0403167101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins K, Gorovsky MA. Tetrahymena thermophila. Curr Biol. 2005;15:R317–R318. doi: 10.1016/j.cub.2005.04.039. [DOI] [PubMed] [Google Scholar]
- 11.Li S, Yin L, Cole ES, Udani RA, Karrer KM. Progeny of germ line knockouts of ASI2, a gene encoding a putative signal transduction receptor in Tetrahymena thermophila, fail to make the transition from sexual reproduction to vegetative growth. Dev Biol. 2006;295:633–646. doi: 10.1016/j.ydbio.2006.03.048. [DOI] [PubMed] [Google Scholar]
- 12.Terry LJ, Shows EB, Wente SR. Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science. 2007;318:1412–1416. doi: 10.1126/science.1142204. [DOI] [PubMed] [Google Scholar]
- 13.Martindale DW, Allis CD, Bruns PJ. Conjugation in Tetrahymena thermophila. A temporal analysis of cytological stages. Exp Cell Res. 1982;140:227–236. doi: 10.1016/0014-4827(82)90172-0. [DOI] [PubMed] [Google Scholar]
- 14.Shang Y, Song X, Bowen J, Corstanje R, Gao Y, Gaertig J, Gorovsky MA. A robust inducible-repressible promoter greatly facilitates gene knockouts, conditional expression, and overexpression of homologous and heterologous genes in Tetrahymena thermophila. Proc Natl Acad Sci U S A. 2002;99:3734–3739. doi: 10.1073/pnas.052016199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ward JG, Herrick G. Effects of the transcription inhibitor actinomycin D on postzygotic development of Tetrahymena thermophila conjugants. Dev Biol. 1996;173:174–184. doi: 10.1006/dbio.1996.0015. [DOI] [PubMed] [Google Scholar]
- 16.Staehelin LA. The plant ER: a dynamic organelle composed of a large number of discrete functional domains. Plant J. 1997;11:1151–1165. doi: 10.1046/j.1365-313x.1997.11061151.x. [DOI] [PubMed] [Google Scholar]
- 17.van der Bliek AM, Redelmeier TE, Damke H, Tisdale EJ, Meyerowitz EM, Schmid SL. Mutations in human dynamin block an intermediate stage in coated vesicle formation. J Cell Biol. 1993;122:553–563. doi: 10.1083/jcb.122.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mozdy AD, Shaw JM. A fuzzy mitochondrial fusion apparatus comes into focus. Nat Rev Mol Cell Biol. 2003;4:468–478. doi: 10.1038/nrm1125. [DOI] [PubMed] [Google Scholar]
- 19.Wolfe J, Hunter B, Adair WS. A cytological study of micronuclear elongation during conjugation in Tetrahymena. Chromosoma. 1976;55:289–308. doi: 10.1007/BF00292827. [DOI] [PubMed] [Google Scholar]
- 20.Muhlberg AB, Warnock DE, Schmid SL. Domain structure and intramolecular regulation of dynamin GTPase. Embo J. 1997;16:6676–6683. doi: 10.1093/emboj/16.22.6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harder Z, Zunino R, McBride H. Sumo1 conjugates mitochondrial substrates and participates in mitochondrial fission. Curr Biol. 2004;14:340–345. doi: 10.1016/j.cub.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 22.Cribbs JT, Strack S. Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep. 2007;8:939–944. doi: 10.1038/sj.embor.7401062. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Experimental procedures and ten figures are available at: