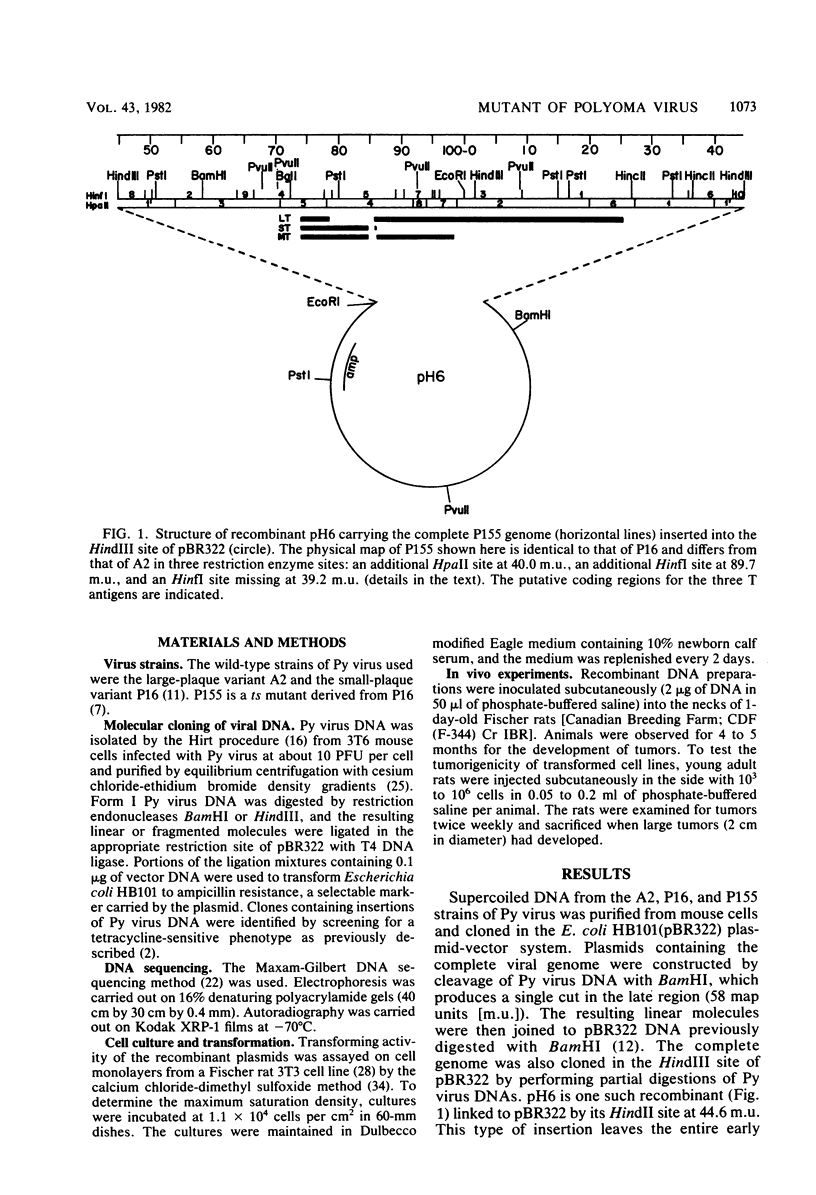
Abstract
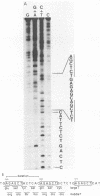
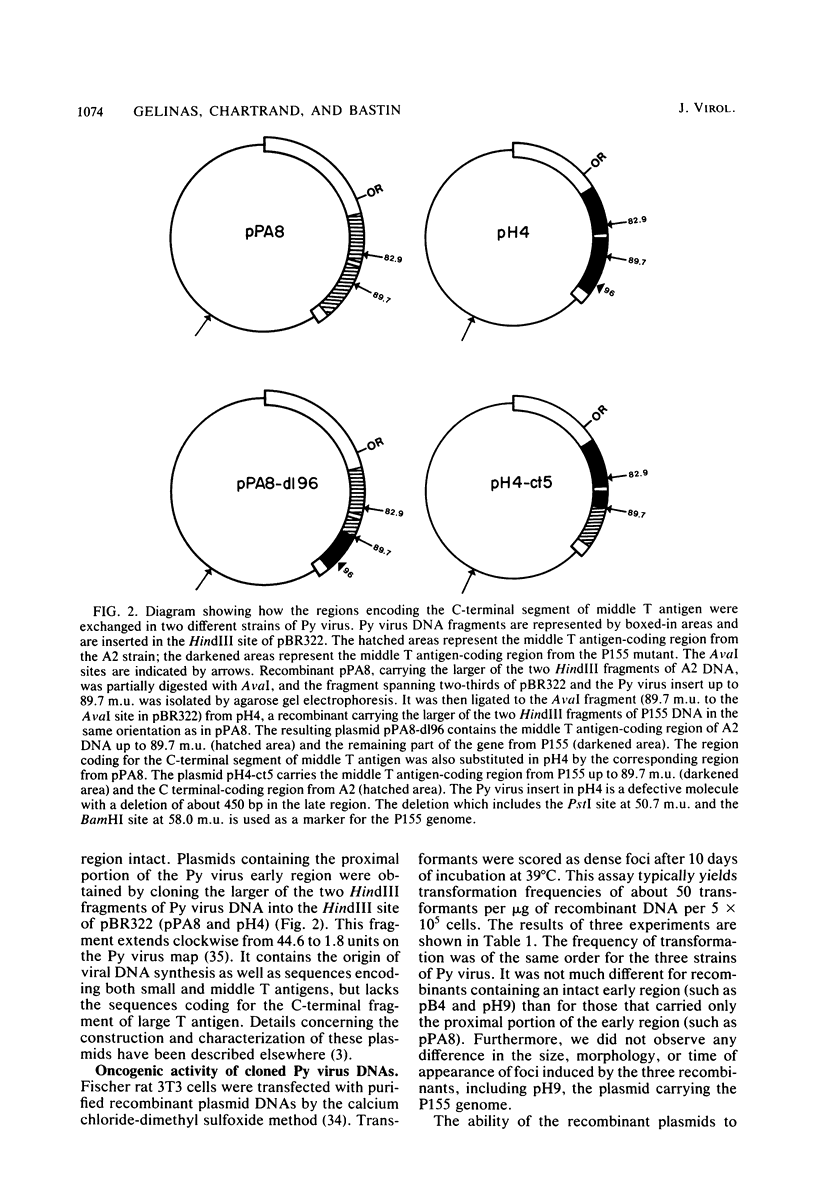
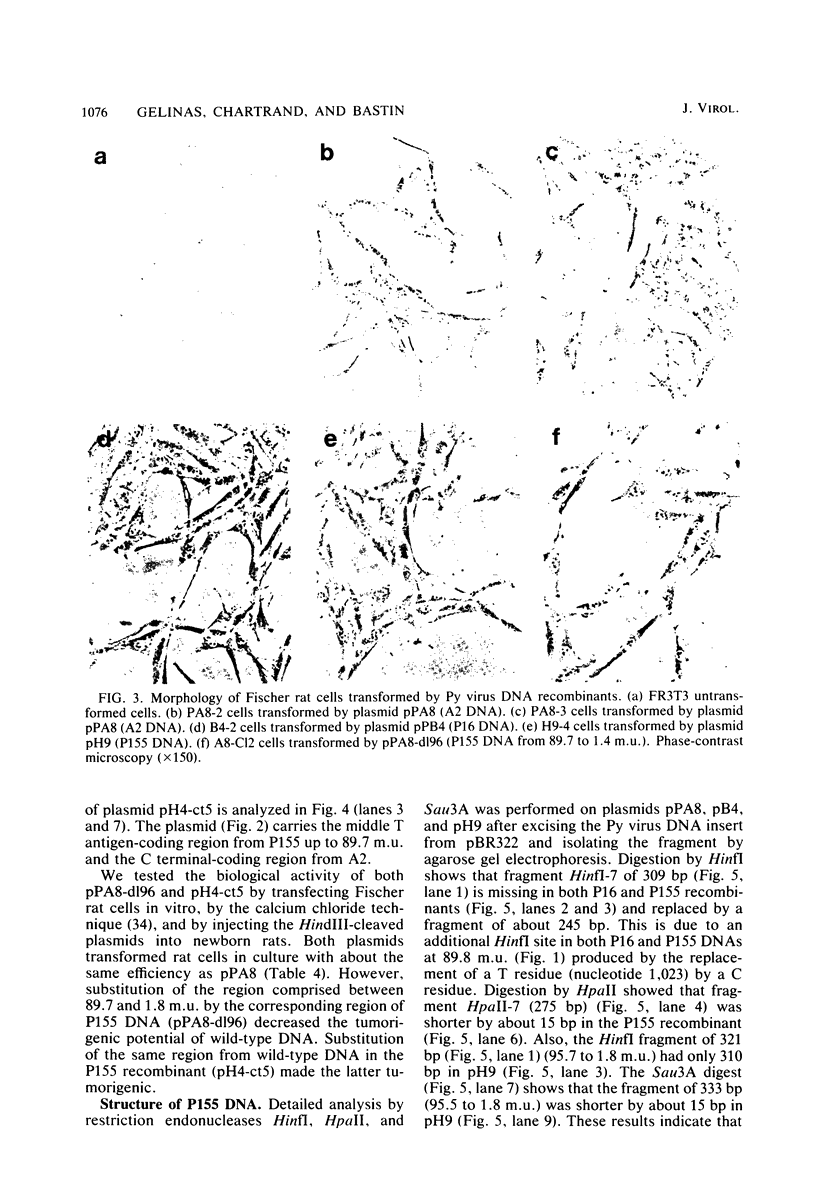
Cloned DNA from the P155 mutant of polyoma virus transforms cells in culture as efficiently as wild-type DNA, but has a much lower tumorigenic potential when injected into newborn rats. Like cells transformed by wild-type DNA, cells transformed by the mutant DNA grow in low serum concentrations, form colonies in agar suspension, and grow to high saturation densities compared with untransformed cells. They are, however, much less tumorigenic since they transplant 100- to 2,000-fold less efficiently than cells transformed by wild-type DNA. Substitution of the region between 89.7 and 1.8 map units by the corresponding region of P155 DNA decreased the tumorigenicity of wild-type DNA. When this region was isolated from wild-type DNA and substituted in P155 DNA, the tumorigenicity of the latter increased to values comparable to those of wild-type DNA. This showed that the lesion affecting tumorigenicity occurred between 89.7 and 1.8 map units on the polyoma virus genome. Sequence analysis in this region revealed a 12-base-pair deletion between nucleotides 1,347 and 1,360. This identified P155 as an mlt mutant, i.e., a mutant with a deletion from a region which encodes parts of the large and middle T antigens.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barrett J. C., Crawford B. D., Mixter L. O., Schechtman L. M., Ts'o P. O., Pollack R. Correlation of in vitro growth properties and tumorigenicity of Syrian hamster cell lines. Cancer Res. 1979 May;39(5):1504–1510. [PubMed] [Google Scholar]
- Bastin M., Bourgaux-Ramoisy D., Bourgaux P. Biological properties of polyoma DNA fragments cloned in plasmid pBR322. J Gen Virol. 1980 Sep;50(1):179–184. doi: 10.1099/0022-1317-50-1-179. [DOI] [PubMed] [Google Scholar]
