SUMMARY
Parkinson’s disease (PD) is a common neurodegenerative disorder caused by loss of midbrain dopaminergic neurons, the pathogenetic mechanisms of which remain unclear. Mitochondrial dysfunction, which has long been implicated in sporadic PD, has recently been highlighted as a key pathological cause, particularly with the identification of mutations in the PTEN-induced putative kinase (pink1), parkin and htrA2 (also known as omi) genes that are linked to PD. Studies in Drosophila melanogaster have shown that pink1 and parkin act in a common genetic pathway that maintains mitochondrial integrity, but other upstream or downstream components of this pathway are currently unknown. Using ectopic expression in the Drosophila eye as an assay, we have investigated the involvement of the mitochondrial protease encoded by omi in the Pink1/Parkin pathway and found that it acts genetically downstream of pink1 but functions independently of Parkin. Using the same approach, we also found that Rhomboid-7, a mitochondrial protease not previously implicated in PD, acts as an upstream component of this pathway, and showed that it is required to cleave the precursor forms of both Pink1 and Omi. These data further elucidate the composition of the Pink1 pathway and suggest that regulated intramembrane proteolysis is involved in its regulation.
INTRODUCTION
Parkinson’s disease (PD) is a common neurodegenerative disorder caused by the progressive loss of midbrain dopaminergic neurons, but the pathological mechanism remains unknown. Mitochondrial dysfunction has recently been highlighted as a potential, common disease mechanism in PD linking environmental, genetic and age-related influences (Abou-Sleiman et al., 2006; Mandemakers et al., 2007). Our understanding of the molecular events in PD pathogenesis has been greatly advanced by the identification and analysis of PD-associated genes (Farrer, 2006). The identification of mutations in PTEN-induced putative kinase (PINK1, which encodes a mitochondrial-targeted kinase) leading to autosomal recessive PD (Valente et al., 2004), and mutations in high temperature requirement A2 (HTRA2, also known as OMI, encoding a mitochondrial serine protease) in sporadic PD patients (Strauss et al., 2005), provide a compelling molecular link to mitochondrial function. Furthermore, an important role for Parkin, an ubiquitin-protein ligase and the most common cause of autosomal recessive PD, has been shown in the maintenance of mitochondrial integrity (Darios et al., 2003; Greene et al., 2003; Muftuoglu et al., 2004; Palacino et al., 2004; Ved et al., 2005). Importantly, genetic analysis of pink1 and parkin in Drosophila has shown that they act together in a common pathway, with parkin acting downstream of pink1 (Clark et al., 2006; Park et al., 2006; Yang et al., 2006). This pathway appears to maintain mitochondrial integrity via a mechanism affecting mitochondrial dynamics (Exner et al., 2007; Poole et al., 2008; Yang et al., 2008).
The striking similarities between the Drosophila mutant phenotypes pink1 and parkin, such as apoptotic muscle degeneration, male sterility, mitochondrial defects and flightlessness, led us to consider other Drosophila mutants with similar characteristics. Flies with mutations in rhomboid-7 (which encodes a mitochondrial intramembrane protease) share all of these phenotypes (McQuibban et al., 2006), suggesting that it may function in a common biological pathway with Pink1 and Parkin. This presents the possibility that mitochondrial Rhomboid could function to regulate the Pink1/Parkin pathway, although no link to Pink1 or Parkin has so far been shown. In addition, the activity of Pink1 has recently been linked to Omi (Plun-Favreau et al., 2007); however, the in vivo relevance of this interaction is currently unclear. Similarly, it is also unknown whether the activity of Omi is related to Parkin.
Thus, we sought to elucidate the functional relationship of Pink1, Parkin, Omi and Rhomboid-7 in Drosophila. We have conducted genetic interaction studies using a combination of overexpression and loss-of-function mutations in the Drosophila compound eye –a system that has proven particularly useful for genetic dissection of neurodegenerative mechanisms (Warrick et al., 1999; Fernandez-Funez et al., 2000; Shulman and Feany, 2003; Bilen and Bonini, 2005; Bilen and Bonini, 2007). Our findings indicate that omi acts downstream of pink1 but in an independent pathway from parkin; whereas the rhomboid-7 gene interacts with pink1, parkin and omi, acting upstream of pink1. Furthermore, we find that Rhomboid-7 is required for the processing of full-length forms of both Pink1 and Omi, presumably by regulated intramembrane proteolysis. Importantly, we only find the Rhomboid-7-dependent processed form of Pink1 in the cytoplasm, where it has recently been shown to protect from 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-induced mitochondrial dysfunction (Haque et al., 2008). These findings suggest an important role for Rhomboid-7-mediated proteolysis in the regulation of the Pink1/Parkin/Omi molecular pathway of PD.
RESULTS
Omi acts downstream of Pink1 and independently from Parkin
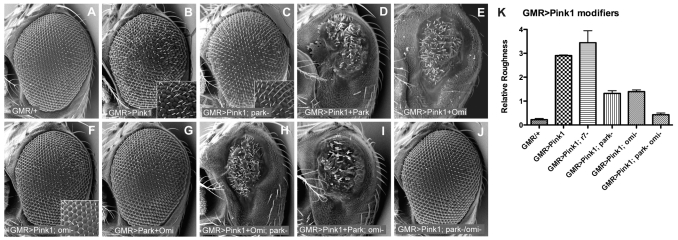
The Drosophila compound eye forms a stereotypical lattice of ~800 ommatidia (Fig. 1A). Overexpression of Drosophila pink1 causes a disorganization of the ommatidial array and roughening of the external eye morphology (Fig. 1B). The physiological relevance of this phenotype to the normal function of the Pink1/Parkin pathway can be inferred since the rough eye, resulting from pink1 overexpression, is significantly suppressed by removal of Parkin, which acts downstream of Pink1 (Fig. 1C,K). Furthermore, overexpression of both pink1 and parkin results in a severe rough eye phenotype, which greatly exceeds that conferred by pink1 or parkin overexpression alone (Fig. 1D and supplementary material Fig. S1B). These data are consistent with the finding that the pink1 overexpression phenotype is derived from amplified signaling through the normal physiological targets of Pink1, which commends this system as a useful tool to test whether certain genes interact with pink1.
Fig. 1. Gentic epistatis analysis in the Drosophila eye indicates that Parkin and Omi function downstream of Pink1.
Scanning electron microscopy (SEM) images of fly eyes expressing indicated transgenes. (A) GMR-GAL4 driver alone reveals the wild-type eye morphology of regularly arrayed ommatidia. Overexpression of pink1 causes a rough eye phenotype (B) that is partially suppressed in parkin mutants (C). (D) Overexpression of both pink1 and parkin shows an enhanced phenotype. (E) Overexpression of both pink1 and omi also shows a strong interaction. (F) omi mutants suppress the pink1 rough eye. (G) Parkin and Omi do not genetically interact. (H) parkin mutants do not suppress the Pink1/Omi interaction, and (I) omi mutants do not suppress the Pink1/Parkin interaction. (J) Loss of both parkin and omi completely suppresses the pink1 overexpression phenotype. (K) Mean + s.d. from more than 50 flies of specified genotypes, scored blind for relative roughness (see Methods).
The PD-linked mitochondrial serine protease Omi is phosphorylated in a Pink1-dependent manner (Plun-Favreau et al., 2007); however, a functional relationship between Omi and the Pink1/Parkin pathway has not been demonstrated. Thus, we sought to test whether Omi functions in the Pink1 pathway, and to determine its epistatic relationship with pink1 and parkin. To test for a genetic interaction between omi and pink1, we assessed the effects of combinations of omi and pink1 overexpression and loss-of-function. Overexpression of omi alone does not cause disorganization of the eye surface, but does result in a loss of pigmentation (supplementary material Fig. S1C,E). Dual overexpression of omi and pink1 causes a dramatic reduction in eye size (Fig. 1E), indicating a strong genetic interaction. Moreover, the pink1 overexpression phenotype is largely suppressed by loss of omi function (Fig. 1F,K). Conversely, the loss of pigment caused by omi overexpression is not suppressed in a pink1 mutant background (supplementary material Fig. S1F). To verify that suppression of the pink1 phenotype by loss of omi is not due to non-specific prevention of apoptosis, we expressed the pan-caspase inhibitor P35, and found that this does not suppress the pink1 overexpression phenotype (supplementary material Fig. S1H). Together these data indicate that omi acts downstream of pink1.
Given the strong genetic interaction between pink1 and omi, we surprisingly find that co-expression of parkin and omi has no such synergistic interaction (Fig. 1G). To better understand the epistatic relationship between these factors we tested whether parkin or omi loss-of-function mutations disrupted the synergistic interactions with pink1. We found that synergistic disruption of the eye by co-expression of pink1 and omi is not altered by loss of parkin (Fig. 1H). Similarly, the pink1/parkin interaction is not affected in omi mutants (Fig. 1I). These findings indicate that parkin does not act between, or downstream of, the pink1/omi interaction and that omi does not act in the pink1/parkin interaction, suggesting parkin that and omi function in parallel pathways downstream of pink1. Bifurcation of the pathway also explains why suppression of pink1 rough eye is incomplete when either parkin or omi are removed. To further support this interpretation, we found that when parkin and omi are both removed, the pink1 overexpression phenotype is completely suppressed (Fig. 1J,K). Taken together, these results are consistent with both parkin and omi acting downstream of pink1, but in independent pathways.
Rhomboid-7 genetically interacts with pink1, parkin and omi
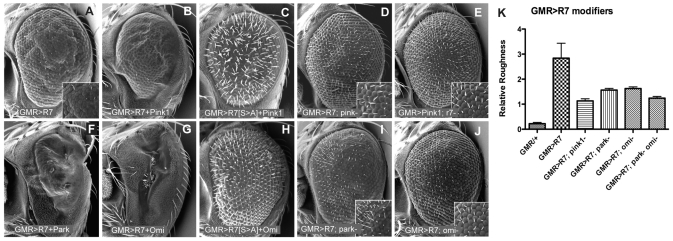
The mutant phenotypes of Drosophila rhomboid-7 showed striking similarity to those of pink1 and parkin mutants, thus, we hypothesized that Rhomboid-7 may function in the Pink1/Parkin pathway. To explore a potential functional relationship between Rhomboid-7 and Pink1, we performed a series of genetic interaction experiments utilizing Drosophila eyes. Overexpression of rhomboid-7 causes a rough eye phenotype, with widespread loss of bristles (Fig. 2A); expression of both rhomboid-7 and pink1 together enhances this phenotype, causing a much smaller eye with distorted morphology and complete loss of bristles (Fig. 2B). Importantly, this strong interaction is significantly reduced when the Rhomboid-7 catalytic serine residue is mutated to an alanine residue (Fig. 2C). To address the epistatic relationship of rhomboid-7 and pink1, we combined overexpression genotypes with loss-of-function mutant backgrounds. Interestingly, we found that the rhomboid-7 overexpression phenotype was significantly suppressed in a pink1 mutant background; notably the normal abundance of bristles was restored (Fig. 2D,K). In contrast, the pink1 overexpression phenotype (disorganization and roughening) was not suppressed in a rhomboid-7 mutant background (Fig. 2E; Fig. 1K) indicating that rhomboid-7 acts upstream of pink1.
Fig. 2. Genetic epistatis assays reveal that Rhomboid-7 is an upstream regulator of the Pink1 pathway.
(A) Overexpression of rhomboid-7 causes a rough eye phenotype. Overexpression of both rhomboid-7 and pink1 enhances the rough eye (C), through an interaction that requires the catalytic serine of Rhomboid-7 (C). The rhomboid-7 phenotype is suppressed in pink1 mutants (D), whereas the pink1 phenotype is not suppressed in rhomboid-7 mutants (E). rhomboid-7 overexpression is also enhanced by co-expression with parkin (F) and omi (G). Synergistic interaction of Rhomboid-7 and Omi requires the catalytic serine of Rhomboid-7 (H). The rhomboid-7 phenotype is also suppressed by loss of parkin (I) and omi (J). (K) Mean + s.d. from more than 50 flies of specified genotypes, scored blind for relative roughness (see Methods).
Consistent with parkin and omi acting as downstream effectors of pink1, we find that parkin and omi also act downstream of rhomboid-7. Overexpression of rhomboid-7 with either parkin or omi shows a strong genetic interaction, causing a severe loss of eye tissue (Fig. 2F,G). Furthermore, the rhomboid-7 overexpression phenotype is partially suppressed in either parkin or omi mutant backgrounds (Fig. 2I–K). Taken together, these data place rhomboid-7 upstream of pink1 in a common pathway with both parkin and omi (Fig. 4E). Also, because additional evidence indicates that parkin and omi act independently downstream of pink1, we find that omi and parkin mutants do not suppress rhomboid-7/parkin and rhomboid-7/omi synergistic phenotypes, respectively (supplementary material Fig. S1I,J).
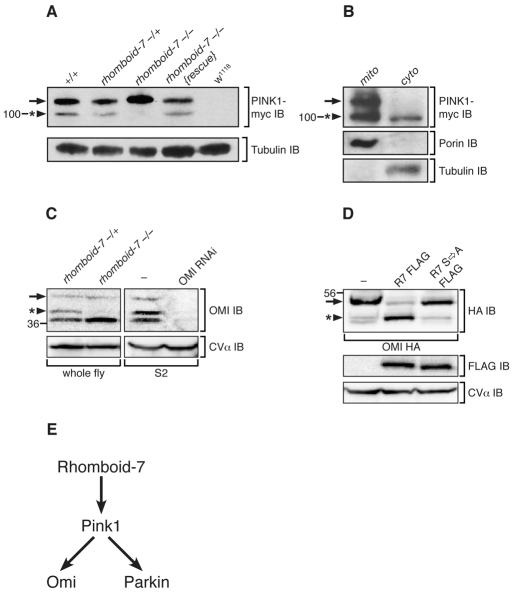
Fig. 4. Rhomboid-7 is required for cleavage of both Pink1 and Omi.
(A) Western blot analysis of Pink1-myc expression from whole fly lysates with the indicated genotypes. Pink1 appears as two forms, a long form (arrow) and a short form (arrowhead with asterisk) that is absent in rhomboid-7 mutants. Tubulin expression is used as a loading control. (B) Differential centrifugation of adult fly cells into mitochondrial (mito) and cytoplasmic (cyto) fractions. Porin and tubulin indicate the separation of mitochondria and cytosol, respectively. (C) Left panel: western blot analysis of endogenous Omi from whole fly lysates, showing a long form of Omi (arrow) and a shorter form (arrowhead with asterisk), which is absent in rhomboid-7 mutant flies. Right panel: western blot analysis of S2 cells treated with double-stranded RNA specific for Omi. The mitochondrial inner-membrane protein CV-α is used as a loading control. (D) Western blot analysis of Omi-HA expression in S2 cells with FLAG-tagged Rhomboid-7 and the Rhomboid-7 S256A catalytic mutant. (E) Schematic of the Pink1 pathway in Drosophila.
Rhomboid-7 is required to process Pink1 and Omi in vivo
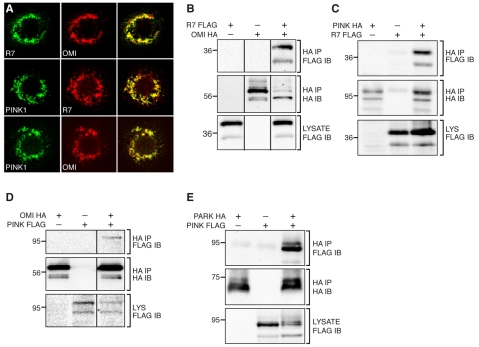
The genetic interactions between rhomboid-7, pink1, parkin and omi suggest there may be some physical and functional interaction; therefore, we investigated the molecular relationship between these factors. Consistent with their known mitochondrial function, Rhomboid-7, Pink1 and Omi co-localized with each other (Fig. 3A), and with the alpha subunit of complex V (CV-α) of the inner membrane ATPase (supplementary material Fig. S2A) in Drosophila S2 cells, suggesting that they could physically interact. To test this, we conducted immunoprecipitation assays using cultured Drosophila cells. We find that both Omi and Pink1 are able to physically interact with Rhomboid-7, as both proteins are detected with Rhomboid-7 in their respective immunoprecipitation assays (Fig. 3B,C). Interestingly, we also detected an interaction between Pink1 and Omi (Fig. 3D), consistent with a recent report demonstrating that Omi is phosphorylated in a Pink1-dependent manner (Plun-Favreau et al., 2007). In addition, we also find that Pink1 and Parkin co-purify in immunoprecipitates when they are expressed in cultured Drosophila cells (Fig. 3E). To demonstrate the specificity of the protein interactions in our exogenous immunoprecipitation assay, we investigated other potential interactions and found no association between Rhomboid-7 and the CV-α subunit of the inner membrane ATPase, nor with the outer membrane protein Porin (supplementary material Fig. S2D,E).
Fig. 3. Rhomboid-7, Pink1 and Omi physically interact in the mitochondria.
(A) Co-expression of Rhomboid-7 and Omi (top row); Pink1 and Rhomboid-7 (middle row); and Pink1 and Omi (bottom row) in Drosophila S2 cells shows respective co-localization in mitochondria. (B) Immunoprecipitation of Rhomboid-7-FLAG with Omi-HA in S2 cells. (C) Immunoprecipitation of Rhomboid-7-FLAG with Pink1-HA in S2 cells. (D) Immunoprecipitation of Pink1-FLAG with Omi-HA in S2 cells. (E) Immunoprecipitation of Parkin-HA with Pink1-FLAG in S2 cells.
The physical interaction of Rhomboid-7 with Pink1 and Omi, coupled with our genetic interaction and epistasis data, suggest that Pink1 and Omi may be targets of Rhomboid-7 intramembrane proteolysis. To test the processing of Pink1 in vivo, we utilised a 9myc-tagged pink1 transgenic line that drives expression from its endogenous promoter, and that has previously been shown to function normally (Clark et al., 2006). Consistent with previous reports, we find Pink1 exists as two distinct isoforms (Fig. 4A), a full-length form and a C-terminal portion that has been cleaved at the putative transmembrane domain (Muqit et al., 2006). To test whether Rhomboid-7 functions in the processing of Pink1, we assessed the protein isoforms present in a rhomboid-7 mutant background. Strikingly, we find that in the absence of Rhomboid-7, the shorter isoform of Pink1 is no longer found (Fig. 4A and supplementary material Fig. S2F). Notably, production of this short isoform is restored upon reintroduction of a wild-type rhomboid-7 transgene. This result demonstrates that Rhomboid-7 is required for the processing of full length Pink1. Given the important roles for Pink1, both inside and outside of the mitochondria, we assessed the subcellular localization of the two Pink1 isoforms. While both forms of Pink1 are found in mitochondria, we found that only the Rhomboid-7-dependent shorter isoform of Pink1 is constitutively present in the cytoplasm both in vivo and in vitro (Fig. 4B; supplementary material Fig. S2C).
To determine whether Omi is a Rhomboid-7 substrate, we assayed Omi protein patterns in vivo. Detection of endogenous Omi from wild-type flies revealed three isoforms (Fig. 4C), consistent with a full-length form and two processed forms, as previously described (Challa et al., 2007). In rhomboid-7 null mutant flies, one of the shorter forms of Omi is lost (Fig. 4C and supplementary material Fig. S2F), indicating that Rhomboid-7 is required for this processing event. Additionally, when omi is co-expressed with rhomboid-7 in cultured cells, the amount of the shorter form of Omi increases relative to the longer form, whereas a catalytically inactive Rhomboid-7 has no effect on Omi processing (Fig. 4D). Further, a recent report has determined that the cleavage site that corresponds to the Rhomboid-dependent isoform lies within the putative transmembrane domain (Khan et al., 2008). These data strongly suggest that the Rhomboid-7 intramembrane protease performs this cleavage. Interestingly, the cleavage of both Pink1 and Omi by Rhomboid-7 appears to be functionally important since their synergistic genetic interactions in our eye assay are significantly attenuated upon mutation of Rhomboid-7’s catalytic serine (Fig. 2C,H).
Since Rhomboid-7 appears to be required for the processing of two transmembrane proteins of the mitochondrial inner membrane, Pink1 and Omi, we sought to determine whether Rhomboid-7 is a non-selective protease for mitochondrial inner-membrane proteins. Not many proteins are known to exist as a mitochondrial inner-membrane tethered form and undergo proteolysis to produce a soluble form, but one such protein is the dynamin-related GTPase optic atrophy 1 (OPA1) (Satoh et al., 2003). Thus, we assessed whether Rhomboid-7 was required for cleavage of the Drosophila ortholog of OPA1 (dOpa1) in vivo. Although we detect multiple isoforms of dOpa1, loss of Rhomboid-7 does not affect the relative abundance of these isoforms (supplementary material Fig. S2B), indicating that Rhomboid-7 is not required for the processing of dOpa1.
In conclusion, our results indicate that Rhomboid-7 cleaves two different components of the Pink1 pathway and that this activity is functionally important for signaling, at least in our assay system.
DISCUSSION
A significant advance in elucidating the pathologic mechanisms of PD was made with the genetic analysis of Drosophila pink1 and parkin, which revealed that they act in a common pathway to maintain mitochondrial homeostasis (Clark et al., 2006; Park et al., 2006; Yang et al., 2006); however, other components of this pathway have not yet been demonstrated in vivo. Using genetic approaches in the fly eye we sought to identify additional components of the Pink1 pathway. We have shown that the mitochondrial intramembrane protease Rhomboid-7 acts upstream of Pink1 and, furthermore, found that Omi acts functionally downstream of Pink1 but in a pathway that is independent from Parkin (Fig. 4E).
We also found that Rhomboid-7 is required to cleave the full-length forms of Pink1 and Omi; a processing event that is required for their respective genetic interactions. This is largely in agreement with a recent study showing that the presenilin-associated, rhomboid-like (PARL) protease, the mammalian ortholog of Rhomboid-7, promotes the cleavage of vertebrate Omi (Chao et al., 2008); however, we note some differences with this report. We find that Drosophila Rhomboid-7 and Omi can physically interact, and that Rhomboid-7 is both necessary and sufficient to process one Omi isoform in vitro and in vivo. In contrast, vertebrate PARL does not directly interact with OMI and requires HAX1, a Bcl-2 family protein not found in Drosophila, to process it. Our findings further extend the repertoire of Rhomboid-7 to include Pink1; however, it remains to be determined whether PARL acts similarly to Drosophila Rhomboid-7 regarding the processing of vertebrate PINK1.
Recently, Omi has been shown to be phosphorylated in a Pink1-dependent manner (Plun-Favreau et al., 2007), although the functional importance of this interaction was unclear. Our studies confirm the interaction of Pink1 with Omi and, further, demonstrate that Omi functions downstream of Pink1. Surprisingly, however, we found no evidence that omi genetically interacts with parkin. Rather, our epistatis experiments indicate that parkin and omi act as independent downstream effectors of pink1. Although emerging evidence indicates that the Pink1/Parkin pathway is likely to affect mitochondrial dynamics, the function of the Pink1/Omi pathway is currently unclear.
Our subcellular fractionation results corroborate previous reports in which the processed form of Pink1 is found in the cytoplasm (Haque et al., 2008; Takatori et al., 2008), whereas cleaved Drosophila Omi is not constitutively found there (Challa et al., 2007; Khan et al., 2008). Although Omi released into the cytosol is known to induce apoptosis (Vande Walle et al., 2008), in contrast, Omi was recently shown to cooperate with Pink1, likely to be within mitochondria, to promote cell survival (Plun-Favreau et al., 2007). Significantly, cytoplasmic Pink1 has been shown to protect against toxic insult in neurons (Haque et al., 2008), indicating that it has additional and important unidentified functions outside of the mitochondria. Together these results suggest that mitochondrial and cytoplasmic forms of Pink1 and Omi may have distinct functions, and further, that the Rhomboid-dependent cleavage that we have identified may play a important role in regulating the abundance and localization of these isoforms.
Interestingly, while the Pink1/Parkin pathway has recently been linked to mitochondrial dynamics (Exner et al., 2007; Poole et al., 2008; Yang et al., 2008) – the process of fission and fusion reactions that control mitochondrial morphology – the function of the mitochondrial rhomboids in yeast, flies and mammals has also previously been linked to mitochondrial dynamics (McQuibban et al., 2003; Cipolat et al., 2006; McQuibban et al., 2006). Further work will be needed to determine the role and/or significance of these PD signaling pathway components with mitochondrial dynamics, but our findings further indicate the significance of this function for the Pink1 pathway.
In conclusion, our molecular and genetic studies in Drosophila demonstrate that mitochondrial Rhomboid-7 participates in the Pink1 pathway by regulating levels of the mitochondrial inner membrane-bound and cleaved forms of Pink1 and Omi. However, further work will be required to determine whether the vertebrate pathway is similarly composed, and how this pathway acts to maintain neuronal integrity.