- Bastin M. Molecular cloning in plasmid pBR322 giving altered expression of the tetracycline resistance gene. J Gen Microbiol. 1981 Mar;123(1):187–191. doi: 10.1099/00221287-123-1-187. [DOI] [PubMed] [Google Scholar]
- Bendig M. M., Thomas T., Folk W. R. Viable deletion mutant in the medium and large T-antigen-coding sequences of the polyoma virus genome. J Virol. 1980 Mar;33(3):1215–1220. doi: 10.1128/jvi.33.3.1215-1220.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin T. L. Host range mutants of polyoma virus. Proc Natl Acad Sci U S A. 1970 Sep;67(1):394–399. doi: 10.1073/pnas.67.1.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury K., Light S. E., Garon C. F., Ito Y., Israel M. A. A cloned polyoma DNA fragment representing the 5' half of the early gene region is oncogenic. J Virol. 1980 Nov;36(2):566–574. doi: 10.1128/jvi.36.2.566-574.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhart W. Complementation and transformation by temperature-sensitive mutants of polyoma virus. Virology. 1969 May;38(1):120–125. doi: 10.1016/0042-6822(69)90133-0. [DOI] [PubMed] [Google Scholar]
- Efstratiadis A., Posakony J. W., Maniatis T., Lawn R. M., O'Connell C., Spritz R. A., DeRiel J. K., Forget B. G., Weissman S. M., Slightom J. L. The structure and evolution of the human beta-globin gene family. Cell. 1980 Oct;21(3):653–668. doi: 10.1016/0092-8674(80)90429-8. [DOI] [PubMed] [Google Scholar]
- Farabaugh P. J., Schmeissner U., Hofer M., Miller J. H. Genetic studies of the lac repressor. VII. On the molecular nature of spontaneous hotspots in the lacI gene of Escherichia coli. J Mol Biol. 1978 Dec 25;126(4):847–857. doi: 10.1016/0022-2836(78)90023-2. [DOI] [PubMed] [Google Scholar]
- Feunteun J., Sompayrac L., Fluck M., Benjamin T. Localization of gene functions in polyoma virus DNA. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4169–4173. doi: 10.1073/pnas.73.11.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M., Griffin B. E., Lund E., Robberson D. L. Polyoma virus--a study of wild-type, mutant and defective DNAs. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):45–52. doi: 10.1101/sqb.1974.039.01.009. [DOI] [PubMed] [Google Scholar]