METHODS
Fly stocks
Drosophila melanogaster stocks were raised on cornmeal agar media under standard conditions. The glass multiple reporter (GMR)-GAL4 driver line was obtained from the Bloomington Drosophila Stock Center, and the same line was used throughout for consistency. rhomboid-7ΔP null mutants (McQuibban et al., 2006), parkin25 null mutants and parkin combined with an upstream activating sequence (UAS-parkin) have been described previously (Greene et al., 2003). pink1B9 null mutant flies, and UAS-pink1 and UAS-omi sequences were a gift from J. Chung (KAIST) (Park et al., 2006), and the pink1-9myc transgenic line was provided by M. Guo (UCLA) (Clark et al., 2006). UAS-rhomboid-7 was constructed using the Gateway system (Carnegie Institute of Washington). Loss of omi/htrA2 was achieved by using omi/htrA2 null mutants, provided by N. Tapon (CRUK), or by using UAS-omi-RNAi from M. Miura (Univ. Tokyo) (Igaki et al., 2007); both methods gave equivalent phenotypes.
Scanning electron microscopy and eye quantification
Scanning electron microscopy (SEM) was performed according to a standard protocol (Sullivan et al., 2000). All animals of a given genotype displayed essentially identical phenotypes and randomly selected representative images are shown. Quantification of the partial rough eye phenotypes was achieved in the following way: specified genotypes were collected by one researcher, coded and given to another researcher for blind scoring. Scoring of relative roughness was conducted in comparison to known, uncoded ‘standard’ flies, i.e. GMR/+ flies, or flies overexpressing pink1 (GMR>Pink1) or rhomboid-7 (GMR>R7), respectively, as appropriate. We assigned GMR/+ flies a relative roughness score of 0 and GMR>Pink1 or GMR>R7 flies a score of 3. ‘Test’ flies were viewed under a dissecting microscope side-by-side with a few sample flies of ‘standard’ genotypes. Each individual fly was given a relative roughness score in comparison with the known standards:
0 – perfect WT eye with no aberrations; perfect facet/bristle alignment
1 – not perfect WT, slightly rough/disorganized
2 – slightly less rough than GMR>Pink1/R7 standards; greater degree of organization
3 – same/equivalent roughness to GMR>Pink1/R7 standards
4 – more rough than GMR>Pink1/R7 standards.
Groups of flies of the same genotype as the ‘standards’ were also included in blind scoring. For consistency, one researcher scored all of the individual flies. Scores were collected for at least 50 individual flies before the blinding was removed. In addition, a second researcher assessed groups of coded flies (modifiers and appropriate controls) and ranked whole groups for roughness, relative to each other. The ranked groups were in exact agreement with the detailed number of scored individual flies.
Immunohistochemistry
Transfected Drosophila S2 cells seeded on concanavalin A-coated coverslips were fixed for 15 minutes in 4% paraformaldehyde in PBS, permeabilized for 15 minutes in 0.1% TX-100 in PBS, blocked overnight in 10% goat serum in PBS, and subsequently incubated at room temperature with primary and secondary antibodies in 10% goat serum in PBS (1–2 hours each) with extensive washing in PBS between incubations. The following primary antibodies were used: mouse M2 anti-flag (Sigma), rabbit anti-HA (Sigma), mouse anti-CVα (Mitosciences). Alexa Fluor 546- and 488-conjugated secondary antibodies (Invitrogen) were used to detect the primary antibodies using a Zeiss LSM 510 confocal microscope.
Immunoprecipitation, fractionation and western blotting
S2 cells were transiently transfected with FuGENE6 (Roche). Cells were harvested 48 hours after transfection and lysed in IP buffer (150 mM NaCl, 10 mM Tris-HCl, 1 mM EDTA, 1% TX-100, 0.5% NP-40) with protease inhibitor cocktail (Bioshop Canada) and PMSF. Following centrifugation at 1000 g, lysates were precleared with Protein G Sepharose beads (Invitrogen), incubated with primary antibody overnight, followed by precipitation of tagged proteins with Protein G Sepharose beads. Whole adult flies were fractionated by differential centrifugation. Following an initial 1500 g spin, the mitochondria were sedimented at 10–103 g while the cytosol was purified from remaining organelles, as supernatant, following a 100–103 g spin. The following primary antibodies were used: mouse M2 anti-flag (Sigma), mouse HA7 anti-HA (Sigma), rabbit anti-HA (Sigma), mouse anti-myc (Santa Cruz Biotechnology), mouse anti-Tubulin (Sigma), rabbit anti-Porin/VDAC (Sigma), and rabbit anti-Omi (generous gift from Masayuki Miura).
DNA contructs
To make cell culture expression constructs, cDNAs for Drosophila Rhomboid-7, Omi (catalytically inactive S266A) and Pink1 were obtained from the Canadian Drosophila Microarray Centre and recombined using the Gateway system (Invitrogen) into pAWF and pAWH for C-terminal FLAG and HA epitope tagging, respectively. See: http://www.ciwemb.edu/labs/murphy/Gatewayvectors.html.
Supplementary Material
ACKNOWLEDGEMENTS
We thank J. K. Chung, N. Tapon, M. Guo and M. Miura for fly stocks, and the Centre for Light and Electron Microscopy, University of Sheffield, for assistance. We thank D. Strutt and J. Parker for critical reading of the manuscript. A.J.W. also thanks L. Pallanck for critical reading of the manuscript and his continued support and encouragement. This work is supported by grants from the Parkinson’s Disease Society, UK (4063), and the Wellcome Trust (081987) to A.J.W., the Canadian Institutes of Health Research (303157) to G.A.M., and a MRC Studentship to V.M.-W.H. The MRC Centre for Developmental and Biomedical Genetics is supported by Grant G070091.
Footnotes
COMPETING INTERESTS
The authors declare no competing financial interests.
AUTHOR CONTRIBUTIONS
A.J.W. and J.R.L. contributed equally to this work; A.J.W., G.A.M. and J.R.L. conceived of, and conducted, experiments; V.M.-W.H. and R.F. conducted experiments; R.C. provided unpublished reagents; A.J.W. and G.A.M. wrote the paper with editorial input from J.R.L.
SUPPLEMENTARY MATERIAL
Supplementary material for this article is available at http://dmm.biologists.org/content/1/2-3/168/suppl/DC1
REFERENCES
- Abou-Sleiman PM, Muqit MM, Wood NW. (2006). Expanding insights of mitochondrial dysfunction in Parkinson’s disease. Nat. Rev. Neurosci. 7, 207–219 [DOI] [PubMed] [Google Scholar]
- Bilen J, Bonini NM. (2005). Drosophila as a model for human neurodegenerative disease. Annu. Rev. Genet. 39, 153–171 [DOI] [PubMed] [Google Scholar]
- Bilen J, Bonini NM. (2007). Genome-wide screen for modifiers of ataxin-3 neurodegeneration in Drosophila. PLoS Genet. 3, 1950–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challa M, Malladi S, Pellock BJ, Dresnek D, Varadarajan S, Yin YW, White K, Bratton SB. (2007). Drosophila Omi, a mitochondrial-localized IAP antagonist and proapoptotic serine protease. EMBO J. 26, 3144–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao JR, Parganas E, Boyd K, Hong CY, Opferman JT, Ihle JN. (2008). Hax1-mediated processing of HtrA2 by Parl allows survival of lymphocytes and neurons. Nature 452, 900. [DOI] [PubMed] [Google Scholar]
- Cipolat S, Rudka T, Hartmann D, Costa V, Serneels L, Craessaerts K, Metzger K, Frezza C, Annaert W, D’Adamio L, et al. (2006). Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1-dependent cristae remodeling. Cell 126, 163–175 [DOI] [PubMed] [Google Scholar]
- Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, Yoo SJ, Hay BA, Guo M. (2006). Drosophila Pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 441, 1162–1166 [DOI] [PubMed] [Google Scholar]
- Darios F, Corti O, Lucking CB, Hampe C, Muriel MP, Abbas N, Gu WJ, Hirsch EC, Rooney T, Ruberg M, et al. (2003). Parkin prevents mitochondrial swelling and cytochrome c release in mitochondria-dependent cell death. Hum. Mol. Genet. 12, 517–526 [DOI] [PubMed] [Google Scholar]
- Exner N, Treske B, Paquet D, Holmstrom K, Schiesling C, Gispert S, Carballo-Carbajal I, Berg D, Hoepken HH, Gasser T, et al. (2007). Loss-of-function of human Pink1 results in mitochondrial pathology and can be rescued by parkin. J. Neurosci. 27, 12413–12418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer MJ. (2006). Genetics of Parkinson disease: paradigm shifts and future prospects. Nat. Rev. Genet. 7, 306–318 [DOI] [PubMed] [Google Scholar]
- Fernandez-Funez P, Nino-Rosales ML, de Gouyon B, She WC, Luchak JM, Martinez P, Turiegano E, Benito J, Capovilla M, Skinner PJ, et al. (2000). Identification of genes that modify ataxin-1-induced neurodegeneration. Nature 408, 101–106 [DOI] [PubMed] [Google Scholar]
- Greene JC, Whitworth AJ, Kuo I, Andrews LA, Feany MB, Pallanck LJ. (2003). Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc. Natl. Acad. Sci. USA 100, 4078–4083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque ME, Thomas KJ, D’Souza C, Callaghan S, Kitada T, Slack RS, Fraser P, Cookson MR, Tandon A, Park DS. (2008). Cytoplasmic Pink1 activity protects neurons from dopaminergic neurotoxin MPTP. Proc. Natl. Acad. Sci. USA 105, 1716–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igaki T, Suzuki Y, Tokushige N, Aonuma H, Takahashi R, Miura M. (2007). Evolution of mitochondrial cell death pathway: Proapoptotic role of HtrA2/Omi in Drosophila. Biochem. Biophys. Res. Commun. 356, 993–997 [DOI] [PubMed] [Google Scholar]