- Griffin B. E., Ito Y., Novak U., Spurr N., Dilworth S., Smolar N., Pollack R., Smith K., Rifkin D. B. Early mutants of polyoma virus (dl8 and dl23) with altered transformation properties: is polyoma virus middle T antigen a transforming gene product? Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):271–283. doi: 10.1101/sqb.1980.044.01.031. [DOI] [PubMed] [Google Scholar]
- Griffin B. E., Maddock C. New classes of viable deletion mutants in the early region of polyoma virus. J Virol. 1979 Sep;31(3):645–656. doi: 10.1128/jvi.31.3.645-656.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss P., Lai C. J., Dhar R., Khoury G. Splicing as a requirement for biogenesis of functional 16S mRNA of simian virus 40. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4317–4321. doi: 10.1073/pnas.76.9.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gélinas C., Bouchard L., Bastin M. Tumorigenic activity of cloned polyoma virus DNA in newborn rats. Experientia. 1981 Oct 15;37(10):1074–1075. doi: 10.1007/BF02085017. [DOI] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Ito Y., Brocklehurst J. R., Dulbecco R. Virus-specific proteins in the plasma membrane of cells lytically infected or transformed by pol-oma virus. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4666–4670. doi: 10.1073/pnas.74.10.4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Spurr N., Griffin B. E. Middle T antigen as primary inducer of full expression of the phenotype of transformation by polyoma virus. J Virol. 1980 Jul;35(1):219–232. doi: 10.1128/jvi.35.1.219-232.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis A. M., Jr, Cook J. L. Presence of allograft-rejection resistance in simian virus 40-transformed hamster cells and its possible role in tumor development. Proc Natl Acad Sci U S A. 1980 May;77(5):2886–2889. doi: 10.1073/pnas.77.5.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson G., Berg P. Construction and analysis of viable deletion mutants of polyoma virus. J Virol. 1979 Nov;32(2):523–529. doi: 10.1128/jvi.32.2.523-529.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marotta C. A., Wilson J. T., Forget B. G., Weissman S. M. Human beta-globin messenger RNA. III. Nucleotide sequences derived from complementary DNA. J Biol Chem. 1977 Jul 25;252(14):5040–5053. [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L. K., Fried M. Construction of the genetic map of the polyoma genome. J Virol. 1976 Jun;18(3):824–832. doi: 10.1128/jvi.18.3.824-832.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent C., Bastin M. Transformation of rat fibroblasts by cloned defective polyoma DNA. Arch Int Physiol Biochim. 1981 Sep;89(3):225–233. doi: 10.3109/13813458109069470. [DOI] [PubMed] [Google Scholar]
- Radloff R., Bauer W., Vinograd J. A dye-buoyant-density method for the detection and isolation of closed circular duplex DNA: the closed circular DNA in HeLa cells. Proc Natl Acad Sci U S A. 1967 May;57(5):1514–1521. doi: 10.1073/pnas.57.5.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risser R., Rifkin D., Pollack R. The stable classes of transformed cells induced by SV40 infection of established 3T3 cells and primary rat embryonic cells. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):317–324. doi: 10.1101/sqb.1974.039.01.042. [DOI] [PubMed] [Google Scholar]
- Seif I., Khoury G., Dhar R. A rapid enzymatic DNA sequencing technique: determination of sequence alterations in early simian virus 40 temperature sensitive and deletion mutants. Nucleic Acids Res. 1980 May 24;8(10):2225–2240. doi: 10.1093/nar/8.10.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seif R., Cuzin F. Temperature-sensitive growth regulation in one type of transformed rat cells induced by the tsa mutant of polyoma virus. J Virol. 