- Khan FS, Fujioka M, Datta P, Fernandes-Alnemri T, Jaynes JB, Alnemri ES. (2008). The interaction of DIAP1 with dOmi/HtrA2 regulates cell death in Drosophila. Cell Death Differ. 15, 1073–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandemakers W, Morais VA, De Strooper B. (2007). A cell biological perspective on mitochondrial dysfunction in Parkinson disease and other neurodegenerative diseases. J. Cell Sci. 120, 1707–1716 [DOI] [PubMed] [Google Scholar]
- McQuibban GA, Saurya S, Freeman M. (2003). Mitochondrial membrane remodelling regulated by a conserved rhomboid protease. Nature 423, 537–541 [DOI] [PubMed] [Google Scholar]
- McQuibban GA, Lee JR, Zheng L, Juusola M, Freeman M. (2006). Normal mitochondrial dynamics requires rhomboid-7 and affects Drosophila lifespan and neuronal function. Curr. Biol. 16, 982–989 [DOI] [PubMed] [Google Scholar]
- Muftuoglu M, Elibol B, Dalmizrak O, Ercan A, Kulaksiz G, Ogus H, Dalkara T, Ozer N. (2004). Mitochondrial complex I and IV activities in leukocytes from patients with parkin mutations. Mov. Disord. 19, 544–548 [DOI] [PubMed] [Google Scholar]
- Muqit MM, Abou-Sleiman PM, Saurin AT, Harvey K, Gandhi S, Deas E, Eaton S, Payne Smith MD, Venner K, Matilla A, et al. (2006). Altered cleavage and localization of Pink1 to aggresomes in the presence of proteasomal stress. J. Neurochem. 98, 156–169 [DOI] [PubMed] [Google Scholar]
- Palacino JJ, Sagi D, Goldberg MS, Krauss S, Motz C, Wacker M, Klose J, Shen J. (2004). Mitochondrial dysfunction and oxidative damage in parkin-deficient mice. J. Biol. Chem. 279, 18614–18622 [DOI] [PubMed] [Google Scholar]
- Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, Bae E, Kim J, Shong M, Kim JM, et al. (2006). Mitochondrial dysfunction in Drosophila Pink1 mutants is complemented by parkin. Nature 441, 1157–1161 [DOI] [PubMed] [Google Scholar]
- Plun-Favreau H, Klupsch K, Moisoi N, Gandhi S, Kjaer S, Frith D, Harvey K, Deas E, Harvey RJ, McDonald N, et al. (2007). The mitochondrial protease HtrA2 is regulated by Parkinson’s disease-associated kinase Pink1. Nat. Cell Biol. 9, 1243–1252 [DOI] [PubMed] [Google Scholar]
- Poole AC, Thomas RE, Andrews LA, McBride HM, Whitworth AJ, Pallanck LJ. (2008). The Pink1/Parkin pathway regulates mitochondrial morphology. Proc. Natl. Acad. Sci. USA 105, 1638–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh M, Hamamoto T, Seo N, Kagawa Y, Endo H. (2003). Differential sublocalization of the dynamin-related protein OPA1 isoforms in mitochondria. Biochem. Biophys. Res. Commun. 300, 482–493 [DOI] [PubMed] [Google Scholar]
- Shulman JM, Feany MB. (2003). Genetic modifiers of tauopathy in Drosophila. Genetics 165, 1233–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss KM, Martins LM, Plun-Favreau H, Marx FP, Kautzmann S, Berg D, Gasser T, Wszolek Z, Muller T, Bornemann A, et al. (2005). Loss of function mutations in the gene encoding Omi/HtrA2 in Parkinson’s disease. Hum. Mol. Genet. 14, 2099–2111 [DOI] [PubMed] [Google Scholar]
- Sullivan W, Ashburner M, Hawley RS. (2000). Drosophila Protocols. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Takatori S, Ito G, Iwatsubo T. (2008). Cytoplasmic localization and proteasomal degradation of N-terminally cleaved form of Pink1. Neurosci. Lett. 430, 13–17 [DOI] [PubMed] [Google Scholar]
- Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, et al. (2004). Hereditary early-onset Parkinson’s disease caused by mutations in Pink1. Science 304, 1158–1160 [DOI] [PubMed] [Google Scholar]
- Vande Walle L, Lamkanfi M, Vandenabeele P. (2008). The mitochondrial serine protease HtrA2/Omi: an overview. Cell Death Differ. 15, 453–460 [DOI] [PubMed] [Google Scholar]
- Ved R, Saha S, Westlund B, Perier C, Burnam L, Sluder A, Hoener M, Rodrigues CM, Alfonso A, Steer C, et al. (2005). Similar patterns of mitochondrial vulnerability and rescue induced by genetic modification of alpha-synuclein, parkin, and DJ-1 in Caenorhabditis elegans. J. Biol. Chem. 280, 42655–42668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrick JM, Chan HY, Gray-Board GL, Chai Y, Paulson HL, Bonini NM. (1999). Suppression of polyglutamine-mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat. Genet. 23, 425–428 [DOI] [PubMed] [Google Scholar]
- Yang Y, Gehrke S, Imai Y, Huang Z, Ouyang Y, Wang JW, Yang L, Beal MF, Vogel H, Lu B. (2006). Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc. Natl. Acad. Sci. USA 103, 10793–10798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Ouyang Y, Yang L, Beal MF, McQuibban A, Vogel H, Lu B. (2008). Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc. Natl. Acad. Sci. USA 105, 7070–7075 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.