1977 Dec;24(3):721–728. doi: 10.1128/jvi.24.3.721-728.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S. I., Freedman V. H., Risser R., Pollack R. Tumorigenicity of virus-transformed cells in nude mice is correlated specifically with anchorage independent growth in vitro. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4435–4439. doi: 10.1073/pnas.72.11.4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. E., Smith R., Griffin B., Fried M. Protein kinase activity associated with polyoma virus middle T antigen in vitro. Cell. 1979 Dec;18(4):915–924. doi: 10.1016/0092-8674(79)90204-6. [DOI] [PubMed] [Google Scholar]
- Smolar N., Griffin B. E. DNA sequences of polyoma virus early deletion mutants. J Virol. 1981 Jun;38(3):958–967. doi: 10.1128/jvi.38.3.958-967.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeda E., Arrand J. R., Smolar N., Griffin B. E. Sequence from early region of polyoma virus DNA containing viral replication origin and encoding small, middle and (part of) large T antigens. Cell. 1979 Jun;17(2):357–370. doi: 10.1016/0092-8674(79)90162-4. [DOI] [PubMed] [Google Scholar]
- Soeda E., Arrand J. R., Smolar N., Walsh J. E., Griffin B. E. Coding potential and regulatory signals of the polyoma virus genome. Nature. 1980 Jan 31;283(5746):445–453. doi: 10.1038/283445a0. [DOI] [PubMed] [Google Scholar]
- Stow N. D., Wilkie N. M. An improved technique for obtaining enhanced infectivity with herpes simplex virus type 1 DNA. J Gen Virol. 1976 Dec;33(3):447–458. doi: 10.1099/0022-1317-33-3-447. [DOI] [PubMed] [Google Scholar]
- Van Heuverswyn H., Fiers W. Nucleotide sequence of the Hind-C fragment of simian virus 40 DNA. Comparison of the 5'-untranslated region of wild-type virus and of some deletion Mutants. Eur J Biochem. 1979 Oct;100(1):51–60. doi: 10.1111/j.1432-1033.1979.tb02032.x. [DOI] [PubMed] [Google Scholar]